
Aberrant miRNA-mRNA regulatory network in polycystic ovary syndrome is associated with markers of insulin sensitivity and inflammation
Introduction
As one of the most prevalent multifactorial endocrine diseases, polycystic ovary syndrome (PCOS) is also a common cause of ovarian dysfunction and infertility in women of reproductive age. PCOS is related to metabolic aberrations such as dyslipidemia and may also increase the risk of obesity, insulin resistance, and endometrial carcinoma (1,2). It is characterized by polycystic ovaries in morphology, chronic anovulation, and hyperandrogenism. Hyperandrogenism is one of the clinical diagnostic criteria for PCOS. The dihydrotestosterone (DHT)-induced prenatally androgenized (PNA) mouse model is commonly used to mimic hyperandrogenemia and is an important model for the serological, pathological, and “omics” studies of PCOS.
The specific etiology of PCOS remains unexplored. In addition to genetic predisposition, environmental factors, and lifestyle factors, epigenetic modification, or regulation is also thought to promote the development of PCOS (2). Our previous study illustrated that the epigenetic silencing of LPCAT1 and PCYT1A was associated with lipidomic alterations in PCOS (3). As one of the main epigenetic modifiers, microRNAs (miRNAs) regulate gene expression. miRNAs are small non-coding RNAs that negatively regulate gene expression in post-transcriptional processing and participate in the regulation of various diseases including diabetes, inflammatory disease (4), and cancer (5). Increasingly, the available evidence indicates a role for the aberrant regulation of miRNA in PCOS. In human adipose tissue, granulosa cells (GCs), and peripheral blood leukocytes, miRNA expression may be altered, this has been found to be related to PCOS through the targeting of specific molecules and the regulation of pathways related to hormones and apoptosis (6,7). However, the aberrant expression of miRNAs in patients with PCOS and hyperandrogenemia has not been fully elucidated, and there have been few studies on the regulatory function of the miRNA-mRNA axis.
To investigate the molecular mechanism of the hyperandrogenic phenotype of PCOS, the PNA mouse model was used for our research. To identify the differentially expressed genes (DEGs) and differentially expressed microRNAs (DEmiRs) in the ovarian tissues of PNA and control mice, we performed RNA-seq and miRNA-seq, respectively. We then constructed a regulatory network between the miRNAs and negatively regulated target mRNAs and identified the biological pathways that were significantly associated with DEGs and DEmiRs. The expression of the selected miRNAs and target genes was validated in animal models and clinical samples using RT-qPCR. In addition, we treated GCs with DHT in vitro to examine whether androgens had a direct effect on cells. We aimed to investigate whether miRNAs dysregulation is potentially involved in the pathogenesis of PCOS in mice. We present the following article in accordance with the ARRIVE reporting checklist and STROBE reporting checklist (available at https://dx.doi.org/10.21037/atm-21-1288).
Methods
DHT induces the construction of PNA mice
Animal experiments in our research were approved by the School of Biomedical Engineering, Shanghai Jiao Tong University, Experimental Animals Welfare & Ethical Committee (no. 2019005), in compliance with Laboratory Animal Center of Shanghai Jiao Tong University guidelines for the care and use of animals. The Institute of Cancer Research mice (ICR mice) were purchased from Shanghai Jiesijie Laboratory Animal Co., Ltd. and housed at the Laboratory Animal Center of Shanghai Jiao Tong University in a suitable condition with light duration, constant temperature, and humidity with ad libitum access to food and water.
Females were mated with males and the copulatory plugs were checked daily. Day 1 of gestation meant the first day for plug development. On days 16–17 of gestation, 70 µL sesame oil having 350 µg DHT or only sesame oil vehicles were injected subcutaneously in pregnant dams daily. The offspring from the DHT-induced group were defined as PNA mice, and the control group’s offspring were defined as, in the same way, control mice. All offspring were placed in animal rooms in an environment of specific-pathogen-free. The experimental materials were harvested after the mice were euthanized using the anesthetic chloral hydrate.
Confirmation of PNA mice model establishment
To confirm the establishment of the PCOS model, we randomly selected six PNA mice and six control mice and assessed estrus duration, serum testosterone level, and the number of secondary follicles at 8 weeks of age. Vaginal smears were undergone daily and last for at least one complete estrous cycle to assess estrous duration. To assess the serum testosterone level, we collected the serum from each mouse in diestrus by removing the eyeball after anesthetizing with chloral hydrate. The serum testosterone level was assessed by the enzyme-linked immunosorbent assay (ELISA) kit (Beckman Coulter, Inc., USA) and microplate reader (BioTek, Inc., Winooski, USA).
After collecting blood from experimental mice, the ovaries were dissected, fixed in 4% paraformaldehyde at 4 °C overnight. After embedded in paraffin, sections at 5-µm thickness were cut and mounted onto 3-aininopropyltricttiosilane-coated glass slides. Then the hematoxylin and eosin (HE) stain were performed on the sections to observe the growth of follicles and count the number of follicles. The assessor was blinded with respect to group assignment.
GCs isolation and DHT treatment in vitro
Ovaries dissected from female ICR mice at 6 weeks of age were incubated in DMEM basic medium containing 10% fetal bovine serum, 2 mM glutamine, 1 mM sodium pyruvate, 100 µg/mL streptomycin, and 100 IU/mL penicillin (Gibco BRL/Invitrogen, Carlsbad, CA, USA). We used a syringe needle puncturing the ovaries quickly to release the GCs and oocytes. After filtered out the oocytes, isolated GCs in experimental group were cultured in a medium containing DHT (20649, MCE, China) accompanied by the control group was in a no DHT-supplementary medium. The groups were all incubated at 37 °C and 5% CO2. After incubated for 48 hours, the samples were centrifuged to remove the supernatant and then resuspended by 1 mL of TRIzol reagents (Invitrogen, Waltham, MA, USA).
