
Co-expression network of long non-coding RNA and mRNA reveals molecular phenotype changes in kidney development of prenatal chlorpyrifos exposure in a mouse model
Introduction
As most kidney diseases’ etiologies and mechanisms are not fully understood, it continues to be a challenge to diagnose and effectively treat for kidney patients at clinics. However, research on physiological and pathological embryonic kidney development may provide potential breakthroughs (1) in certain kidney diseases’ pathogenesis and inform new therapeutic strategies.
Environmental chemical exposure has become a global public health problem in recent years. Many environmental chemicals can be detected in human blood and urine samples (2). Organophosphorus pesticides (OPs) are widely studied chemicals that can be absorbed by humans (3). Researchers have confirmed that prenatal exposure to OPs leads to neurodevelopmental disabilities and impairments (4-6). Chlorpyrifos (CPF) is one of the most widely used OPs in the world (7). Prenatal CPF exposure can cause abnormal neurodevelopment and behavioral abnormalities (8-10). CPF has been found to accumulate in various organs of rats, including the kidney (11).
Further, acute CPF exposure can lead to acute kidney injury (12). Another study showed that CPF has cytotoxic effects on the human embryonic kidney (HEK) 293 cell line. However, very few studies have been conducted on the effects of CPF exposure on kidney development.
Long non-coding ribonucleic acids (lncRNAs) are large ribonucleic acid (RNA) transcripts with a length >200 nt that do not code for proteins (13). Despite the abundance of lncRNAs in the human genome, little is known about their biological characteristics or functional mechanisms (14,15). With the rapid development of high-throughput sequencing technology, many aspects of lncRNAs, including their regulatory functions, have been widely studied. Research has shown that lncRNAs are involved in renal cell carcinoma and kidney fibrosis (16-18). Such findings suggest that lncRNAs have affirmative action in normal kidney functions. LncRNAs also play a role in development mechanisms, such as central nervous system development (19-21). Thus, it was hypothesized that lncRNAs would also play a role in embryonic kidney development.
This study established a prenatal CPF exposure mouse model and performed high-throughput RNA-sequencing of embryonic kidneys. Additionally, under prenatal CPF exposure, the lncRNA-messenger ribonucleic acid (mRNA) networks that regulate kidney development were constructed using a weighted gene co-expression network analysis (WGCNA). We present the article following the ARRIVE reporting checklist (available at http://dx.doi.org/10.21037/atm-20-6632).
Methods
The Research Ethics Committee of The First Affiliated Hospital, College of Medicine, Zhejiang University approved the experimental protocols (No. 2018361). All the experiments were carried out following the National Institute of Health (NIH) Guide for the Care and Use of Laboratory Animals.
Animal samples
Eight-week-old male and female Institute of Cancer Research mice were obtained from Shanghai SLAC Laboratory Animal Co., Ltd (Shanghai, China), and bred in timed mating; 9 pm on the day of vaginal plug detection was considered embryonic day 0.5 (E0.5). CPF, a donation from Professor Liezhong Chen of the Zhejiang Academy of Agricultural Sciences (5 mg; 96% purity), was dissolved in 1 mL of dimethyl sulfoxide (DMSO). The pregnant mice were randomly divided into two groups. A CPF solution (5 mg/kg/day) was subcutaneously injected in pregnant mice from E7.5 to E11.5 (CPF group; n=6), while the control group (n=6) mice were injected with DMSO solution at the same times. Embryonic kidneys at E12.5, E14.5, E16.5, and E18.5 were examined under a Leica S8AP0 microscope (Leica, Frankfurt, Germany) as previously reported (22). All mice were housed in the Animal Center of Zhejiang University following animal-care regulations.
RNA isolation, quantification, and qualification
Total RNA was extracted from the E12.5, E14.5, E16.5, and E18.5 whole mount embryonic kidneys of both the CPF and control groups. A miRNeasy Mini Kit (Qiagen, Hilden, Germany) was used to isolate total RNA. RNA purity was checked using the NanoPhotometer® spectrophotometer (IMPLEN, CA, USA). RNA integrity was assessed using the RNA Nano 6000 Assay Kit of the Bioanalyzer 2100 system (Agilent Technologies, CA, USA). RNA concentration was measured using the Qubit® RNA Assay Kit (Life Technologies, CA, USA) in the Qubit® 2.0 Fluorometer (Life Technologies, CA, USA).
