
STAG2 loss-of-function mutation induces PD-L1 expression in U2OS cells
Introduction
Cohesin is a protein complex well known for its function in sister chromatid cohesion. It also plays roles in chromatin spatial organization, gene expression, and DNA repair (1,2). Cohesin consists of a ring-like tripartite structure composed of SMC1, SMC3 and SCC1, and a peripheral subunit, STAG1 or stromal antigen 2 (STAG2) (3). STAG1 and STAG2 are mutually exclusive components of Cohesin: STAG1 is required for telomere cohesion, and STAG2 for centromere cohesion (4). Recent studies reported that cohesin slides along chromatin and contributes to the chromatin topological associating domains (TADs), which regulate gene expression by mediating genome-wide enhancer-promoter interactions (5).
Cancer genome sequencing found that the genes encoding the cohesin components are recurrently mutated in many types of cancer cells (6). The STAG2 gene is located in the X chromosome and can be deactivated in both males and females by one mutational event (3). STAG2 loss-of-function mutations have been observed in many types of cancer including bladder cancer, Ewing sarcoma, melanoma, myeloid leukemia, and other cancers (3,7-9). Most of STAG2 gene mutations (about 85%) in the cancer genome are truncating and have resulted in the complete absence of protein expression, indicating that STAG2 is a tumor suppressor. However, the mechanism of STAG2 mutation which promotes tumorigenesis is still unknown. An earlier study reported that the loss of SATG2 in cancer cells leads to aneuploidy in tumorigenesis, but subsequent reports did not support that claim (3,10).
Programmed death-ligand 1 (PD-L1), also known as CD274 or B7-H1, is a transmembrane protein that plays a major role in suppressing the immune system by interacting with PD-1 on the T cell’s surface (11). PD-L1 is up-regulated in many types of cancer cells and it contributes to immune evasion during tumorigenesis (12). Several cancer-related genes like Myc, regulate PD-L1 expression in cancer cells undergoing tumorigenesis by manipulating the host immune system (13). The antibodies blocking the PD-1 and PD-L1 axis are used to unleash tumor-reactive T cells and induce a durable anti-tumor response in the clinical environment (14). Clinical data indicate that high PD-L1 expression in tumor cells might be correlated with a response to anti-PD-1/PD-L1 treatment (15). In this study, we observed that PD-L1 expression is up-regulated in STAG2-deficient U2OS cells generated by clustered regularly interspaced short palindromic repeated (CRISPR)-Cas9. This observation may explain how STAG2’s loss-of-function mutation contributes to tumorigenesis and suggests that the cancer patients with STAG2 mutation may be responsive to anti-PD-L1 treatment.
Methods
Bioinformatics analysis with COSMIC databases
COSMIC database (http://cancer.sanger.ac.uk/cosmic) was used to explore the distribution of STAG2 mutations in cancer (16). The filtering conditions were restricted to the following: genes (STAG2); range [1–1,232]; coordinate-system (amino-acid); tissue (bone); screen type (all); somatic status (confirmed somatic, previously reported, variant of unknown origin); sample type (cultured, tumor sample).
Design of STAG2 gRNA and STAG2 gRNA-Cas9 plasmid construct
The p2U6-pCGA-Cas9 plasmid and p2U6-PKM-kana-Cas9 plasmid were generously provided by Dr. Zhili Rong (17). The STAG2 gRNA was designed by using the Zhang lab online program available at http://crispr.mit.edu. The forward gRNA sequence was 5'-actttgaggatttattggag-3' and the reverse gRNA sequence 5'-gcgaattcctggaaatggga-3'.
Cell culture and generation of genetically modified cell subclones
The human osteosarcoma cell line (U2-OS) was grown in McCoy’s 5A medium, supplemented with a 10% heat-inactive fetal bovine serum (FBS) (Gibco, Rockville, MD, USA) with streptomycin (100 µg/mL) and penicillin (100 U/mL) at 37 °C with 5% CO2. The STAG2 gRNA-Cas9 plasmid construct was transfected into the cells using Lipofectamine 2000 (Invitrogen, CA, USA) following the manufacturer’s protocol. Briefly, the cells were seeded into six-well plates at a density of 1×105 cells/well and transfected with 4 µg plasmids per well. After a 48-hour incubation and selection with puromycin (2 µg/mL) (Sigma), the normal medium was changed after 3 days. The cell suspension was diluted to a suitable concentration by low-density dilution and added to a 96-well plate (1 cell/well), cultured and passaged. Finally, a STAG2 knockout monoclonal cell line was selected. After the cells had grown to an appropriate number, genomic DNA was extracted for polymerase chain reaction (PCR) identification (forward primer: 5'-cagagtaccagtcttccaaggca-3'; reverse primer: 5'-ccatcctacctggagaggcaacg-3').