Clinical characteristics and isolation of GCs
We recruited 12 participants including six female PCOS patients and six matched non-PCOS women for this study at the Yuncheng Center Hospital. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013) and was approved by the Ethics Committees on Human Research of Yuncheng Center Hospital (Approve ID: KYLL2019073). The PCOS patients were diagnosed by the Rotterdam ESHRE/ASRM (2003) criteria (8). And the patients had no other etiologies with similar symptoms and had not been treated with drugs for curing hyperandrogenism in the past 6 months. The control groups were healthy fertile women without symptoms of ovulatory dysfunction or androgen excess. Besides, all participants provided written informed consent.
The gonadotropin-releasing hormone agonist (GnRHa) was injected into participants at the beginning of the middle luteal phase. Serum estradiol and ultrasonography were performed to monitor follicle size. When more than three 16 mm-size follicles were monitored, participants were injected with human chorionic gonadotropin (HCG). Oocytes were then collected under the ultrasound 36 hours later. The isolated GCs were quickly immersed in liquid nitrogen for more than 20 min and then preserved at −80 °C.
Total RNA extraction
Extract total RNA from tissue samples and GCs samples using TRIzol reagents (Invitrogen, Waltham, MA, USA) following the guidance of the Manual. The total RNA was quantified by Nanodrop 2000 (Thermo Scientific, Waltham, MA, USA) and the quality was evaluated via running 1% formaldehyde denaturing gel electrophoresis. The gel was stained with 10,000× 4S Red Plus Nucleic Acid Stain (Sango biology, Shanghai, China) and photographed under UV illumination using Bio-Rad Chemi Doc XRS imaging system (Bio-Rad, Hercules, CA, USA).
Transcriptome sequencing and data analysis
The RNA-seq libraries were constructed with five hundred nanograms of total RNA using the KAPA Stranded RNA-Seq Library Preparation Kit (KAPA Biosystem, Wilmington, MA, USA). The Agilent 2100 Bioanalyzer system (Agilent, USA) was used to assess the quality of libraries.
The sequencing data were firstly performed quality inspection and data preprocessing to obtain the clean reads. Then the reads were aligned to the UCSC Mouse Genome Browser (assembly with HISAT2, version 2.0.5) and Stringtie (version 1.3.3) was used to define the read counts of each gene based on Ensembl gene-level annotations (Mouse GRCm38/mm10) (9). The final unnormalized counts were assembled into an R (version 3.6.0) count-matrix as an input for DESeq2 (version 1.24.0) and edgeR (version 3.26.5) (10,11). Expression changes with of |log2FC| ≥1 and P value <0.05 were thought to be significant. The transcriptome sequencing was presented in the dataset GSE156961.
Small RNA sequencing and data analysis
The small RNA-seq libraries were constructed with one microgram of total RNA using TruSeq® Small RNA Library Prep Kit (NEB, San Diego, California, USA) following the manufacturer’s protocol. The Agilent 2100 Bioanalyzer (Agilent, USA) was used to assess the quality of libraries.
Briefly, after data quality inspection and preprocessing, the clean reads were mapped onto the reference mouse genome mm10 using the bowtie2 (version 2.1.0) (12). The mapped tags were then aligned against miRBase (version 22.1), and read counts were calculated by customized script (13). Expression level change with an absolute value of |log2FC| ≥1, RPM ≥0.1, and P value <0.05 were considered significant and used for further analysis. The target genes of DEmiRs were predicted through the miRTarbase database. Then functional analysis was performed by clusterProfiler (14). Raw sequence data of miRNA-seq were uploaded to the GEO database (https://www.ncbi.nlm.nih.gov/geo/) and presented in the dataset GSE168166.
PPI network analysis of DEGs and DEmiRs in PCOS
Based on potential predicted target genes of DEmiRs and the negative-regulated mRNAs, the PPI network of DEmiR-DEG pairs was constructed through STRING database (version 11.0). The regulation network was visualized by Cytoscape (version 3.8.2).
Validation of the expression by RT-qPCR and statistical analyses
Five hundred nanograms of total RNA was used for reverse transcription using the PrimeScript™ RT reagent Kit (RR037, TAKARA), which was also used for reverse transcription with miRNA-specific stem-loop primers. Quantitative real-time PCR (RT-qPCR) was carried out with a Luna® Universal qPCR Master Mix (New England Biolabs, USA) at the StepOne Plus qPCR instrument (Applied Biosystems). Primer sequences are summarized in Table S1.
Statistical analyses were performed using GraphPad Prism version 7. software (GraphPad Software, Inc., San Diego, California, USA). Statistically significant differences between groups were determined by using a two-sided Student’s t-test and P value of <0.05 was considered statistically significant.
Results
Histological, physiological, and serological characterization of PNA mice
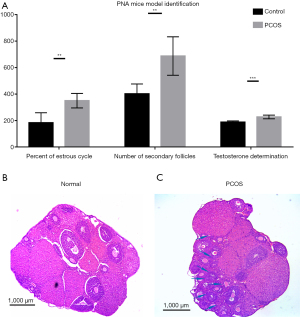
The statistical data for model characterization showing the duration of estrus, the serum testosterone concentration, and the number of secondary follicles are shown in Figure 1A. PNA mice had significantly higher serum testosterone level (P<0.001) and more secondary follicles (P<0.01) than the control mice. Moreover, the estrous duration was longer in the PNA mice than in the control mice (P<0.01). Histological results showed that the typical morphology of polycystic ovaries, with many fewer corpora lutea, was present in the ovaries of the DHT-induced PNA mice, indicating that ovulation occurred rarely. In contrast, numerous corpora lutea were present in the ovaries of the control mice, which was consistent with recent ovulation. Furthermore, in the ovaries of the PNA mice, which were different from the control mice, follicles with an atretic cyst-like appearance were observed (Figure 1B,1C).
