
COL11A1 promotes esophageal squamous cell carcinoma proliferation and metastasis and is inversely regulated by miR-335-5p
Introduction
As a malignant tumor that develops in esophageal epithelial tissue, the prevalence of esophageal cancer is quite high in China. In 2015, around 477,000 newly diagnosed cases were reported with early 375,000 deaths (1,2). Esophageal cancer is divided into 2 main histological subtypes: adenocarcinoma and squamous cell carcinoma. Esophageal squamous cell carcinoma (ESCC) is the dominant subtype in China (3). While modern diagnostic tools, perioperative care, radiation, and chemotherapy have improved, the risk of postoperative metastasis and recurrence of esophageal malignancy remains high, and the 5-year overall rate of survival is poor (4,5). Early identification and treatment of esophageal cancer can significantly improve the prognosis. Thus, understanding the molecular processes underlying ESCC is critical for the improvement of detection methods and for treating individuals suffering from esophageal cancer.
The progress of high-throughput technology has resulted in the identification of new biomarkers involved in the development of ESCC. Wei et al. discovered a long non-coding RNA (lncRNA), namely, esophageal epithelial intergenic associated transcript (EPIST) using RNA sequencing, as a tumor suppressor in ESCC (6). Tong et al. used Illumina high-throughput sequencing and RNA-Seq profiling in 12 ESCC clinical samples from humans, identifying Rab25 as a tumor suppressor gene having anti-invasive and antiangiogenic effects in ESCC (7). You et al. discovered that through its interaction with EZH2, the lncRNA HERES controlled both canonical and noncanonical Wnt signaling pathways (8). Bioinformatic analytical tools have been developed and are useful for reanalyzing Gene Expression Omnibus (GEO) datasets. For example, Dong et al. reanalyzed the GSE38129, and GSE20347 datasets and identified TPX2, CDK1, and CEP55 as key genes for ESCC diagnosis and treatment (9); and Yang et al. performed analysis on the GSE29001, GSE20347, GSE100942, and GSE38129 datasets and found that nuclear division cycle 80 was intimately associated with the survival of ESCC individuals (10).
Research has associated the abnormal expression of type I collagen and type IV collagen genes with cancer progression (11). The collagen type XI alpha 1 (COL11Al) gene is housed in the lp21 zone of the chromosome and carries 68 exons (12). Recent studies have shown that COL11Al essentially functions in tumor occurrence and development (13,14). Under normal circumstances, most tissues have a very low or non-existent expression of COL11A1. However, COL11A1 was found to be upregulated, in head and neck squamous cell carcinoma, pancreatic and ovarian cancers, whereas silent COL11A1 inhibited tumor cells proliferation, migration, and invasion in ovarian cancer along with head and neck squamous cell carcinoma (15-17). At present, indepth studies of COL11A1 tumorigenesis and development are scant, and the roles and mechanisms of COL11A1 in esophageal cancer is obscure.
The current work performed integrated bioinformatics analysis using the GSE29968, GSE32424, and GSE130078 datasets and identified COL11A1 as the hub gene in ESCC progression. In vitro functional investigations were then conducted for determining the purpose of COL11A1 in ESCC progression. The effect mechanism of COL11A1 on cells was studied, and a validation experiment was carried out in nude mice. The present study may provide new understanding of COL11A1 in the pathophysiology of ESCC.
We present the following article in accordance with the MDAR reporting checklist (available at https://dx.doi.org/10.21037/atm-21-4951).
Methods
Microarray data information
The publically accessible GEO databank, a genomics data repository for next-generation sequencing and comprising sequence- and array-based data of gene profiles, was used for profiling studies of ESCC gene expression. Three datasets GSE130078, GSE32424, GSE29968 and were picked out of the GEO repository. Three tissues of ESCC and three typical tissues of the esophagus were a part of the GSE29968 dataset, which was obtained from the GPL10999 Illumina Genome Analyzer IIx. The GSE32424 dataset includes 7 ESCC tissues and 5 normal tissues of the esophagus and was derived from the GPL10999 Illumina Genome Analyzer IIx. The dataset GSE130078 includes 23 ESCC tissues and 23 normal tissues of the esophagus and was generated from the GPL11154 Illumina HiSeq 2000. The various steps of the research were conducted in line with the Helsinki Declaration (2013 revision).