Western blotting
Proteins were extracted using the RIPA solution from the harvested cells and quantified by the bicinchoninic acid (BCA) assay (Beyotime, Beijing, China). Twenty µg of protein were separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to PVDF membranes. The primary antibodies against STAG2 (sc-81852, Santa Cruz, USA) GAPDH (10494-1-AP, Proteintech, USA), CCND1 (ab134175, Abcam), CCNB1 (ab32053, Abcam), CDK1 (ab133327, Abcam), CDK4 (ab108357, Abcam), RB (ab181616, Abcam), P21 (ab109520, Abcam), PI3K110a (C73F8, Cell Signaling Technology), p-PI3K (T607) (E1A3241-2, EnoGene, China), T-mTOR (ab317341, Abcam), p-mTOR [2481] (ab137133, Abcam), p-mTOR [2448] (ab109268, Abcam), p-AKT (S473) (ab81283, Abcam), t-AKT (C67E7, Cell Signaling Technology), N-Cadherin (610920, BD Transduction Laboratories), and E-cadherin (610182, BD Transduction Laboratories) were diluted according to the instructions for antibodies in Western blotting.
RNA extraction, reverse transcription and Q-PCR
Total RNA was extracted with TRIzol Reagent (Life Technologies, NY) following the manufacturer’s instructions. cDNA was synthesized from 1 µg of total RNA using the M-MuLV First Strand cDNA Synthesis Kit (Sangon Biotech, China). The following primers were used to detect the expression of PD-L1 and GAPDH (internal control): PD-L1 forward primer: 5'- TGGCATTTGCTGAACGCATTT-3', PD-L1 reverse primer, 5'-TGCAGCCAGGTCTAATTGTTTT-3', GAPDH forward primer, 5'-AGAAGGCTGGGGC-TCATTTG-3', and GAPDH reverse primer 5'-AGGGG-CCATCCACAGTCTTC-3'. Data were analyzed using the comparative Ct method (2−ΔΔCt).
RNA-seq assay and bioinformatics analysis
The RNA-sequencing was performed by Novogene (Beijing, China). The library was prepared for sequencing using the standard Illumina protocols. Differentially expressed gene (DEG) analysis of the KO group and the control group (each group has two biologic replicates per condition) was performed using the DESeq R package (1.10.1). DESeq provides statistical routines for determining differential expression in digital gene expression data using a model based on the negative binomial distribution. The P values were adjusted using the Benjamini and Hochberg approach for controlling the false discovery rate (FDR). Genes with an adjusted P value of less than 0.05 found by DESeq were assigned as differentially expressed. Gene Ontology (GO) enrichment analysis of the DEGs was performed by the GOseq R package, in which gene length bias was corrected. GO terms with a corrected P value of less than 0.05 were considered significantly enriched by differential expressed genes. Pathway enrichment was determined using the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway annotation. Pathways were considered significantly enriched with an adjusted P value of less than 0.05.
Analysis of PD-L1 expression by FACS-analysis
To detect PD-L1 expression, cells were incubated with or without anti PD-L1 antibody (diluted 1:1,000, ab205921, Abcam), for 30 min at 4 °C. Cells were then incubated with the Donkey anti-Rabbit IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 568 (diluted 1:100 in PBS/2% FBS) for 30 min at 4 °C. After repeated washing, cells were resuspended in 500 µL of 0.01 M PBS and analyzed immediately on a FACS flow cytometer (FACSCalibur, BD Biosciences, San Jose, CA, USA).
Statistics
All values were presented as mean ± SD as experiments independently were carried out in triplicate. Data were analyzed using one-way or two-way ANOVA or Student’s t-test (GraphPad Prism), depending upon the number of groups and variances. The difference was considered statistically significant when the probability was less than 0.05 (P<0.05).