Identification of DEGs and functional enrichment analyses
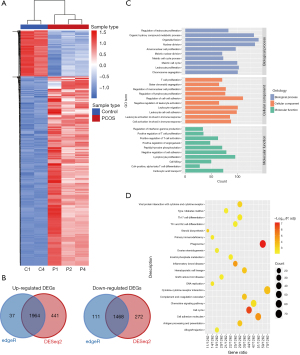
To identify the significant DEGs between the PNA mice and the control mice, we used RNA-seq to assess the transcriptome ovary profiles in PNA mice (n=3) and in the control mice (n=2). Detailed information of sequencing data was shown in Table S2. Two methods (DESeq2 and edgeR) were used to identify the DEGs accurately, and the research was focus on the common DEGs. The difference in expression patterns, as presented in the heatmap, between the PNA mice and the control mice, was illustrated by hierarchical clustering analysis (Figure 2A). In total, 3,432 DEGs were identified, 1,964 upregulated genes and 1,468 downregulated genes (P<0.05, |log2FC| >1; Figure 2B and available online: https://cdn.amegroups.cn/static/public/atm-21-1288-01.xlsx). We used Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses to determine if the enriched DEGs contained any regulatory genes. Certain GO biological processes, cellular components, and molecular functions were significantly enriched, including leukocyte proliferation (GO:0070661), cell activation involved in immune response (GO:0002263), and lymphocyte differentiation (GO:0030098) (Figure 2C). Furthermore, the KEGG enrichment analysis, demonstrated that the DEGs were related to the phagosome, the cell cycle, cytokine-cytokine receptor interactions, Th17 cell differentiation, and the B cell receptor signaling pathway (Figure 2D). These pathways indicated that the various aspects of inflammation and the immune response were related to the occurrence and development of PCOS. Previous studies indicated that PCOS was associated with systemic low-grade inflammation (LGI) (15). In recent research, increased intrafollicular inflammatory mediators were found in the periovulatory follicles of women with PCOS, which showed that chronic LGI may be a precursor of ovarian dysfunction in PCOS (16).
Identification of DEmiRs and functional enrichment analyses
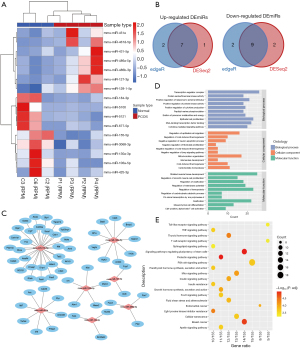
We used the miRNA-seq analysis of ovaries from PNA mice (n=4) and the control mice (n=3) to determine the significant DEmiRs and compared the differentially expressed miRNAs. Detailed sequencing data information is summarized in Table S3. We then identified 16 DEmiRs using DESeq2 and edgeR, which are commonly used methods, between the PNA mice and the control mice; these comprised seven upregulated miRNAs and nine downregulated miRNAs (P<0.05, |log2FC| >1; Table S4). There were significantly different expression patterns, as shown by the hierarchical clustering analysis, between the PNA mice and the control mice (Figure 3A,3B). We then characterized the regulatory roles of DEmiRs by predicting the potential miRNA target genes. In total, 59 target genes were predicted for the 16 DEmiRs, as shown in Figure 3C. Of the 16 DEmiRs, miR-106a-5p, miR-155-5p, and miR-451a had the highest number of potential target genes. Moreover, GO annotation and KEGG enrichment analyses were performed for the 59 target genes of those 16 DEmiRs. Enrichment occurred in several biological processes related to cell proliferation and immune response, including muscle tissue development (GO:0060537), cytokine-mediated signaling pathway (GO:0019221), cellular response to insulin stimulus (GO:0032869), and positive regulation of protein kinase activity (GO:0045860) (Figure 3D). In addition, the KEGG analyses showed enrichment mainly in some metabolic function and inflammation-related pathways, such as the PI3K-AKT signaling pathway, signaling pathways regulating pluripotency of stem cells, the prolactin signaling pathway, and the insulin signaling pathway (P<0.05, Figure 3E).
Identification of the negatively regulated miRNA-gene pairs in PCOS
MiRNAs are involved in the post-transcriptional regulation of their target genes. They can negatively regulate the expression of target genes through the inhibition of mRNA translation or the degradation of mRNA. To investigate the regulatory functions of miRNAs and mRNA in PCOS, the potential target mRNAs were mapped to our RNA-seq results and the differently expressed target mRNAs were obtained. Based on the negative regulation of mRNA by miRNA, we obtained 12 differently expressed target mRNAs from the 3,432 DEGs identified. The regulatory relationship between the DEmiRs and their potential target mRNAs is presented in Table 1. There were only 12 upregulated differently expressed target mRNAs and three downregulated differently expressed miRNAs. The KEGG functional enrichment analyses were enriched in EGFR tyrosine kinase inhibitor resistance, insulin resistance, the T cell receptor signaling pathway, and the mTOR signaling pathway (Figure 4A). We found that miR-106a-5p and miR-155-5p had the highest number of differential target genes that were strongly associated with the development and process of PCOS. The KEGG analysis showed that the target genes of these two miRNAs (Stat3, Gsk3b, and Nr1h3) were mostly concentrated within the pathway of insulin resistance. Moreover, Cd28 and Il10rb were reported to be involved in T cell activation and inflammation (17,18), suggesting that miR-106a-5p and miR-155-5p may be involved in the regulation of inflammation and insulin resistance in PCOS.
