
Integrated metabolomics and lipidomics profiling of hippocampus reveal metabolite biomarkers in a rat model of chronic unpredictable mild stress-induced depression
Introduction
An increasing body of evidence has revealed that long-term exposure to stress can have harmful effects on the structure and function of the brain, thereby leading to depression. Animals that are chronically stressed demonstrate functional and morphological damage in different brain regions such as the prefrontal cortex (PFC), the hippocampus, and the amygdala (1). The effects of these stress-induced changes are many, such as impacting learning, memory, and emotional responses. Stress has been associated with pathological processes of depression (2). Therefore, to thoroughly understand the potential pathophysiology of chronic unpredictable mild stress (CUMS)-induced depressive-like behavior is urgently needed.
CUMS is a well-accepted and the most validated animal model of depressive-like behavior (3). Following prolonged exposure to stress, the depressive behavior of rats is reflected in reduced preference for sweet solutions (evaluated with the sucrose preference test), and increased immobility in the forced swimming test.
Metabolomics and lipidomics have been used to analysis the extensive metabolites and lipids that exist in the periphery (e.g., serum and urine) and the central nervous system (e.g., PFC, the hippocampus and the amygdala) (4,5). The hippocampus is a vital limbic structure that is associated with mood and cognition, additionally, the hippocampus could regulate the hypothalamus-pituitary-adrenal (HPA) axis, making it more vulnerable to stress and depression. As a result, the hippocampus is often selected for depression studies (6,7). CUMS is known to decrease hippocampal neurogenesis, and various studies have suggested that brain-derived neurotrophic factor (BDNF) expression decreases in both the hippocampus of animals and the serum of humans in depression (8-10). Thus, studying the hippocampus of the rat with CUMS-induced depressive-like behavior may provide a deeper understanding.
Liquid chromatography (LC) with mass spectrometry (MS) is the preferred technique for metabolomics and lipidomics because of high separation efficiency, sensitivity, selectivity and throughput (4,11). Advancements in analytical technology and data processing have allowed for thousands of molecular species to be profiled and for a comparison of the composition between control and diseased groups, or diseased and treatment groups. More and more clinical research centers have been used MS-based metabolomics and lipidomics to study the transformation of neuropsychiatric disorders and the discovery of biomarkers (12,13).
In our paper, we describe a workflow (14) for conducting integrated lipidomic and metabolomic analyses of frozen hippocampal samples from CUMS-induced depressive and control rats (Figure 1). First, the CUMS rat model was established to obtain hippocampal samples. Second, we described the methods used for the extraction of lipids using the liquid-liquid extraction procedure and metabolites by protein precipitation from the hippocampal samples. Finally, we processed the data of lipidomic and metabolomic and identified the potential metabolites biomarkers by mapping them into metabolic pathways. We aimed to employ an integrated metabolomic and lipidomic approach coupled with principal component analysis (PCA), partial least squares-discriminate analysis (PLS-DA), and orthogonal partial least-squares discriminant analysis (OPLS-DA) to determine the potential metabolites and lipids in the hippocampus of the CUMS-induced rats compared to control. The discovery of metabolites and lipids biomarkers might aid researchers in understanding the underlying stress-related pathophysiological mechanisms of depression and provides a promising opportunity to generate novel potential targets for tentative antidepressants.
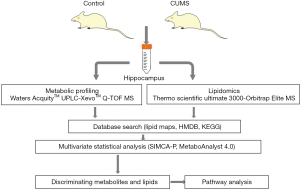
Methods
Materials and instruments
DL-o-Chlorophenylalanine, as an internal standard (IS), was from GL Biochem (Shanghai) Ltd. Formic acid was obtained from CNW, Shanghai, China. Chromatographic grade acetonitrile and methanol were purchased from the Merck Company, Darmstadt, Germany. Waters AcquityTM UPLC system combined with the use of Waters XevoTM Q-TOF mass analyzer (Waters Corporation, Milford, MA, USA) and Waters Acquity UPLC HSS T3 column (2.1 mm × 100 mm, 1.8 µm, Waters Corporation, Milford, MA, USA) were used to perform metabolomic analysis. Ultimate 3000 (Dionex, Sunnyvale, CA, USA) was coupled to a Thermo Orbitrap Elite with a heated electrospray ion source (HESI-II) (Thermo Fisher Scientific, Waltham, MA, USA) and Kinetex C18 column (100 mm × 2.1 mm, 1.9 µm, Dionex, Sunnyvale, CA, USA) were used to do lipidomic analysis.