Results
Loss of STAG2 inhibits cell proliferation and induces G2/M phase arrest
To determine the distribution of STAG2 mutations in cancer, we analyzed the STAG2 mutation profiles of 10 primary cancer tissues in the COSMIC database. The STAG2 mutation frequencies in Urinary carcinoma and bone cancer were higher than 5%, which is consistent with the previous reports (Figure 1A). The most common mutation in OS is R216* in exon 8, which is a truncating mutation and results in loss of expression (Figure 1B). To investigate how the loss of STAG2 contributes to tumorigenesis, we deleted the exon 8 in U2OS cells by CRISPR-Cas9 technique to mimic the mutation of R216* (Figure 1C). The mRNA transcriptional level of the STAG2 gene is normal, but the protein is undetectable in the sub-clone cells (Figures 1D,S1).
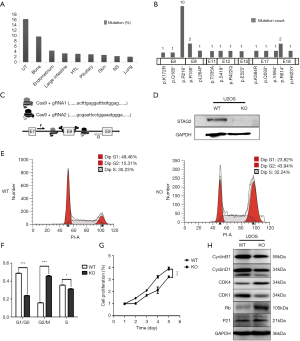
Due to the role of cohesin in cell cycling, we first evaluated the cell cycling of STAG2 deficient cells. As depicted in Figure 1E,F, the SATG2 deficient cells displayed an increase in the percentage of G2/M phase compared to the parental cells, indicating that loss of STAG2 induces G2/M phase arrest. We next evaluated the effect of STAG2-loss on cell proliferation and growth by CCK-8 assay. As shown in Figure 1G, STAG2 loss significantly reduced the proliferation rate of U2-OS cells after a two-day culture. Expression of the cell cycling proteins like CCNB1, CCND1 and CDK1 decreased in STAG2 mutated cells, whereas expressions of CDK4 and RB increased (Figure 1H). These data points indicate that STAG2 loss leads to a prolonged cell cycle and slow cell growth, which is consistent with the previous report (18).
Loss of STAG2 increased cell migration ability and drug resistance
Cancer malignancy is closely related to the migration ability of tumor cells. To test the relationship between SATG2 deletion and cell migration, we performed Transwell migration assays. More STAG2-loss cells migrated to the lower compartment than the parental cells after a 14-hour culture, indicating that the STAG2 loss promotes cell migration (Figure 2A,B). To further explore the mechanism of STAG-loss on cell migration, we checked the level of expression for E-cadherin and N-cadherin, two proteins involved in epithelial-mesenchymal transition (EMT). The E-cadherin expression in STAG2-deficient cells significantly decreased compared with the intact STAG2 cells, while N-cadherin expression increased (Figure 2C). Down-regulation or loss of E-cadherin and increased expression of N-cadherin usually lead to enhanced tumor invasion and migration status. Thus, the switch of E- and N-cadherin expression is consistent with the Transwell assay results, supporting the idea that STAG2-loss enhances cancer invasion.
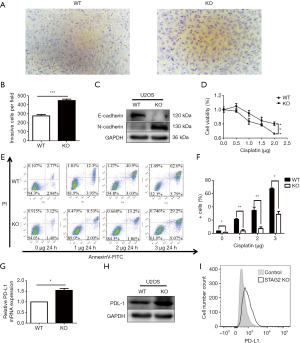
Chemo-resistance represents a significant cause of poor prognosis for OS patients. To explore whether STAG2 plays a role in modulating chemo-resistance, a CCK-8 assay was performed to examine the effect of STAG2 loss on U2OS cells treated with cisplatin, one of the most commonly used drugs in OS chemotherapy. STAG2 loss cells displayed stronger resistance to cisplatin treatment than control cells (Figure 2D). To further confirm that STAG2-loss induced resistance mediated through the inhibition of apoptosis, cells were incubated with cisplatin, and apoptosis was examined using cytoflow with AnnexinV-FITC/PI staining. We found that STAG2 deletion significantly reduced cisplatin-mediated cell apoptosis (Figure 2E,F). These data points suggest that STAG2-loss induced chemo-resistance is mediated through the inhibition of apoptosis.