Table 1
| miRNA | Log2FC | Expression change | Target gene | Log2FC | Expression change |
|---|---|---|---|---|---|
| miR-106a-5p | −1.72 | Down |
|
8.39 | Up |
|
|
2.25 | Up | |||
|
|
1.36 | Up | |||
|
|
1.09 | Up | |||
| miR-155-5p | −1.14 | Down |
|
1.30 | Up |
|
|
2.04 | Up | |||
|
|
1.14 | Up | |||
|
|
2.60 | Up | |||
|
|
2.22 | Up | |||
|
|
3.74 | Up | |||
|
|
1.04 | Up | |||
| miR-184-3p | −1.07 | Down |
|
1.08 | Up |
PCOS, polycystic ovary syndrome; PNA, prenatally androgenized.
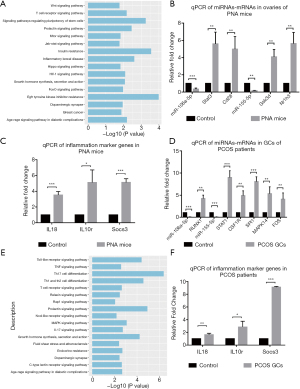
MiR-155 has been shown to regulate insulin sensitivity in vitro (19) and in vivo in mice (20). To illustrate the changes in the expression of miRNAs and target mRNAs, RT-qPCR was performed on samples from the ovaries of six PNA mice and six control mice to validate the negative regulation of miR-106a-5p and miR-155-5p and their target genes Stat3, Cd28, Gsk3b, and Nr1h3, which are involved in insulin resistance and inflammation. In addition, three inflammation-related genes, Il18, Il10r, and Socs3 (21), were selected and validated as inflammation markers. The results indicated that the expression of miR-106a-5p and miR155-5p was downregulated in PNA mice compared with the control mice, and that this was accompanied by the upregulation of the target genes Stat3, Cd28, Gsk3b, and Nr1h3. The validation confirmed that the inflammatory marker genes were significantly upregulated. These results are shown in Figure 4B,4C.
Clinical characteristics and the expression of miRNAs in GCs
To explore the alternation of miR-106a-5p and miR-155-5p in human GCs, the clinical GCs were used to assess the change in expression. All participants were unrelated Han people from the same geographical region, with no significant difference in their average age, and agreed to participate in this research. The clinical characteristics of the participants and the serum levels of seven sex hormones are shown in Table 2. The characteristics of patients with PCOS were not significantly different, except for a higher BMI and an increased level of LH concentration (P<0.05). The GCs isolated from the abovementioned women were used in our validation study.
Table 2
| Clinical characteristics | PCOS (n=6) | Normal (n=6) | P value |
|---|---|---|---|
| Age (year) | 30.33±3.98 | 31.5±5.05 | 0.666 |
| BMI (kg/m2) | 24.92±2.46 | 21.17±2.42 | 0.022 |
| W (cm) | 77±4.52 | 73.33±2.73 | 0.120 |
| H (cm) | 93.5±3.67 | 89.83±2.28 | 0.063 |
| FSH (mIU/mL) | 5.218±1.01 | 3.973±1.23 | 0.084 |
| E2 (pg/mL) | 42.11±16.04 | 43.06±18.07 | 0.925 |
| P (ng/mL) | 0.7683±0.33 | 0.56±0.26 | 0.258 |
| PRL (ng/mL) | 9.918±3.86 | 8.807±0.83 | 0.506 |
| LH (mIU/mL) | 7.442±4.44 | 2.095±0.98 | 0.016 |
| T (ng/mL) | 1.785±1.04 | 1.63±0.41 | 0.742 |
| β-HCG (mIU/mL) | 1.017±0.54 | 0.425±0.68 | 0.127 |
| Fasting glucose (mmol/L) | 5.085±0.40 | 5.035±0.29 | 0.808 |
All data are presented as the mean ± SEM. P<0.05 was considered significant. PCOS, polycystic ovary syndrome; BMI, body mass index; W, waist circumference; H, hip circumference; FSH, follicle stimulating hormone; E2, estrogen; P, progesterone; PRL, prolactin; LH, luteinizing hormone; T, testosterone; β-HCG, Human chorionic gonadotropin β.
To compare the regulatory role of miRNAs-target mRNAs in inflammation in the GCs of patients with PCOS and in PNA mice, we performed RT-qPCR analysis of the expression of miR-106a-5p and miR-155-5p in GCs from six patients with PCOS and six patients in the control group. In the GCs from women with PCOS, the expression of miR-106a-5p and miR-155-5p was significantly downregulated; this was similar to the changes observed in the PNA mice (Figure 4D) and supported a similar regulatory role for miR-106a-5p and miR155-5p in PCOS.
Identification of the target genes of selected miRNAs and validation in clinical GCs
MiR-155 has a multifunctional role in autoimmune disease and other chronic inflammatory diseases (22,23). To investigate the regulatory roles of miRNAs-target mRNAs in the inflammation in PCOS, we predicted the potential target genes of miR-106a-5p and miR-155-5p in the human genome using the miRTarbase database, which identified 264 target genes for these two miRNAs (available online: https://cdn.amegroups.cn/static/public/atm-21-1288-02.xlsx). The potential target mRNAs were mapped to our previous RNA-seq data of GCs from women with PCOS and the control group (24). Finally, we screened out one upregulated target gene of miR-106a-5p (RUNX1) and five upregulated target genes of miR-155-5p (CSF1R, FOS, MAPK14, SPI1, and STAT1) from the DEGs identified in our previous data (Table 3). RT-qPCR analysis of these genes was performed to detect the expression relationship of miR-106a-5p and miR-155-5p and the target genes in GCs in six patients with PCOS and six healthy volunteers. Similarly, to illustrate the regulatory relationship between the miRNAs and inflammation, three inflammation markers (IL18, IL10R, and SOCS3) were also evaluated. Compared with the control group, the expression of miR-106a-5p and miR155-5p was significantly downregulated in the PCOS sample, in addition to the upregulation of the target genes RUNX1, CSF1R, FOS, MAPK14, SPI1, and STAT1 (Figure 4D). The KEGG analysis indicated that the six genes were also concentrated within the inflammation and immune-related pathways, such as Th17 cell differentiation, Th1 and Th2 cell differentiation, the TNF signaling pathway, and the Toll-like receptor signal pathway (Figure 4E). In addition, we observed higher expression of the inflammatory marker genes (IL18, IL10R, and SOCS3) in the PCOS samples than in the control group (Figure 4F), which indicated the activation and increase of inflammation in PCOS.