Processing of data and identification of differentially expressed genes (DEGs)
EdgeR computer program (18) was employed for analyzing the unprocessed microarray data from the three datasets acquired from the GEO repository to attempt the detection of DEGs in normal esophageal tissue compared to ESCC tissue. The cutoff criteria for DEG screening were a log fold change (FC) >1.5 and a false discovery rate (FDR) value <0.05. By employing the Interactive Gene Expression Analysis Kit (iGEAK), overlapping DEGs between GSE29968, GSE130078, and GSE32424 were subsequently discovered (19).
Functional analysis of DEGs
The Kyoto Encyclopedia of Genes and Genomes (KEGG) databank (http://www.genome.jp/kegg/) and the Gene Ontology (GO) Resource were employed for the classification and grouping of DEGs. To perform GO enrichment, and KEGG, Reactome Database, WikiPathways, and miRNA pathway analysis of the DEGs, an ontology-based tool, g:Profiler (https://biit.cs.ut.ee/gprofiler/gost), was employed. GO enrichment study evaluated which GO terms were over or underrepresented within a particular set of genes, inclusive of cellular components, biological processes, and molecular functions. To identify metabolic and functional pathways, the KEGG database, an integrated database resource, is commonly employed.
Protein-protein interaction (PPI) network construction and identification of hub genes
The PPI of the DEGs and each pair of PPI were attained using STRING (http://string-db.org), having a reliability threshold of a combined score of >0.4 chosen as a pair of meaningful interaction. A PPI network was built and visualized using Cytoscape computer program (http://www.cytoscape.org/; version 3.4.0,) based on the interaction pair information. For further analysis, the Cytoscape plug-in Network Analyzer was utilized, and topological parameters of the network of PPI, such as the degree of the node, were determined for finding hub genes within the network of PPI. Following that, making use of the mentioned parameters, the PPI network was searched for important modules using Molecular Complex Detection (MCODE) assessment in Cytoscape.: node score cutoff =0.2, K-Core =2, and degree cutoff =2.
Hub gene expression level and survival assessment
The expression of 10 hub genes was visualized using Gene Expression Profiling Interactive Analysis (GEPIA) as a boxplot. P<0.05 was deemed statistically significant. Survival assessment of the DEGs was executed through making use of the GEPIA database (20) and Kaplan-Meier curves were plotted using data on 364 ESCC patients considering P<0.05 as statistically significant.
Cell lines and culture
Human esophageal cancer cell lines Eca-109, KYSE-30, and KYSE-510, and human esophagus epithelial cells (HEEC) were bought from Guangzhou Cellcook Cell Biotech Co., Ltd. (Guangzhou, China) and cultivated in Gibco RPMI 1640 milieu (Thermo Fisher Scientific, Waltham, MA, USA) with 10% Gibco fetal bovine serum (FBS). The American Type Culture Collection (ATCC, Manassas, VA, USA) provided the HEEC, which were maintained in Gibco DMEM media supplied with 10% Gibco FBS. The cells were stored at 37 °C in a 5% CO2 ambient.
Real-time RT-PCR
Trizol reagent (Invitrogen, Waltham, MA, USA) was utilized for extracting total RNA from epithelial cells cancerous cells of the human esophagus The PrimeScript 1st strand cDNA Synthesis Kit (Takara Biomedical Technology, Beijing, China) was then used to reverse transcription 100 ng total RNA. During reverse transcription, Bulge-Loop miRNA primers (Guangzhou RiboBio Co., Ltd., Guangzhou, China) were added. The LightCycler 480 II Real-time PCR Instrument (Roche Diagnostics GmbH, Mannheim, Germany) was then used to run real-time PCR by employing the LightCycler 480 SYBR Green I Master Mix (Roche Diagnostics GmbH). COL11A1 primers: forward: 5'-TGGTGATCAGAATCAGAAGTTCG-3', reverse: 5'-AGGAGAGTTGAGAATTGGGATTC-3'. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH): forward: 5'-AAAGGGTGTAACGCAACTA-3', reverse: 5'-AAAGGGTGTAACGCAACTA-3'. U6: forward: 5'-ATGGACTATCATATGCTTACCGTA-3', reverse: 5'-GTAAGTAGTTTATGTGGCCAGTCG-3'. Internal controls respectively employed for mRNA and miRNA were GAPDH and U6. The approach of 2(−ΔΔCt) was employed for calculating the relative expressions.