Study animals
Eight-week-old male Sprague-Dawley rats (weight 210±30 g) were provided by Jining Medical University and distributed to control or CUMS the two groups with six animals each. All rats were kept under standard laboratory conditions and were allowed to acclimate to the environment for five days. All animal procedures followed the Regulations of Experimental Animal Administration issued by the State Committee of Science and Technology of the People’s Republic of China. In addition, our study was approved by the Medical Ethics Committee of Jining First People’s Hospital, Jining Medical University, and the ethical number was 20170016.
CUMS procedure
CUMS was induced based on our earlier study (15). Briefly, the CUMS treatment was performed according to the following conditions for four weeks: (I) food and water deprivation for 24 h, (II) cage tilting (45°) for 24 h, (III) crowded housing for 24 h, (IV) kept in an empty water bottle (Wahaha, Hangzhou, Zhejiang, China) for 4 h, (V) one minute of tail clamping, (VI) 20 min of noise, (VII) forced swimming for 10 min, and (VIII) day/night reversal (12 h/12 h). To ensure that the rats received unpredictable stress, these above procedures were randomly scheduled (one per day).
Behavioral tests—the sucrose preference test (SPT) and forced swim test (FST) were conducted to assess depressive-like states of the rats. Results from the behavioral tests were presented as means ± SD and GraphPad version 6.0 software was used to performed t-test.
Sucrose preference test (SPT)
The SPT was described in our previous study (15). All rats were placed separately and habituated to taste a 1% sucrose solution for 48 h in two bottles on each side before the SPT test. After 14 h of water deprivation, two pre-weighed bottles, one containing tap water and another containing a 1% sucrose solution, were placed in each cage to rat. The side (left and right) that the bottles were placed on was randomly selected to avoid spatial bias. Two bottles were weighed again after 1h, and the weight difference was used as the rat intake from each bottle. Sucrose preference was measured as 1% sucrose solution intake /total liquid consumption.
Forced swim test (FST)
The FST was performed following our previous study with minor modifications (15). Briefly, all rats were separately placed in a Plexiglas cylinder (45 cm height, 15 cm diameter) containing with water (25±2 °C) up to 35 cm deep for a 15 min swim test. Then, all rats were dried and placed back to their cages. Twenty-four hours later, all rats were forced to swim again for 5 min. The two test sessions were both videotaped and the duration of immobility in the 5 min test was recorded by an experienced observer blinded to the design of experimental in our study.
Preparation of hippocampus tissue samples
One day later, all rats were euthanized with 1% sodium pentobarbital (50 mg/kg), all rats brains were quickly resected, and the hippocampus was rapidly dissected on the ice surface, then washed with 0.9% physiological saline, and immediately frozen in liquid nitrogen. All hippocampal tissue samples of rats were placed at –80 °C until further needed.
Sample pretreatment for metabolomics and lipidomics
For metabolite extraction, the hippocampal tissue was cut into pieces and weighed 50 mg to a 2-mL tube homogenizer, then 800 µL of methanol (with IS, 5 µg/mL) was added to the 2-mL tube, homogenized evenly, and centrifuged at 12,000 rpm for 15 min at 4 °C. At last, 200 µL supernatant was used for UPLC-MS analysis.
For lipid extraction, the hippocampal tissue sample (50 mg) was homogenized with a 1.5 mL organic solvent mixture consisting of chloroform/methanol (2:1, v/v), mixed, vortexed for 1 min, and centrifuged at 3,000 rpm for 15 min. Then, 800 µL of the supernatant was moved to another tube and dried under gentle flow nitrogen gas at 40 °C. Finally, 200 µL mixture of isopropanol/methanol (1:1, v/v) was used to reconstitute, vortexed for 1 min, and then placed in a vial for further analysis.
Data acquisition for metabolomic and lipidomic analysis
For metabolomics, all hippocampal tissue samples of rats was separated on an Acquity UPLC HSS T3 column (2.1 mm × 100 mm, 1.8 µm), eluted with the mixture of mobile phase A (water containing 0.1% formic acid) and mobile phase B (acetonitrile containing 0.1% formic acid) at a flow rate of 0.3 mL/min with the column temperature kept at 40 °C. The gradient conditions of mobile phase as following: 0/95, 2/95, 12/5, 15/5, 17/95, and 20/95 (min/% solvent A). Six µL was used for analysis. Mass spectrometry analysis was conducted on both the positive and negative electrospray ionization (ESI) modes and the mass range was from 50 to 1,500 m/z.