Table 3
| miRNA | Log2FC | Expression change | Target gene | Log2FC | Expression change |
|---|---|---|---|---|---|
| hsa-miR-106a-5p | −1.72 | Down |
|
1.46 | Up |
| hsa-miR-155-5p | −1.14 | Down |
|
2.44 | Up |
|
|
1.00 | Up | |||
|
|
1.36 | Up | |||
|
|
2.89 | Up | |||
|
|
1.14 | Up |
GCs, granulosa cells; PCOS, polycystic ovary syndrome.
To investigate the regulatory relationship of miR-106a-5p and miR155-5p in PCOS, we mapped the sequences of the 12 target genes identified from the analysis of the PNA mice compared with the control mice against the human genome and obtained 12 homologous genes. Subsequently, the six target genes from the GCs identified in women with PCOS compared with the healthy volunteers group and the 12 homologous genes were used to construct a PPI network, which was then studied using KEGG analysis. There was a high correlation between these genes, and STAT1, STAT3, and MAPK14 had the highest counts of the node. The top eight most significant pathways were mainly related to the immune response and metabolism-related pathways: Th17 cell differentiation; the IL-17 signaling pathway; Th1 and Th2 cell differentiation; insulin resistance; the T cell receptor signaling pathway; the MAPK signaling pathway; the prolactin signaling pathway, and the FoxO signaling pathway (Figure 5A). Through the pathways identified, it was confirmed that miR-106a-5p and miR155-5p and their target genes were associated with inflammation and insulin sensitivity.
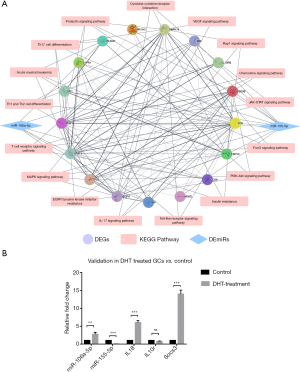
The effect of DHT on expression of miR155-5p and miR-106a-5p and related inflammation markers in GCs in vitro
To determine whether DHT was directly associated with the changes in miRNA and mRNA expression, we performed the RT-qPCR analysis on miR-106a-5p, miR-155-5p, and the inflammation markers in DHT-treated GCs (n=6) and the control cells (n=6). After an initial series of concentration and time gradient experiments, a DHT treatment of 1 µmol/L DHT-treated for 48 h was selected. This treatment significantly decreased the expression of the miR-155-5p, but increased the expression of miR-106a-5p. After 48 h, Il18 and Socs3 were significantly upregulated in the 1 μmol/L DHT treatment group. However, the expression of Il10r was not significantly altered (Figure 5B). The above results indicated that DHT directly caused the upregulation of miR-155-5p expression and increased the expression of some inflammation-related genes. In PCOS, the aberrant expression of miRNAs-mRNAs and the increased expression of inflammation markers may be affected by excessive androgens, other comprehensive factors, and more complex regulation.
Discussion
Epigenetic modifiers, such as small RNAs, are involved in gene translation and/or stabilization in various crucial physiological and pathological processes, including cell differentiation, development, homeostasis, and metabolism (25). Recently, the specific expression pattern of miRNAs in different body fluids was reported (26), which revealed significantly different expression levels in women with PCOS and healthy women, suggesting that miRNAs play an important role in the occurrence and development of PCOS. Women with PCOS had a lower expression of miR-145, which was proposed to stimulate cell proliferation via the modulation of the IRS1/MAPK/ERK cascade in human GCs from patients with PCOS (27). The expression of miR-126-5p and miR-29a-5p were downregulated in the GCs of patients with PCOS. This was reported to induce apoptosis in GCs in PCOS through klotho-associated signaling (28). However, the specific effects of miRNAs in PCOS with hyperandrogenism on the relevant gene expression and downstream regulation and metabolism remains relatively unexplored. MiR-106a, a member of the miR-17 family, has been shown to be aberrantly regulated in various tumors. It is reported that miR-106a is upregulated in most cancers, however, in renal carcinoma, astrocytoma, and glioma, it is significantly downregulated (29). The aberrant expression of miR-106a is prominently associated with increased invasiveness and metastasis of tumors (30). In 2020, miR-106a-5p was first reported to confer a risk of PCOS (31). In our present study, miR-106a-5p was found to be significantly downregulated in PNA mice compared with the control mice and in GCs from women with PCOS compared with the control group; this was accompanied by a significant increase in the target genes Stat3 and Cd28 in PNA mice and the target gene RUNX1 in women with PCOS. In a previous study, an increase in the expression of IL-6 and IL-11 was associated with the AKT/STAT3 pathway in a rat model of PCOS (32). Cd28 is involved in T-cell activation and the induction of cell proliferation (33). RUNX1 is one of the key regulatory proteins in vertebrates and is generally involved in embryonic development, hematopoiesis, angiogenesis, tumorigenesis, and the immune response, such as the NF-κβ signaling pathway (34). Further, the target gene App was reported to be involved in the phosphorylation signaling in Alzheimer’s disease (35). Vegfa is related to angiogenesis and is reported to be a negative regulator of miR-185-5p in PCOS (36).