Oligonucleotide transfection
Empty pcDNA3.1 vector, COL11A1 pcDNA3.1 overexpression vector, specific small interfering RNA (siRNA) against COL11A1, NC siRNA, microRNA-335-5p (miR-335-5p) mimics, and mimics NC were prepared with the aid of Guangzhou RiboBio Co., Ltd. (Guangzhou, China). In the Opti-MEM medium, oligonucleotides were transfected into the cells employing Lipofectamine 2000 (Invitrogen) for various time intervals. The cells were then collected for future research.
Colony formation
KYSE-30/510 cells were counted and plated in dishes containing 6 wells at a density of 1,000 cells/well during the logarithmic growth period. The cells were kept in full media for 2 weeks and allowed to grow. The colonies were then fixed, counted using a stereo microscope after staining by utilizing 0.5% w/v crystal violet solution. At least 50 cells should make up a colony.
Cell Counting Kit-8 (CCK-8) assessment
A CCK-8 test was exerted for detecting cell growth (Dojindo Laboratories, Kumamoto, Japan). Each of the 96-well plates was planted with 1×104 cells. Following transfection of cells for 24, 48, 72, and 96 hours, CCK-8 reagent was applied to individual wells. After that, the cells were exposed to incubation in an incubator for another 2 hours at 37 °C. A Multiskan FC Microplate Photometer was employed for measuring optical density (OD) at 450 nm (Thermo Fisher Scientific).
Cell migration and invasion assessments
Both the transwell migration and invasion assessments were carried out in transwell inserts containing 24 wells with an 8 µm pore size (Corning, Corning, NY, USA). A total of 2×104 transfected cells were placed in the top compartment with a growth medium in the absence of FBS for the migration assay. The upper Matrigel-coated compartment was filled with transfected cells for the invasion assay. The lower chambers in both experiments were infused with the appropriate culture media along with the chemoattractant; precisely FBS. A hundred percent methanol was used to preserve the cells that transferred to or entered the lower chamber, stained with 0.5% crystal violet solution. The cells were quantified after observing them under a microscope after 24 hours at 37 °C.
In vivo study
Male athymic BALB/c nude mice, 4–5 week-old, were kept individually in ventilated cages in a holding room with 40–60% humidity. Animals were provided with sterilized or disinfected food, water, and bedding. The mice were subcutaneously injected with 1×107 transfected cancer cells into the left flank (n=6 for each group). Tumor volume was detected once a week (volume = 0.5 × length × width2). Five weeks after inoculation, the mice were euthanized. All animal experiments were carried out in accordance with the Chinese Society of Experimental Animals (Chinese Journal of Experimental Animals). The Affiliated Cancer Hospital of Nanjing Medical University’s Animal Welfare Committee granted project permission (No. 202120584) to conduct the experiments.
Dual-luciferase reporter assessment
To make wild-type (WT) or mutant (MUT) luciferase reporter vectors, cloning of the WT or MUT COL11A1 3' untranslated region (3'UTR) sequence was done into the pmirGLO vector (Promega Corporation, Madison, WI, USA). pmirGLO-COL11A1-WT and pmirGLO-COL11A1-MUT were subsequently co-transfected with miR-335-5p mimics or mimic controls in KYSE-30 cells utilizing Lipofectamine 2000 reagent (Invitrogen). A dual-luciferase reporter assay kit (Promega Corporation) was implemented for the evaluation of the luciferase activity 48 hours later.