PD-L1 over-expression on tumor cells inhibits host-adaptive immunity and is sufficient for immune evasion (12). To evaluate whether STAG2-loss is involved in the immune evasion of tumor cells, we checked PD-L1 expression on STAG2-loss cells. The mRNA level of CD274 gene encoding PD-L1 in STAG2 deletion cells increased 1.5-fold when compared to the parental cells (Figure 2G). The protein level of PD-L1 expression was checked by Western Blot and Cytoflow (Figure 2H,I). STAG2-loss cells displayed higher PD-L1 expression, which indicates that STAG2 loss may be involved in the immune evasion of cancer cells.
Genes involved in extracellular matrix organization and immune response were altered in STAG2-loss cells
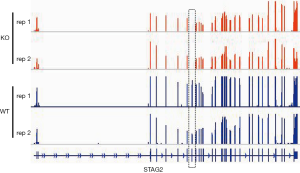
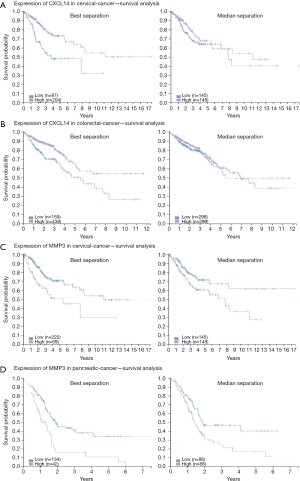
To elucidate alteration of cell behavior caused by STAG2 loss, we performed expression profiling by RNA-seq. There are 576 genes which were significantly up-regulated in STAG2-loss cells and 485 genes which were significantly down-regulated when compared with the parental cells (Figure 3A). Some of the genes like MMP3 encoding Matrix metallopeptidase 3 and CXCL14 encoding C-X-C motif chemokine ligand 14 play crucial roles in tumorigenesis and malignancy. The data from the Human Protein Atlas showed that a higher MMP3 and lower CXCL14 expression is associated with a poor prognosis (Figure S2) (19). Gene Ontology analysis showed that the genes involved in cell adhesion and extracellular matrix organization are significantly altered, indicating that STAG2-loss promotes cell invasion by regulating genes encoding extracellular protein (Figure 3B).
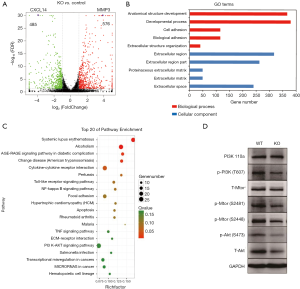
KEGG pathway enrichment analysis was applied to identify the pathways regulated by STAG2-loss. The top 20 pathways which were significantly enriched are shown in Figure 3C. The pathways on systemic lupus erythematosus and alcoholism which include 20 genes encoding histone variants are on the top of the list. Most of the genes encoding histone variants are up-regulated, which are consistent with a recent finding showing that altered expression histone variants are associated with cancer (20). The pathways including AGE/RAGE signaling, Chagas disease, cytokine-cytokine receptor interaction, Pertussis, Toll-like receptor signaling, NF-kappa B signaling, Rheumatoid arthritis, Malaria, TNF signaling are related with the immune system, suggesting that STAG2-loss is involved in cancer evasion from the host immune system.
STAG2-loss also regulates PI3K/AKT signaling pathway, which is involved in many types of cancer initiation and progress. We checked the signaling pathway in SATG2-loss cells by Western blot (Figure 3D). PI3 kinase kept the same expression level but phosphorylation decreased in STAG2-loss cells. Phosphorylation of mTOR was also reduced, while there was no detectable change of total protein levels. Total AKT protein level decreased, which also reduced the phosphorylated forms. These data points showed that PI3K/AKT signaling was reduced in STAG-loss cells.
Discussion
There are recurrent STAG2 truncating mutations in OS cells, and mutations result in the loss of STAG2 (7-9). However, as one of the essential complex components in cell division, the mechanism by which STAG2 deletion promotes cell tumorigenesis remains unclear. Here we reported three effects of STAG2-loss on cancer cells: (I) reduced cell growth and proliferation; (II) increased cell invasion and migration; (III) enhanced immune evasion of cancer cells. The enhanced ability of immune evasion may partially explain how STAG2-loss promotes tumorigenesis.