MiR-155 is considered a multifunctional regulator of inflammation. There are previous reports of increases in the aberrant expression of miR-155 in various activated immune cells indicating the importance of miR-155 in the immune response. MiR-155 is sensitive to many inflammatory stimuli, such as tumor necrosis factor-alpha, interferons, damage-associated molecular patterns, hypoxia, and Interleukin-1a (4). In particular, in PCOS, miR-155 is strongly related to inflammation, insulin resistance, obesity, and steroidogenesis (20,37,38), reflecting the main features of the disease. In this study, miR-155-5p was found to be downregulated in PCOS and the negatively correlated target genes, Gsk3b, Nr1h3, STAT1, MAPK14, CSF1R, SPI1, and FOS, were upregulated. The target genes were also concentrated in the insulin-sensitive pathway of ovaries and GCs, supporting that miR-155 may be involved in the regulation of inflammation and insulin sensitivity. Recently, it was reported that miR-155-5p directly regulated Gsk3b and participated in the regulation of steroid metabolism (39), and that Nr1h3 was related to autophagy. STAT1 was reported to be related to inflammation and insulin sensitivity in adipose tissue and obesity (40). MAPK14 has a wide effect on regulating inflammation and metabolism, and was shown to be regulated by miRNA epigenetics in PCOS in our previous study (24). CSF1R was a key gene in leukoencephalopathy (41). SPI1 was reported as an ETS transcription factor involved in the androgen-mediated transcriptional regulation of NR4A1 in PCOS (42). FOS was reported to mediate the regulation of neurotrophins in the placenta of female offspring of mothers with PCOS (43). The target gene Arntl was reported to affect PCOS by altering the circadian clock (44). Trp53inp1 is involved in the cell stress response, and acts as a dual regulator of transcription and autophagy (45). Maf was involved in the inhibition of Th17 cells and the induction of IL-10 production (46). S1pr1 was involved in the many processes related to immune activation and is expected to be a target for cancer treatment (47).
PCOS is considered a low-grade inflammatory endocrine disease accompanied by a variety of metabolic symptoms, such as hyperandrogenism and insulin resistance. Previous studies showed that there was an increase in indicators of insulin resistance and inflammation in PCOS mice (48,49). Our study indicated that the downregulation of miR-155-5p and miR-106a-5p could inhibit the degradation of mRNA or stabilize mRNA, which indirectly increases the expression of the target genes. The functional analysis of the target genes indicated that the miRNAs-mRNAs regulatory network could be involved in inflammation in PCOS. The RT-qPCR results illustrated that in PNA mice or PCOS GCs, the expression of miR-155-5p, miR-106a-5p, and targeting mRNAs was consistent with our analyses. Moreover, the inflammatory marker genes were detected to be upregulated in PNA mice and clinical samples, supporting our hypothesis that the miRNAs-mRNA axis was related to the inflammatory response in PCOS. In cell experiments, a significant downregulation of miR-155-5p was observed in DHT-treated GCs in addition to the upregulation of the Il18/IL18 and Socs3/SOCS3. However, miR-106a-5p was upregulated, which indicated that an excess of androgens was not the direct cause of the downregulation of miR-106a-5p. The in vitro experimental results showed that excessive DHT was a direct cause of the abnormal expression of miR-155-5p and could lead to a high expression of inflammatory marker genes.
Due to the limitation of the sample size, some extended experiments need supplementing in the following research. The detection of the target protein is necessary for studying the regulation of miRNA on protein abundance intuitively. Quantitative detection of insulin resistance and inflammation in PNA mice also helps to understand the regulatory mechanism of PCOS. Knocking out or blocking the expression of target miRNAs is helpful to understand the specific mechanism of key miRNAs involving the regulatory network. Combining with the clinical studies on the pathogenesis of PCOS is essential, including the regulatory and endocrine function of GCs and follicular fluids, which could play a role in generating new treatment strategies and determining diagnostic and prognostic indicators in PCOS. In addition, despite the modeling methods of construct PCOS model mice were various, no one can completely mimic the human symptoms of PCOS. The physiological state of hyperandrogenemia could be simulated with high efficiency by DHT-induced PNA mice, but the level of luteinizing hormone (LH) in the serum was decreased, instead of being increased, which was not in accord with clinical features of PCOS patients. Moreover, the anti-Mullerian hormone (AMH) expression level was no different between the PNA mice and the control mice, which indicated that the PNA mice were lack of a neuroendocrine phenotype (50). Therefore, researchers usually choose a suitable modeling method according to the phenotype or symptom of the study requirements.
Conclusions
This research indicated that miR-106a-5p and miR-155-5p are important miRNAs in the ovaries and GCs in PCOS, and are related to insulin sensitivity and inflammation in the ovaries. We observed that the inflammation and insulin-related genes were in significantly increased in expression in the ovaries of PNA mice, with similar manifestations observed in the GCs from patients with PCOS. In vitro, excessive androgens, such as DHT, alter the expression of specific miRNAs such as miR-106a-5p and miR-155-5p, which affects the related genes, and may participate in the inflammation in PCOS. Overall, this study offers new insight into the epigenetics and pathophysiology of PCOS, supporting and extending the knowledge of the regulatory role of miRNAs and their target pathways in the immune and metabolic mechanisms of PCOS, and offers new insights into the metabolic disorders associated with PCOS.
Acknowledgments
We would like to thank all doctors, patients, and their family members for their kindness to support our study.
Funding: This study was supported by the Natural Science Foundation of Shanghai (19ZR1476100), National Infrastructures for Translational Medicine (Shanghai) (TMSK-2020-109), Interdisciplinary Program of Medical Engineering Cross Fund (YG2019GD02, YG2019QNB23, YG2019QNA49 and YG2019QNA52) and Laboratory Innovative Research Program of Shanghai Jiao Tong University (JCZXSJB2019002).