For lipidomic, all hippocampal tissue samples of rats were performed on a Kinetex C18 column (100 mm × 2.1 mm, 1.9 µm) with a flow rate of 0.4 mL/min and the temperature of column was set at 45 °C. The mobile phase A: acetonitrile-water (60:40, v/v) with 10 mM ammonium acetate and mobile phase B: isopropanol-water (10:90, v/v) with 10 mM ammonium acetate and 0.1% formic acid. The gradient conditions were: 0/70, 2/70, 20/0, 40/0, 40.01/70, and 45/70 (min/% solvent A). The volume of sample injection was 4 µL. The mass spectrometer was performed in positive and negative ion modes, and the scan range in both modes was 200 to 1,500 m/z.
Statistical analysis
The Masslynx 4.1 software (Waters, Milford, MA, USA) was used to feature extraction and preprocess the original data of metabonomics. Then, the data was normalized and observations (samples), retention time (RT), peak intensity, and mass were edited into a two-dimensional data matrix with the Microsoft Excel version 2010 software.
The Thermo Lipid Search v 4.0.20 Software (Thermo Fisher Scientific, Waltham, MA, USA) was performed to analysis the original data of the lipidomics for all hippocampal tissue samples. Then, the data was normalized and all the metabolomic and lipidomic data were analyzed separately with version 13.0 of SIMCA-P software (Umetrics, Umea, Sweden) where multivariate analyses of PCA, PLS-DA and OPLS-DA were performed. As the basis of multivariate modeling, PCA is especially helpful, but abnormal value detection and the discovery of patterns and trends, abnormal treatment of sample, instrumental drift, artifacts, and other variation of experimental will make the result is unrelated to the scientific question of interest (16). Thus, for class separation, i.e., the CUMS and controls in our study, the obtained principal components are not necessarily aligned with the best predictive components. In this case, a supervised method is needed, which could make full use of any prior data to refocus the analysis on the study aim, so as to better quantify the prediction of class membership. Herein, in our study, the supervised OPLS-DA analysis was used to better identify potential metabolites and lipids that contribute to the sample classification and removal uncorrelated changes in the spectrum (17). The OPLS score plots and variable importance for projection (VIP) statistics was applied to select important variables which were responsible for separation of group. When the VIP value of the variables was greater than 1.0, the variable can be selected as candidates. Then, these variables were further carried out by SPSS version 17.0 software with a two-tailed t-test. When both VIP >1 and t-test P values <0.05 met the thresholds, features could be reported as significant. In addition, in our study, extra standards of fold-change >5 or fold-change <0.2 were used because large lipids were identified according to the VIP >1 and P<0.05. After the above analysis, potential indicators were collected and further identified with the Human Metabolome Database and Lipid Search software. Subsequent pathway analysis used the public databases-MetaboAnalyst 4.0 and the Kyoto Encyclopedia of Genes and Genomes, with Raw P<0.05, and Impact >0 being defined as significant, for possible help in the biochemical interpretation of the metabolites.
Results
Behavioral tests
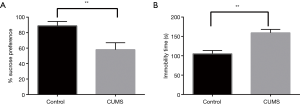
After four weeks of the CUMS procedures, the percentage sucrose preference of the control rats was 88.75%±5.63% (mean ± SD, n=6), where as the CUMS rats showed a reduced sucrose preference of 57.92%±9.05% (mean ± SD, n=6) in SPT (P<0.01). In the FST, CUMS showed a longer immobility time of 159.17±9.11 s (mean ± SD, n=6) (P<0.01) than the controls at 105.33±8.26 s (mean ± SD, n=6), the results of which are shown in Figure 2.