RNA pull-down assessment
Trypsinization and homogenization of the cells were carried out using Beyotime Biotechnology’s lysis buffer (Wuxi, China). Using streptavidin-coated magnetic beads, RNA was pulled down from cell lysates. The expression of COL11A1 mRNA in bound fractions was measured with the aid of qPCR and normalized to input controls. RNeasy mini kit (Qiagen, Shanghai, China) was utilized to purify the bound mRNA.
Statistical analysis
The entire outcome was documented as a mean ± SD, and tests were carried out in triplicate, at the very least. The data was analyzed and the graph was plotted using GraphPad Prism. To assess if there existed significant differences among groups, the Student’s t-test or one-way ANOVA is employed. Statistical significance was referred to as a P value of lower than 0.05.
Results
Identifying the DEGs of ESCC
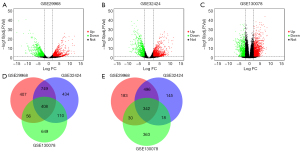
Overall, 2,681 DEGs were identified from GSE29968, comprising 1,051 downregulated genes and 1,620 upregulated genes (Figure 1A). A total of 2,702 DEGs were identified from GSE32424, inclusive of 1,001 downregulated genes and 1,701 upregulated genes (Figure 1B). Whereas within GSE130078, a sum of 1,976 DEGs was detected, comprising 753 downregulated genes and 1223 upregulated genes (Figure 1C). The iGEAK tool analysis found a total of 750 genes overlapping among GSE29968, GSE32424, and GSE130078, including 408 upregulated and 342 downregulated genes (Figure 1D,1E).
DEGs functional and pathway enrichment assessment
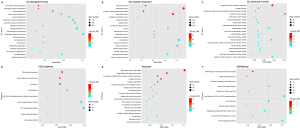
As an outcome of the GO enrichment assessment, DEGs were found to be associated with: (I) extracellular structures organization, extracellular matrix (ECM) organization, tissue development, and organ development in biological processes (Figure 2A); (II) ECM, extracellular region, collagen-carrying ECM, and extracellular space in the cellular component (Figure 2B); and (III) ECM structural constituents, collagen binding, conferring tensile strength in molecular function (Figure 2C). The DEGs were abundant in, protein absorption and digestion, the interaction of ECM-receptor, amoebiasis, and focal adhesion, according to KEGG pathway analysis (Figure 2D). The DEGs were found to be abundant in ECM organization, ECM degradation, the assembly of collagen fibrils and other multimeric structures, and the synthesis of collagen, according to Reactome pathway analysis (Figure 2E). The DEGs were also shown to be enriched in miRNA targets in the membrane receptors, ECM, matrix metalloproteinases, the miR-509-3p alteration of YAP1/ECM axis, and the vitamin D receptor pathway, according to WikiPathways study (Figure 2F).
Construction of PPI network and identification of hub gene
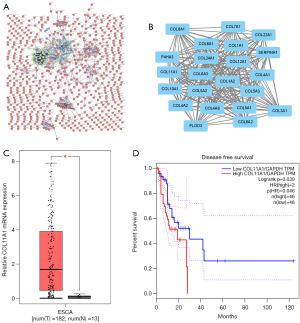
To obtain PPI information, the potential DEGs were uploaded onto the web tool STRING, and the outcome was reported as a PPI network (Figure 3A). Using MCODE analysis in Cytoscape, significant modules were extracted from the network of PPI. For further investigation, it was identified the most crucial module (Figure 3B). This module consisted of 22 nodes and 228 edges, and 22 of the genes were classified as hub DEGs.
Hub gene expression level and survival analysis
Among these hub genes, we chose COL11A1 for further analysis. COL11A1 was increased in ESCC tissue when compared to normal esophageal tissue, as seen in Figure 3C. Furthermore, the survival analysis revealed that raised COL11A1 expression has an association with shorter disease-free survival in ESCC patients (Figure 3D), implying that COL11A1 plays an oncogenic function in ESCC.