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist and STROBE reporting checklist. Available at https://dx.doi.org/10.21037/atm-21-1288
Data Sharing Statement: Available at https://dx.doi.org/10.21037/atm-21-1288
Peer Review File: Available at https://dx.doi.org/10.21037/atm-21-1288
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/atm-21-1288). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The trial was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by Ethics Committees on Human Research of Yuncheng Center Hospital (No. KYLL2019073) and informed consent was taken from all individual participants. Animal Experiments were performed under a project license (No. 2019005) granted by School of Biomedical Engineering, Shanghai Jiao Tong University, Experimental Animals Welfare & Ethical Committee, in compliance with Laboratory Animal Center of Shanghai Jiao Tong University guidelines for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Li S, Chu Q, Ma J, et al. Discovery of Novel Lipid Profiles in PCOS: Do Insulin and Androgen Oppositely Regulate Bioactive Lipid Production? J Clin Endocrinol Metab 2017;102:810-21. [PubMed]
- Azziz R, Woods KS, Reyna R, et al. The prevalence and features of the polycystic ovary syndrome in an unselected population. J Clin Endocrinol Metab 2004;89:2745-9. [Crossref] [PubMed]
- Mao Z, Li T, Zhao H, et al. Methylome and transcriptome profiling revealed epigenetic silencing of LPCAT1 and PCYT1A associated with lipidome alterations in polycystic ovary syndrome. J Cell Physiol 2021;236:6362-75. [Crossref] [PubMed]
- Mahesh G, Biswas R. MicroRNA-155: A Master Regulator of Inflammation. J Interferon Cytokine Res 2019;39:321-30. [Crossref] [PubMed]
- Wu KL, Tsai YM, Lien CT, et al. The Roles of MicroRNA in Lung Cancer. Int J Mol Sci 2019;20:1611. [Crossref] [PubMed]
- Chen B, Xu P, Wang J, et al. The role of MiRNA in polycystic ovary syndrome (PCOS). Gene 2019;706:91-6. [Crossref] [PubMed]
- Tu J, Cheung AH, Chan CL, et al. The Role of microRNAs in Ovarian Granulosa Cells in Health and Disease. Front Endocrinol (Lausanne) 2019;10:174. [Crossref] [PubMed]
- . Rotterdam ESHRE/ASRM-Sponsored PCOS Consensus Workshop Group. Revised 2003 consensus on diagnostic criteria and long-term health risks related to polycystic ovary syndrome. Fertil Steril 2004;81:19-25. [Crossref]
- Pertea M, Kim D, Pertea GM, et al. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc 2016;11:1650-67. [Crossref] [PubMed]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [Crossref] [PubMed]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010;26:139-40. [Crossref] [PubMed]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods 2012;9:357-9. [Crossref] [PubMed]
- Kozomara A, Birgaoanu M, Griffiths-Jones S. miRBase: from microRNA sequences to function. Nucleic Acids Res 2019;47:D155-62. [Crossref] [PubMed]
- Yu G, Wang LG, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012;16:284-7. [Crossref] [PubMed]
- Barrea L, Marzullo P, Muscogiuri G, et al. Source and amount of carbohydrate in the diet and inflammation in women with polycystic ovary syndrome. Nutr Res Rev 2018;31:291-301. [Crossref] [PubMed]
- Adams J, Liu Z, Ren YA, et al. Enhanced Inflammatory Transcriptome in the Granulosa Cells of Women With Polycystic Ovarian Syndrome. J Clin Endocrinol Metab 2016;101:3459-68. [Crossref] [PubMed]
- Beckermann KE, Hongo R, Ye X, et al. CD28 costimulation drives tumor-infiltrating T cell glycolysis to promote inflammation. JCI Insight 2020;5:138729 [Crossref] [PubMed]
- Shouval DS, Biswas A, Kang YH, et al. Interleukin 1β Mediates Intestinal Inflammation in Mice and Patients With Interleukin 10 Receptor Deficiency. Gastroenterology 2016;151:1100-4. [Crossref] [PubMed]
- Ying W, Riopel M, Bandyopadhyay G, et al. Adipose Tissue Macrophage-Derived Exosomal miRNAs Can Modulate In Vivo and In Vitro Insulin Sensitivity. Cell 2017;171:372-384.e12. [Crossref] [PubMed]
- Lin X, Qin Y, Jia J, et al. MiR-155 Enhances Insulin Sensitivity by Coordinated Regulation of Multiple Genes in Mice. PLoS Genet 2016;12:e1006308 [Crossref] [PubMed]
- Germolec DR, Shipkowski KA, Frawley RP, et al. Markers of Inflammation. In: DeWitt JC, Rockwell CE, Bowman CC. editors. Immunotoxicity Testing: Methods and Protocols. New York: Springer New York, 2018:57-79.