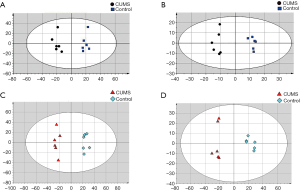
Multivariate statistical analysis of the metabolomics and lipidomics data
OPLS analysis (Figure 3) was performed using the data from UPLC-MS. A total of 35 discriminating metabolites between the control and CUMS groups were identified (VIP >1, P<0.01; Table 1). A total of 171 discriminating lipids were finally identified (VIP >1, P<0.01, fold change >5 or fold change <0.2; online: http://cdn.amegroups.cn/static/application/d11e2ecf3dd55cf06dfa95b094b15467/10.21037atm.2019.11.21-1.pdf) due to large lipids. These metabolites included 6 ceramide (Cer), two cardiolipin (CL), 2 diacylglycerols (DGs), 1 fatty acid(FA), 25 lyso-PCs (LPCs), 13 lyso-phosphatidylethanolamines (LPEs), 7 lyso-prostaglandin (LPGs), 4 lyso-phosphatidylinositols (LPIs), 3 lyso-phosphatidylserines (LPSs), 2 monogalactosyl monoglyceride (MGMGs), 3 hydroxylated fatty acyls (OAHFAs), 4 phosphatidic acids (PAs), 20 phosphatidylcholines (PCs), 16 phosphatidylethanolamines (PEs), 9 phosphatidylglycerols (PGs), 1 ph-sphingomyelin (phSM), 15 phosphatidylinositols (PIs), 26 phosphatidylserines (PSs), 5 sphingomyelins (SMs), 1 sophorolipid (So), 5 sulfoquinovosyldiglycerides (SQDGs), and 1 triglyceride (TG).
Full table
Pathway analysis
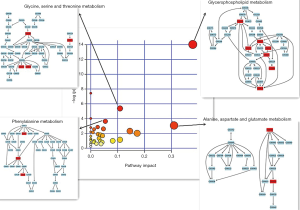
Potential target pathway analysis demonstrated that the metabolites and lipids identified by statistical analysis based on SICMA-P and GraphPad software are accountable for glycerophospholipids, glycine, serine, and threonine metabolism; phenylalanine metabolism; and alanine, aspartate and glutamate metabolism (impact-value >0.00, P<0.05; Figure 4) between the CUMS group and the control group. These were the key different metabolic pathways associated with metabolic changes of the hippocampus in the control and CUMS groups. They were closely connected to each other, and the metabolites involved in the above three pathways were associated with amino acid metabolism.
Discussion
Depression is a multi-factorial disorder that presents a different pathophysiological profile in each individual (18-20). CUMS-induced depression rat models are currently considered ideal tools for elucidating depressive behaviors (21). In our study, the objectives of metabolomics and lipidomics were to identify more and more metabolites and lipids that exist in hippocampal tissue samples, helping to learn the alterations of the metabolites and lipids in hippocampus of rats between the CUMS and control groups and further to understand the status in physiological and biochemical of biosystem (22,23). Therefore, we aimed to employ untargeted metabolomics- and lipidomics-based advanced UPLC-MS technology to discover potential metabolites and lipids biomarkers in the hippocampus of CUMS rats compared to the controls. UPLC-MS was employed, given its advantages of the breadth of coverage and speed of identification of many compounds. The discovery of more metabolites and lipids biomarkers might aid researchers in understanding the underlying stress-related pathophysiological mechanisms of depression and provide new effective therapeutic strategies.
A total of 171 discriminating lipids and 35 discriminating metabolites between the control and CUMS-induced depressive groups were identified and the identified metabolites and lipids play essential roles in amino acid metabolism, energy, lipid metabolism, membrane structure, and signaling. In our study, the potential metabolites and lipids biomarkers between the control and CUMS group were detected, but more importantly, metabolic pathways in which these metabolites and lipids are involved was identified via metabolic pathway analysis. Glycerophospholipid metabolism; glycine, serine, and threonine metabolism; phenylalanine metabolism; and alanine, aspartate, and glutamate metabolism were the key different metabolic pathways associated with metabolic changes in the hippocampus in the control and CUMS groups. Thus, the metabolites, amino acids, and lipid metabolism that were altered may help to understand the mechanism underlying depression.