Inhibitors of the PD-1/PD-L1 axis achieve great successes for the treatment of many types of cancer. PD-L1 can interact with PD-1 on T lymphocytes and inhibit the initial activation of T cells and suppress effector CD8+ T cell cytotoxicity (11,12). Juneja et al. reported that PD-L1 expression in tumor cells could inhibit the CD8+ T cell activities, and protect PD-L1+ tumor cells from an attack of the immune system, which establishes a role of PD-L1 in immune evasion of tumor cells (12). STAG2-loss induced PD-L1 expression may protect tumor cells from attacks by the immune system and promote tumorigenesis. Moreover, the study showed that overexpressed PD-L1 on tumor cells was associated with response to the treatment of inhibitors of the PD-1/PD-L1 immune checkpoint (21), which suggests that patients with STAG2 mutation may be sensitive to the inhibitors PD-1/PD-L1.
How does STAG2-loss affect PD-L1 expression? We observed a 1.5-fold upregulation of PD-L1 mRNA levels. As a component of the cohesin complex, STAG2 mediates interactions between cohesin and CTCF (22), which is critical for chromatin organization and insulator function of the CTCF binding sites. Studies have shown that mutations in the CTCF binding sites in tumor cells lose their insulator functions and up-regulate oncogene expression (23). It is possible that the insulators near the CD274 gene are damaged, which induces an enhancer invasion. A recently published study reported that the expression of the CD274 gene is under the control of 140 kb downstream super-enhancer in cancer cells (24).
On the other hand, up-regulated PD-L1 expression may also be associated with protein degradation. A recent study showed that CCND1 could degrade PD-L1 by regulating ubiquitin ligase (25). Prolonged cell cycling and decreased expression of CCND1 in STAG2-loss cells can repress the degradation of PD-L1. It is possible that in this case, both ways function.
STAG2-loss and low expression are associated with a higher incidence of metastatic disease at the time of diagnosis and a lower probability of survival in Ewing sarcoma patients (7-9). Our Transwell assay showed that the STAG2-loss cells have a stronger ability of migration. At the molecular level, we observed down-regulation of E-cadherin and up-regulation of N-cadherin, which is consistent with the aggressive phenotype of tumors. Many researchers observed that a switch from E-cadherin to N-cadherin expression is associated with epithelial-mesenchymal Transition (EMT). Furthermore, our RNA-seq data showed a significantly changed expression of extracellular matrix-related genes. These data show that STAG2-loss enhances invasive ability of OS cells by altering extracellular matrix and cell migration.
In summary, we observed that STAG2-loss induced PD-L1 expression in OS cells and altered expression of many genes in the immune system, which indicates that the STAG-loss cells obtain immune evasion capability at the expense of the advantage of proliferation to promote tumorigenesis. This finding suggests that STAG2-loss may be a potential biomarker for the treatment of immune checkpoint inhibitors of the PD-1/PD-L1 axis.
Acknowledgements
The authors would like to thank the members of the Hao lab for their technical input and suggestions, along with the staff at the Cancer Research Institute in Southern Medical University who provided support.
Funding: This research was supported by the National Natural Science Foundation of China (81470121 to A Jin), Guangdong Province Natural Science Foundation (2016A030313603 to B Hao).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Brooker AS, Berkowitz KM. The roles of cohesins in mitosis, meiosis, and human health and disease. Methods Mol Biol 2014;1170:229-66. [Crossref] [PubMed]
- Seitan VC, Hao B, Tachibana-Konwalski K, et al. A role for cohesin in T-cell-receptor rearrangement and thymocyte differentiation. Nature 2011;476:467-71. [Crossref] [PubMed]