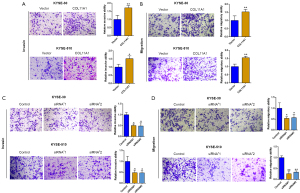
The impact of overexpressing and silencing COL11A1 on esophageal cancer cell proliferation
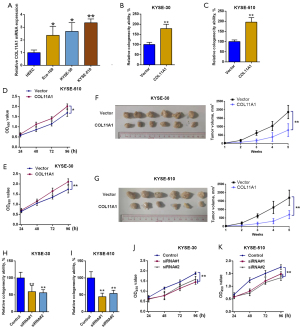
The degree of expressions of COL11A1 in typical esophagus epithelial cells and esophageal cancer cell lines were compared. COL11A1 was upregulated in Eca-109, KYSE-510, and KYSE-30 cells compared with HEEC and the degree of disparity was significant (Figure 4A). To explore the function of COL11A1 in cell proliferation, data on overexpression and silence of COL11A1 was obtained by transfection with COL11A1-pcDNA3.1 overexpression vector and specific siRNA respectively. In both KYSE-510 and KYSE-30 cells, colony formation assays and CCK8 assays revealed that ectopic expression of COL11A1 increased cell proliferation, with colony numbers and CCK-8 OD values increasing significantly (Figure 4B-4E). Consistent with the in vitro study, the COL11A1 overexpressing group manifested a larger tumor volume in comparison to the control group (Figure 4F-4G). Similarly, the silencing of COL11A1 showed the opposite effect (Figure 4H-4K).
The impact of overexpressing and silencing COL11A1 on esophageal cancer cell migration and invasion
Cancer cell migratory and invasive abilities are related to tumor metastasis. As a result, researchers looked into the influence of COL11A1 on migration and invasion of cells. Overexpression of COL11A1 significantly improved cell migration and invasion ability in both KYSE-510 and KYSE-30 cells compared to the control group in transwell migration and invasion assays. Furthermore, knocking down COL11A1 stopped both cancer cells from migrating and invading (Figure 5A-5D).
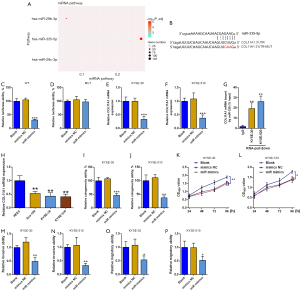
COL11A1 is the target of miR-335-5p
The miRNA pathway assessment showed the potential interplay among COL11A1 and miR-335-5p (Figure 6A). Online software indicated miR-335-5p is the upstream regulator of COL11AL. To confirm the above hypothesis, luciferase reporter assay was conducted. Downregulated luciferase activity occurred when miR-335-5p mimics were cotransfected with pmirGLO-COL11A1 which included wide type sequence. However, when cotransfected with MUT pmirGLO-COL11A1, miR-335-5p mimics demonstrated no influence on luciferase activity (Figure 6A-6D). In addition, miR-335-5p mimics caused a substantial decrease in COL11A1 expression levels in both KYSE-510 and KYSE-30 cells. RNA pull-down assay also demonstrated COL11A1 mRNA bound by miR-335-5p was highly enriched (Figure 6E-6G). These results further validate the existence of interaction among miR-335-5p and COL11A1. Next, the biological role of miR-335-5p in esophageal cancer was explored. QRT-PCR results illustrated a down-regulation of miR-335-5p in the cancerous cells of the esophagus in comparison with normal esophagus epithelial cells. MiR-335-5p mimics exerted anticancer effects, inhibiting colony formation ability, cell proliferation, and invasive and migratory abilities in both KYSE-510 and KYSE-30 cells (Figure 6H-6P).
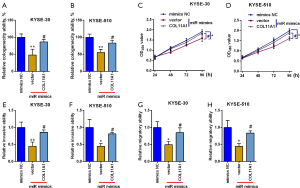
Overexpression of COL11A1 counteracted the influence of miR-335-5p on esophageal cancer
Rescue studies were then executed to look at the miR-335-5p/COL11A1 axis in esophageal cancer. MiR-335-5p mimics alone reduced cologenecity ability and impeded proliferation, invasion, and migration of cells. COL11A1 overexpression counteracted the impact of miR-335-5p mimics, reversing the colony formation, cell proliferation, invasion, and migration (Figure 7A-7H).