Amino acids have gained the increased attention of researchers, and altered amino acid levels have been identified as markers of risk for many diseases such as type 2 diabetes (24,25), schizophrenia (SCH) (26), and Alzheimer’s disease (AD) (27). In our study, the levels of several amino acids and neurotransmitters significantly altered in the hippocampus of rats between the control and CUMS groups, and glycine, serine, and threonine metabolism were altered, as indicated by increased betaine aldehyde, L-threonine, PS (16:0/16:0) and L-tryptophan. Phenylacetic acid, phenyl acetyl glycine, and N-acetyl-L-phenylalanine were the detected metabolites in phenylalanine metabolism and L-glutamine and γ-aminobutyric acid (GABA) were the detected metabolites in alanine, aspartate, and glutamate metabolism. These metabolisms are all categorized as amino acid metabolism (28). Among these interfered amino acids, glutamine, GABA, and glutamate also belong to neurotransmitters and the alterations in glutamatergic or GABAergic neurotransmitters have been associated with disease stages, cognitive symptoms, and triggering depressive-like behavior. In our study, the changes in glutamine and GABA that increased in the hippocampus of the CUMS rat were consistent with the findings reported in earlier studies (29,30). Glutamine and GABA might be potential target for further depression diagnosis and antidepressants development. Glycine, a semi-essential amino acid and a basic nutrient, is part of the endogenous antioxidant glutathione, but more importantly, glycine could be involved in oxygen stress and the cell membrane injury processes of depression (31). Phenylalanine, an essential amino acid, is the precursor of catecholamines, which could be as neurotransmitters and adrenaline-like substances that also play crucial roles in depression. Kynurenic acid (KYNA), one of the related monoaminergic metabolites, was found to be increased in CUMS rats and has been associated with the pathophysiology of depression (31,32). Overall, although the alterations of perturbed amino acids are complicated, our findings showed that disturbances in amino acid metabolism are associated with the pathophysiological mechanism of depression. Now, quantitative measurements of the plasma amino acids of depressive patients are being conducted in our laboratory, which may further help to the diagnosis of depression in clinical and may act as potential targets to help develop new antidepressants.
In this study, perturbations in lipid metabolism were found to be related to the hippocampal response to CUMS in the rats’ model. Phosphatidylethanolamine, phosphatidylcholine, lysoPC (18:1(9Z)), acetylcholine, PA (16:0/16:0), PS (16:0/16:0), and glycerophosphocholine were the detected metabolites in glycerophospholipid metabolism. Glycerophospholipids are the most abundant type of phospholipids (33,34). These lipids are part of biological membranes and are active substances in bile and on the membrane surface. Glycerophospholipids also act as messengers in cell signaling processes and are vital to angiogenesis, neurogenesis, and immunity and other biological processes (35). PCs and PEs are the major lipids distributed in the cell membrane and are involved in the fluidity and integrity of membranes (36). It should be noted that ethanolamine and o-phosphorylethanolamine can act as precursors of PC and PE. Additionally, increased PC and PE were observed in an earlier phospholipidomic research on the CUMS mice brains (37), which was consistent with our findings. All these findings suggested that the dysregulation of glycerophospholipid metabolism may contribute to the occurrence and development of depression.
Our study showed that these metabolites may act as potentially valuable biomarkers for predicting depression. However, several limitations must be mentioned. First, we used a metabolomic and lipidomic approach in our study, and proteomics and genomics are needed to further validate our findings. Second, metabolic changes in the hippocampus of rats were only performed because the hippocampus is more sensitive to stress and depression, but it could not capture the changes of the whole brain, we should take PFC, amygdala and other brain regions (e.g., the amygdala) in consideration for depression study. Finally, the animal model cannot simulate the clinical complex situation very well, and should be combined with clinical research. Thus, we collected a total of 500 samples of healthy and depressive patients to conduct further study.
Conclusions
In our study, an integrated analysis of UPLC-MS-based metabolomics and lipidomics was performed to comprehensively understand the rat hippocampal response to CUMS. The findings underlined the metabolites, lipids and metabolic pathways that were changed in the hippocampus in CUMS when compared to the controls, providing novel insights in the metabolism in hippocampus of rats and revealing the new lipid-related targets. These metabolites and lipids might act as potential biomarkers and contribute to elucidate the pathophysiological mechanisms underlying CUMS-induced depression, but further studies using more independent samples are needed.
Acknowledgments
Funding: Our study was supported by the National Natural Science Foundation of China (81602846 to P Jiang and 31600947 to Y Qiao) and the Taishan Scholar Project of Shandong Province (tsqn201812159 to P Jiang).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Our study was involved in animals and with the approval of the medical ethics committee of the Jining First People’s Hospital, Jining Medical University (No. 20170016).