- Alivernini S, Gremese E, McSharry C, et al. MicroRNA-155-at the Critical Interface of Innate and Adaptive Immunity in Arthritis. Front Immunol 2018;8:1932. [Crossref] [PubMed]
- Li X, Kong D, Chen H, et al. miR-155 acts as an anti-inflammatory factor in atherosclerosis-associated foam cell formation by repressing calcium-regulated heat stable protein 1. Sci Rep 2016;6:21789. [Crossref] [PubMed]
- Mao Z, Li T, Zhao H, et al. Identification of epigenetic interactions between microRNA and DNA methylation associated with polycystic ovarian syndrome. J Hum Genet 2021;66:123-37. [Crossref] [PubMed]
- Sang Q, Yao Z, Wang H, et al. Identification of microRNAs in human follicular fluid: characterization of microRNAs that govern steroidogenesis in vitro and are associated with polycystic ovary syndrome in vivo. J Clin Endocrinol Metab 2013;98:3068-79. [Crossref] [PubMed]
- Weber JA, Baxter DH, Zhang S, et al. The microRNA spectrum in 12 body fluids. Clin Chem 2010;56:1733-41. [Crossref] [PubMed]
- Cai G, Ma X, Chen B, et al. MicroRNA-145 Negatively Regulates Cell Proliferation Through Targeting IRS1 in Isolated Ovarian Granulosa Cells From Patients With Polycystic Ovary Syndrome. Reprod Sci 2017;24:902-10. [Crossref] [PubMed]
- Mao Z, Fan L, Yu Q, et al. Abnormality of Klotho Signaling Is Involved in Polycystic Ovary Syndrome. Reprod Sci 2018;25:372-83. [Crossref] [PubMed]
- Pan YJ, Zhuang Y, Zheng JN, et al. MiR-106a: Promising biomarker for cancer. Bioorg Med Chem Lett 2016;26:5373-7. [Crossref] [PubMed]
- Zhao S, Yang G, Mu Y, et al. MiR-106a is an independent prognostic marker in patients with glioblastoma. Neuro Oncol 2013;15:707-17. [Crossref] [PubMed]
- Jiang X, Li J, Zhang B, et al. Differential expression profile of plasma exosomal microRNAs in women with polycystic ovary syndrome. Fertil Steril 2021;115:782-92. [Crossref] [PubMed]
- Zhuang Z, Pan X, Zhao K, et al. The Effect of Interleukin-6 (IL-6), Interleukin-11 (IL-11), Signal Transducer and Activator of Transcription 3 (STAT3), and AKT Signaling on Adipocyte Proliferation in a Rat Model of Polycystic Ovary Syndrome. Med Sci Monit 2019;25:7218-27. [Crossref] [PubMed]
- Moulana M. Immunophenotypic profile of leukocytes in hyperandrogenemic female rat an animal model of polycystic ovary syndrome. Life Sci 2019;220:44-9. [Crossref] [PubMed]
- Tang X, Sun L, Wang G, et al. RUNX1: A Regulator of NF-kB Signaling in Pulmonary Diseases. Curr Protein Pept Sci 2018;19:172-8. [PubMed]
- Zhang T, Chen D, Lee TH. Phosphorylation Signaling in APP Processing in Alzheimer's Disease. Int J Mol Sci 2019;21:209. [Crossref] [PubMed]
- Wei J, Zhao Y. MiR-185-5p Protects Against Angiogenesis in Polycystic Ovary Syndrome by Targeting VEGFA. Front Pharmacol 2020;11:1030. [Crossref] [PubMed]
- O'Connell RM, Taganov KD, Boldin MP, et al. MicroRNA-155 is induced during the macrophage inflammatory response. Proc Natl Acad Sci U S A 2007;104:1604-9. [Crossref] [PubMed]
- Murri M, Insenser M, Fernández-Durán E, et al. Effects of polycystic ovary syndrome (PCOS), sex hormones, and obesity on circulating miRNA-21, miRNA-27b, miRNA-103, and miRNA-155 expression. J Clin Endocrinol Metab 2013;98:E1835-44. [Crossref] [PubMed]
- Wu F, Huang W, Yang Y, et al. miR-155-5p regulates mesenchymal stem cell osteogenesis and proliferation by targeting GSK3B in steroid-associated osteonecrosis. Cell Biol Int 2021;45:83-91. [Crossref] [PubMed]
- Cox AR, Chernis N, Bader DA, et al. STAT1 Dissociates Adipose Tissue Inflammation From Insulin Sensitivity in Obesity. Diabetes 2020;69:2630-41. [Crossref] [PubMed]
- Tian WT, Zhan FX, Liu Q, et al. Clinicopathologic characterization and abnormal autophagy of CSF1R-related leukoencephalopathy. Transl Neurodegener 2019;8:32. [Crossref] [PubMed]
- Song J, Diao F, Ma X, et al. Androgen upregulates NR4A1 via the TFAP2A and ETS signaling networks. Int J Biochem Cell Biol 2019;113:1-7. [Crossref] [PubMed]
- Wang F, Xie N, Zhou J, et al. Molecular mechanisms underlying altered neurobehavioural development of female offspring of mothers with polycystic ovary syndrome: FOS-mediated regulation of neurotrophins in placenta. EBioMedicine 2020;60:102993 [Crossref] [PubMed]
- Li S, Zhai J, Chu W, et al. Altered circadian clock as a novel therapeutic target for constant darkness-induced insulin resistance and hyperandrogenism of polycystic ovary syndrome. Transl Res 2020;219:13-29. [Crossref] [PubMed]
- Tomasini R, Samir AA, Vaccaro MI, et al. Molecular and functional characterization of the stress-induced protein (SIP) gene and its two transcripts generated by alternative splicing. SIP induced by stress and promotes cell death. J Biol Chem 2001;276:44185-92. [Crossref] [PubMed]
- Xu M, Pokrovskii M, Ding Y, et al. c-MAF-dependent regulatory T cells mediate immunological tolerance to a gut pathobiont. Nature 2018;554:373-7. [Crossref] [PubMed]
- Rostami N, Nikkhoo A, Ajjoolabady A, et al. S1PR1 as a Novel Promising Therapeutic Target in Cancer Therapy. Mol Diagn Ther 2019;23:467-87. [Crossref] [PubMed]
- Roland AV, Nunemaker CS, Keller SR, et al. Prenatal androgen exposure programs metabolic dysfunction in female mice. J Endocrinol 2010;207:213-23. [Crossref] [PubMed]
- Xie Q, Xiong X, Xiao N, et al. Mesenchymal Stem Cells Alleviate DHEA-Induced Polycystic Ovary Syndrome (PCOS) by Inhibiting Inflammation in Mice. Stem Cells Int 2019;2019:9782373 [Crossref] [PubMed]
- Benrick A, Chanclón B, Micallef P, et al. Adiponectin protects against development of metabolic disturbances in a PCOS mouse model. Proc Natl Acad Sci U S A 2017;114:E7187-96. [Crossref] [PubMed]