Discussion
ESCC is a common human malignancy and the 5-year overall rate of survival for individuals with advanced-stage ESCC is relatively low (21). The cause of ESCC carcinogenesis is unknown at this time, which makes it difficult to discover effective treatments for advanced ESCC. Thus, further exploration of molecular mechanisms underlying ESCC pathophysiology is important for developing novel therapies for ESCC. The current work is based on integrated bioinformatics analysis and demonstrated that COL11A1 was upregulated in ESCC tissue and that upregulation of COL11A1 was correlated with poor disease-free survival for ESCC patients, suggesting the oncogenic role of COL11A1 in ESCC. In vitro studies showed the upregulation of COL11A1 in ESCC cell lines. COL11A1 Overexpression stimulated ESCC cell proliferation, invasion, and migration; whereas knockdown of COL11A1 repressed proliferation, invasion, and migration of ESCC cells. Further, TargetScan’s online prediction tool, the luciferase reporter assay, and miRNA pathway analysis together revealed that COL11A1 3'UTR was targeted by miR-335-5p and negatively regulated by miR-335-5p in ESCC cells. miR-335-5p Overexpression slowed the development of ESCC cells. Furthermore, induced COL11A1 expression reduced the inhibitory influence caused by overexpression of miR-335-5p on ESCC cell growth and metastasis.
The role of COL11A1 in regulating tumor progression has been demonstrated in a number of cancers. COL11A1 has been reported to promote ovarian cancer progression in a study by Wu et al. and predicted a poor clinical outcome in patients suffering from ovarian cancer (22). In lung cancer, COL11A1 was observed to undergo upregulation in recurrent lung cancer and to promote lung cancer cell proliferation, metastasis, and chemoresistance (23). Recent studies also revealed that knockdown of COL11A1 could induce oxidation of fatty acid, which subsequently stimulates cell death caused by apoptosis in cisplatin-resistant ovarian cancer (24). GSE17351 analysis found that COL11A1 is indeed a hub gene within the PPI network in ESCC and could be used as a target gene in ESCC treatment (25). It was discovered by Zhang et al. that COL11A1 might be used as a predictive biomarker in ESCC patients (26). The assessment of data on the basis of The Cancer Genome Atlas (TCGA) and GSE53625 has revealed that COL11A1 may be a prospective biomarker for predicting survival outcomes of individuals suffering from ESCC (27). Herein, it was illustrated that COLL11A1 was one of the hub genes, that it was upregulated in ESCC tissue, and that upregulation of COL11A1 has a correlation with poor disease-free survival of patients suffering from ESCC, suggesting the oncogenic role of COL11A1 in ESCC. Besides, functional investigations revealed that COL11A1 overexpression promoted ESCC cell progression whereas COL11A1 knockdown exerted the opposite impacts. Collectively, the achieved outcomes demonstrated that COL11A1 acted as an oncogene to promote ESCC cell progression.
MiRNA pathway analysis revealed the potential interaction between COL11A1 and miR-335-5p, which was additionally confirmed using TargetScan’s database prediction and luciferase reporter assay. MiR-335-5p downregulated COL11A1 expression in ESCC cells by targeting COL11A1 3'UTR. MiR-335-5p behaves as a repressor of tumors in various types of cancers. By targeting rho-associated protein kinase 1 (ROCK1), MiR-335-5p suppressed TGF-beta1-induced epithelial-mesenchymal transition in cancer of the lung (28). MiR-335-5p inhibited colorectal cancer proliferative and metastatic potential by suppressing lactate dehydrogenase B (LDHB), according to Zhang et al. (29). MiR-335-5p has also been illustrated to suppress thyroid carcinoma invasion and metastasis by targeting intercellular adhesion molecule-1 (ICAM-1) (30). The functions of miR-335-5p in ESCC have yet to be discovered. For the first time, we discovered that overexpression of miR-335-5p inhibited ESCC cell proliferation and metastasis, implying that miR-335-5p has suppressive roles in the tumor in ESCC. Furthermore, the rescue tests revealed that enforcing COL11A1 expression reduced the tumor-suppressive impact of miR-335-5p upon ESCC. The obtained findings proposed that miR-335-5p inhibited ESCC development by targeting COL11A1.