References
- Oliveira TG, Chan RB, Bravo FV, et al. The impact of chronic stress on the rat brain lipidome. Mol Psychiatry 2016;21:80-8. [Crossref] [PubMed]
- Menard C, Hodes GE, Russo SJ. Pathogenesis of depression: Insights from human and rodent studies. Neuroscience 2016;321:138-62. [Crossref] [PubMed]
- Szewczyk B, Pochwat B, Muszynska B, et al. Antidepressant-like activity of hyperforin and changes in BDNF and zinc levels in mice exposed to chronic unpredictable mild stress. Behav Brain Res 2019;372:112045. [Crossref] [PubMed]
- Paglia G, Astarita G. Metabolomics and lipidomics using traveling-wave ion mobility mass spectrometry. Nat Protoc 2017;12:797-813. [Crossref] [PubMed]
- Astarita G, Stocchero M, Paglia G. Unbiased lipidomics and metabolomics of human brain samples. Methods Mol Biol 2018;1750:255-69. [Crossref] [PubMed]
- Liu W, Ge T, Leng Y, et al. The role of neural plasticity in depression: from hippocampus to prefrontal cortex. Neural Plast 2017;2017:6871089. [Crossref] [PubMed]
- Gujral S, Aizenstein H, Reynolds C F, et al. Exercise effects on depression: possible neural mechanisms. Gen Hosp Psychiatry 2017;49:2-10. [Crossref] [PubMed]
- Chen B, Dowlatshahi D, Macqueen G M, et al. Increased hippocampal BDNF immunoreactivity in subjects treated with antidepressant medication. Biol Psychiatry 2001;50:260-5. [Crossref] [PubMed]
- Angelucci F, Brenè S, Mathé A A. BDNF in schizophrenia, depression and corresponding animal models. Mol Psychiatry 2005;10:345-52. [Crossref] [PubMed]
- Sen S, Duman R, Sanacora G. Serum BDNF, depression and anti-depressant medications: meta-analyses and implications. Biol Psychiatry 2008;64:527-32. [Crossref] [PubMed]
- Kirwan JA, Kaddurah-Daouk R, Mitchell T, et al. Biobanking for metabolomics and lipidomics in precision medicine. Clin Chem 2019;65:827-32. [Crossref] [PubMed]
- Hu C, Li J, Xu G. Mass Spectrometry-Based Lipidomics for Biomarker Research. In: Preedy V, Patel V. (eds). General Methods in Biomarker Research and their Applications. Biomarkers in Disease: Methods, Discoveries and Applications. Springer, Dordrecht 2015:49-74.
- Jain KK. Technologies for Discovery of Biomarkers. In: The Handbook of Biomarkers. Humana Press, New York, NY 2010:23-72.
- Southam AD, Weber RJ, Engel J, et al. A complete workflow for high-resolution spectral-stitching nanoelectrospray direct-infusion mass-spectrometry-based metabolomics and lipidomics. Nat Protoc 2016;12:310-28. [Crossref] [PubMed]
- Zhang WY, Guo YJ, Han WX, et al. Curcumin relieves depressive-like behaviors via inhibition of the NLRP3 inflammasome and kynurenine pathway in rats suffering from chronic unpredictable mild stress. Int Immunopharmacol 2019;67:138-44. [Crossref] [PubMed]
- Vantaku V, Dong J, Ambati CR, et al. Multi-omics integration analysis robustly predicts high-grade patient survival and identifies CPT1B effect on fatty acid metabolism in bladder cancer. Clin Cancer Res 2019;25:3689-701. [Crossref] [PubMed]
- Zhou L, Wang Z, Hu C, et al. Integrated Metabolomics and Lipidomics Analyses Reveal Metabolic Reprogramming in Human Glioma with IDH1 Mutation. J Proteome Res 2019;18:960-9. [Crossref] [PubMed]
- Bianchi R, Schonfeld IS, Laurent E. Burnout or depression: both individual and social issue. Lancet 2017;390:230. [Crossref] [PubMed]
- Smith K. Mental health: a world of depression. Nature 2014;515:181. [Crossref] [PubMed]
- Roman M, Irwin MR. Novel neuroimmunologic therapeutics in depression: a clinical perspective on what we know so far. Brain Behavl Immun 2019. [Epub ahead of print].