- Balbás-Martínez C, Sagrera A, Carrillo-de-Santa-Pau E, et al. Recurrent inactivation of STAG2 in bladder cancer is not associated with aneuploidy. Nat Genet 2013;45:1464-9. [Crossref] [PubMed]
- Kong X, Ball AR Jr, Pham HX, et al. Distinct functions of human cohesin-SA1 and cohesin-SA2 in double-strand break repair. Mol Cell Biol 2014;34:685-98. [Crossref] [PubMed]
- Zuin J, Dixon JR, van der Reijden MI, et al. Cohesin and CTCF differentially affect chromatin architecture and gene expression in human cells. Proc Natl Acad Sci U S A 2014;111:996-1001. [Crossref] [PubMed]
- Hill VK, Kim JS, Waldman T. Cohesin mutations in human cancer. Biochim Biophys Acta 2016;1866:1-11. [PubMed]
- Brohl AS, Solomon DA, Chang W, et al. The genomic landscape of the Ewing Sarcoma family of tumors reveals recurrent STAG2 mutation. PLoS Genet 2014;10:e1004475. [Crossref] [PubMed]
- Crompton BD, Stewart C, Taylor-Weiner A, et al. The genomic landscape of pediatric Ewing sarcoma. Cancer Discov 2014;4:1326-41. [Crossref] [PubMed]
- Tirode F, Surdez D, Ma X, et al. Genomic landscape of Ewing sarcoma defines an aggressive subtype with co-association of STAG2 and TP53 mutations. Cancer Discov 2014;4:1342-53. [Crossref] [PubMed]
- Solomon DA, Kim T, Diaz-Martinez LA, et al. Mutational inactivation of STAG2 causes aneuploidy in human cancer. Science 2011;333:1039-43. [Crossref] [PubMed]
- Mühlbauer M, Fleck M, Schutz C, et al. PD-L1 is induced in hepatocytes by viral infection and by interferon-alpha and -gamma and mediates T cell apoptosis. J Hepatol 2006;45:520-8. [Crossref] [PubMed]
- Juneja VR, McGuire KA, Manguso RT, et al. PD-L1 on tumor cells is sufficient for immune evasion in immunogenic tumors and inhibits CD8 T cell cytotoxicity. J Exp Med 2017;214:895-904. [Crossref] [PubMed]
- Casey SC, Tong L, Li Y, et al. MYC regulates the antitumor immune response through CD47 and PD-L1. Science 2016;352:227-31. [Crossref] [PubMed]
- Chen DS, Mellman I. Elements of cancer immunity and the cancer-immune set point. Nature 2017;541:321-30. [Crossref] [PubMed]
- Patel SP, Kurzrock R. PD-L1 Expression as a Predictive Biomarker in Cancer Immunotherapy. Mol Cancer Ther 2015;14:847-56. [Crossref] [PubMed]
- Forbes SA, Beare D, Boutselakis H, et al. COSMIC: somatic cancer genetics at high-resolution. Nucleic Acids Res 2017;45:D777-83. [Crossref] [PubMed]
- Zhang X, Wang W, Shan L, et al. Gene activation in human cells using CRISPR/Cpf1-p300 and CRISPR/Cpf1-SunTag systems. Protein Cell 2018;9:380-3. [PubMed]
- Daniloski Z, Smith S.. Loss of Tumor Suppressor STAG2 Promotes Telomere Recombination and Extends the Replicative Lifespan of Normal Human Cells. Cancer Res 2017;77:5530-42. [Crossref] [PubMed]
- Uhlen M, Zhang C, Lee S, et al. A pathology atlas of the human cancer transcriptome. Science 2017;357:eaan2507.
- Vardabasso C, Hasson D, Ratnakumar K, et al. Histone variants: emerging players in cancer biology. Cell Mol Life Sci 2014;71:379-404. [Crossref] [PubMed]
- Shimizu T, Fuchimoto Y, Fukuda K, et al. The effect of immune checkpoint inhibitors on lung metastases of osteosarcoma. J Pediatr Surg 2017;52:2047-50. [Crossref] [PubMed]
- Xiao T, Wallace J, Felsenfeld G. Specific sites in the C terminus of CTCF interact with the SA2 subunit of the cohesin complex and are required for cohesin-dependent insulation activity. Mol Cell Biol 2011;31:2174-83. [Crossref] [PubMed]
- Hnisz D, Weintraub AS, Day DS, et al. Activation of proto-oncogenes by disruption of chromosome neighborhoods. Science 2016;351:1454-8. [Crossref] [PubMed]
- Chen H, Li C, Peng X, et al. A Pan-Cancer Analysis of Enhancer Expression in Nearly 9000 Patient Samples. Cell 2018;173:386-99.e12. [Crossref] [PubMed]
- Zhang J, Bu X, Wang H, et al. Cyclin D-CDK4 kinase destabilizes PD-L1 via cullin 3-SPOP to control cancer immune surveillance. Nature 2018;553:91-5. [Crossref] [PubMed]