Conclusions
In this study, researchers used integrated bioinformatics to identify COL11A1 as an oncogene in ESCC. The mechanistic studies indicated that COL11A1 promoted ESCC cell progression and that the expression of COL11A1 in ESCC was regulated negatively by miR-335-5p. The present exploration was the first one to demonstrate the important performances of the miR-335-5p/COL11A1 axis in regulating ESCC progression. The in vivo functional roles and clinical significance of the miR-335-5p/COL11A1 axis in ESCC still need further exploration.
Acknowledgments
The authors appreciate the academic support from AME Esophageal Cancer Collaborative Group.
Funding: The study was supported by Maternal and Child Health Project of Jiangsu Province Health Committee (Grant No. F201914).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://dx.doi.org/10.21037/atm-21-4951
Data Sharing Statement: Available at https://dx.doi.org/10.21037/atm-21-4951
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/atm-21-4951). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The various steps of the research were conducted in line with the Helsinki Declaration (2013 revision). All animal experiments were carried out in accordance with the Chinese Society of Experimental Animals (Chinese Journal of Experimental Animals) for the care and use of animals. The Affiliated Cancer Hospital of Nanjing Medical University’s Animal Welfare Committee granted project permission (No. 202120584) to conduct the experiments.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016;66:115-32. [Crossref] [PubMed]
- Li M, Wan X, Wang Y, et al. Time trends of esophageal and gastric cancer mortality in China, 1991-2009: an age-period-cohort analysis. Sci Rep 2017;7:6797. [Crossref] [PubMed]
- Liang H, Fan JH, Qiao YL. Epidemiology, etiology, and prevention of esophageal squamous cell carcinoma in China. Cancer Biol Med 2017;14:33-41. [Crossref] [PubMed]
- Huang FL, Yu SJ. Esophageal cancer: risk factors, genetic association, and treatment. Asian J Surg 2018;41:210-5. [Crossref] [PubMed]
- Watanabe M, Otake R, Kozuki R, et al. Recent progress in multidisciplinary treatment for patients with esophageal cancer. Surg Today 2020;50:12-20. [Crossref] [PubMed]
- Wei G, Luo H, Sun Y, et al. Transcriptome profiling of esophageal squamous cell carcinoma reveals a long noncoding RNA acting as a tumor suppressor. Oncotarget 2015;6:17065-80. [Crossref] [PubMed]
- Tong M, Chan KW, Bao JY, et al. Rab25 is a tumor suppressor gene with antiangiogenic and anti-invasive activities in esophageal squamous cell carcinoma. Cancer Res 2012;72:6024-35. [Crossref] [PubMed]
- You BH, Yoon JH, Kang H, et al. HERES, a lncRNA that regulates canonical and noncanonical Wnt signaling pathways via interaction with EZH2. Proc Natl Acad Sci U S A 2019;116:24620-9. [Crossref] [PubMed]
- Dong Z, Zhang H, Zhan T, et al. Integrated analysis of differentially expressed genes in esophageal squamous cell carcinoma using bioinformatics. Neoplasma 2018;65:523-31. [Crossref] [PubMed]
- Yang W, Zhao X, Han Y, et al. Identification of hub genes and therapeutic drugs in esophageal squamous cell carcinoma based on integrated bioinformatics strategy. Cancer Cell Int 2019;19:142. [Crossref] [PubMed]
- Ricard-Blum S. The collagen family. Cold Spring Harb Perspect Biol 2011;3:a004978 [Crossref] [PubMed]