- Liu W, Xue X, Xia J, et al. Swimming exercise reverses CUMS-induced changes in depression-like behaviors and hippocampal plasticity-related proteins. J Affect Disord 2018;227:126-35. [Crossref] [PubMed]
- Cicalini I, Rossi C, Pieragostino D, et al. Integrated lipidomics and metabolomics analysis of tears in multiple sclerosis: an insight into diagnostic potential of lacrimal fluid. Int J Mol Sci 2019;20:E1265. [Crossref] [PubMed]
- Luan H, Meng N, Liu P, et al. Non-targeted metabolomics and lipidomics LC-MS data from maternal plasma of 180 healthy pregnant women. Gigascience 2015;4:16. [Crossref] [PubMed]
- Rebnord EW, Strand E, Midttun Ø, et al. The kynurenine:tryptophan ratio as a predictor of incident type 2 diabetes mellitus in individuals with coronary artery disease. Diabetologia 2017;60:1712-21. [Crossref] [PubMed]
- McCann A, Melvaer Giil L, Ulvik A, et al. Plasma amino acids and incident type 2 diabetes in patients with coronary artery disease. Diabetes Care 2019;42:1225-33. [Crossref] [PubMed]
- Leppik L, Kriisa K, Koido K, et al. Profiling of amino acids and their derivatives biogenic amines before and after antipsychotic treatment in first-episode psychosis. Front Psychiatry 2018;9:155. [Crossref] [PubMed]
- Li Z, Xing Y, Guo X, et al. Development of an UPLC-MS/MS method for simultaneous quantitation of 11 d-amino acids in different regions of rat brain: Application to a study on the associations of d-amino acid concentration changes and Alzheimer's disease. J Chromatogr B Analyt Technol Biomed Life Sci 2017;1058:40-6. [Crossref] [PubMed]
- Wu Y, Li Y, Jia Y, et al. Imbalance in amino acid and purine metabolisms at the hypothalamus in inflammation-associated depression by GC-MS. Mol Biosyst 2017;13:2715-28. [Crossref] [PubMed]
- Yang XH, Song SQ, Xu Y. Resveratrol ameliorates chronic unpredictable mild stress-induced depression-like behavior: involvement of the HPA axis, inflammatory markers, BDNF, and Wnt/β-catenin pathway in rats. Neuropsychiatr Dis Treat 2017;13:2727-36. [Crossref] [PubMed]
- Strawbridge R, Young AH, Cleare AJ. Biomarkers for depression: recent insights, current challenges and future prospects. Neuropsychiatr Dis Treat 2017;13:1245-62. [Crossref] [PubMed]
- Zafra F, Ibáñez I, Bartolomé-Martín D, et al. Glycine transporters and its coupling with NMDA receptors. Adv Neurobiol 2017;16:55-83. [Crossref] [PubMed]
- Cao T, Li N, Cai H. Candidate metabolic biomarkers for schizophrenia in CNS and periphery: Do any possible associations exist? Schizophr Res 2019. [Epub ahead of print]. [Crossref] [PubMed]
- Oresic M, Hanninen VA, Vidal-Puig A. Lipidomics: a new window to biomedical frontiers. Trends Biotechnol 2008;26:647-52. [Crossref] [PubMed]
- Farooqui A A, Horrocks L A, Farooqui T. Interactions between neural membrane glycerophospholipid and sphingolipid mediators: A recipe for neural cell survival or suicide. J Neurosci Res 2007;85:1834-50. [Crossref] [PubMed]
- Rodriguez-Cuenca S, Pellegrinelli V, Campbell M, et al. Sphingolipids and glycerophospholipids – The “ying and yang” of lipotoxicity in metabolic diseases. Prog Lipid Res 2017;66:14-29. [Crossref] [PubMed]
- Stoessel D, Nowell CJ, Jones AJ, et al. Metabolomics and lipidomics reveal perturbation of sphingolipid metabolism by a novel anti-trypanosomal 3-(oxazolo[4,5-b]pyridine-2-yl)anilide. Metabolomics 2016;12:126. [Crossref]
- Hill MN, Miller GE, Carrier EJ, et al. Circulating endocannabinoids and N-acyl ethanolamines are differentially regulated in major depression and following exposure to social stress. Psychoneuroendocrinology 2009;34:1257-62. [Crossref] [PubMed]