- Raglow Z, Thomas SM. Tumor matrix protein collagen XIα1 in cancer. Cancer Lett 2015;357:448-53. [Crossref] [PubMed]
- Mio F, Chiba K, Hirose Y, et al. A functional polymorphism in COL11A1, which encodes the alpha 1 chain of type XI collagen, is associated with susceptibility to lumbar disc herniation. Am J Hum Genet 2007;81:1271-7. [Crossref] [PubMed]
- Vázquez-Villa F, García-Ocaña M, Galván JA, et al. COL11A1/(pro)collagen 11A1 expression is a remarkable biomarker of human invasive carcinoma-associated stromal cells and carcinoma progression. Tumour Biol 2015;36:2213-22. [Crossref] [PubMed]
- Rada M, Nallanthighal S, Cha J, et al. Inhibitor of apoptosis proteins (IAPs) mediate collagen type XI alpha 1-driven cisplatin resistance in ovarian cancer. Oncogene 2018;37:4809-20. [Crossref] [PubMed]
- Sok JC, Lee JA, Dasari S, et al. Collagen type XI α1 facilitates head and neck squamous cell cancer growth and invasion. Br J Cancer 2013;109:3049-56. [Crossref] [PubMed]
- García-Pravia C, Galván JA, Gutiérrez-Corral N, et al. Overexpression of COL11A1 by cancer-associated fibroblasts: clinical relevance of a stromal marker in pancreatic cancer. PLoS One 2013;8:e78327 [Crossref] [PubMed]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010;26:139-40. [Crossref] [PubMed]
- Choi K, Ratner N. iGEAK: an interactive gene expression analysis kit for seamless workflow using the R/shiny platform. BMC Genomics 2019;20:177. [Crossref] [PubMed]
- Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 2017;45:W98-W102. [Crossref] [PubMed]
- Abnet CC, Arnold M, Wei WQ. Epidemiology of esophageal squamous cell carcinoma. Gastroenterology 2018;154:360-73. [Crossref] [PubMed]
- Wu YH, Chang TH, Huang YF, et al. COL11A1 promotes tumor progression and predicts poor clinical outcome in ovarian cancer. Oncogene 2014;33:3432-40. [Crossref] [PubMed]
- Shen L, Yang M, Lin Q, et al. COL11A1 is overexpressed in recurrent non-small cell lung cancer and promotes cell proliferation, migration, invasion and drug resistance. Oncol Rep 2016;36:877-85. [Crossref] [PubMed]
- Nallanthighal S, Rada M, Heiserman JP, et al. Inhibition of collagen XI alpha 1-induced fatty acid oxidation triggers apoptotic cell death in cisplatin-resistant ovarian cancer. Cell Death Dis 2020;11:258. [Crossref] [PubMed]
- He Y, Liu J, Zhao Z, et al. Bioinformatics analysis of gene expression profiles of esophageal squamous cell carcinoma. Dis Esophagus 2017;30:1-8. [Crossref] [PubMed]
- Zhang B, Zhang C, Yang X, et al. Cytoplasmic collagen XIαI as a prognostic biomarker in esophageal squamous cell carcinoma. Cancer Biol Ther 2018;19:364-72. [Crossref] [PubMed]
- Li J, Wang X, Zheng K, et al. The clinical significance of collagen family gene expression in esophageal squamous cell carcinoma. PeerJ 2019;7:e7705 [Crossref] [PubMed]
- Du W, Tang H, Lei Z, et al. miR-335-5p inhibits TGF-β1-induced epithelial-mesenchymal transition in non-small cell lung cancer via ROCK1. Respir Res 2019;20:225. [Crossref] [PubMed]
- Zhang D, Yang N. MiR-335-5p inhibits cell proliferation, migration and invasion in colorectal cancer through downregulating LDHB. J BUON 2019;24:1128-36. [PubMed]
- Luo L, Xia L, Zha B, et al. miR-335-5p targeting ICAM-1 inhibits invasion and metastasis of thyroid cancer cells. Biomed Pharmacother 2018;106:983-90. [Crossref] [PubMed]