Library preparation and sequencing
A total of 3 µg RNA per sample was used as input material for the RNA sample preparations. Ribosomal RNA (rRNA) was removed using an Epicentre Ribo-zero™ rRNA Removal Kit (Epicentre, USA); the rRNA-free residue was cleaned up via ethanol precipitation. Next, sequencing libraries were generated via rRNA-depleted RNA using a NEBNext® Ultra™ Directional RNA Library Prep Kit for Illumina® (NEB, USA) following the manufacturer’s recommendations. Finally, the libraries were sequenced on an Illumina Hiseq 2500 platform. and 125 bp paired-end reads were generated. The sequencing data can be downloaded from the National Center for Biotechnology Information’s Gene Expression Omnibus (accession no. GSE131263).
LncRNA and mRNA data analysis
Raw reads of fastq format were first processed through in-house Perl scripts. Clean reads were obtained upon removing reads containing adapters, poly-N, and low-quality reads from raw reads. The cleaned reads were then mapped to the mouse genome mm10 using Spliced Transcripts Alignment to a Reference (STAR) software (© Alexander Dobin) (23). Cuffdiff2 was used to calculate the transcripts per million (TPM) of both lncRNAs and mRNAs, and the differentially expressed genes (DEGs) were identified with P values <0.05 between the two groups (24).
A WGCNA of the co-expression network of lncRNAs and mRNAs
The WGCNA package in R software (R Foundation for Statistical Computing, Vienna, Austria) was used to analyze lncRNAs and mRNAs’ co-expression networks. Text files of lncRNA and mRNA probes with TPM values, annotated files, and sample information files were created as input files. The input files included eight samples: 4 embryonic kidney samples at different development time points from the CPF group (C12.5, C14.5, C16.5, and C18.8), and 4 from the DMSO control group (D12.5, D14.5, D16.5, and D18.5). The normalized quantified sequencing data were used to conduct a co-expression analysis in R with the WGCNA package. One-step network construction and module detection methodology were used. The samples were clustered using the “hclust” function with the average linkage method. Network construction and module detection were analyzed using the blockwiseModules function in the WGCNA package. The dynamic tree cut method was used in the clustering dendrogram analysis; each module was assigned a unique color and contained a unique set of genes; modules with a correlation coefficient >0.6 were merged. Module eigengene was calculated using the “moduleEigengenes” function. Module-trait associations were computed among eigengenes. Two important parameters were calculated for further analysis: (I) gene significance (GS), which represented the correlation between the expression profile of a gene and the sample trait; and (II) module membership (MM), which represented the correlation of the gene expression profile and the module eigengene. Genes with an absolute value of GS >0.2 and MM >0.8 were selected for further analysis.
Modules selection
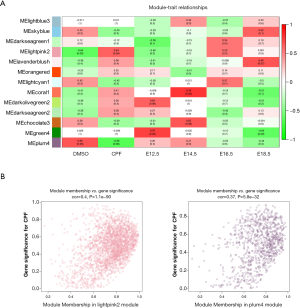
A threshold of 0.5 for the module-trait correlation was set to select the modules with a significant association with prenatal CPF exposure. The lightpink2 and plum4 modules were selected with the proper module-trait correlations. To identify the modules related to different development time points, a LinePlot of the module-trait relationship was created for the development time points, which showed that the module-trait correlation coefficients of the green4 and darkolivegreen2 modules increased with an increase in development time, and the module-trait correlation coefficient of the lavenderblush module decreased with an increase in development time.
GSEA
The mRNAs of selected modules with an absolute value of GS for trait >0.2 and MM >0.8 were included in the gene set enrichment analysis (GSEA), which was performed following a previously published method (25). Six gene set collections of the Molecular Signatures Database v7.1 were analyzed, including hallmark gene sets; c2: chemical and genetic perturbations, c2: all canonical pathways, c5: gene ontology (GO) biological processes, c5: GO molecular functions, and c5: GO cellular components. The resulting gene sets with a P value of <0.01, a false discovery rate (FDR) q value of <0.25, and an absolute value of normalized enrichment score (NES) >1 were identified as meaningful gene sets. The core enrichment genes from a gene set were identified as meaningful genes.
Hub genes identification
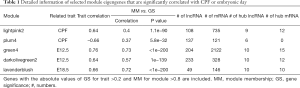
The meaningful mRNAs with the top 10 frequencies from the GSEA were defined as the selected modules’ hub mRNAs. The top 10 differentially expressed lncRNAs with the smallest p values (i.e., the lightpink2 and plum4 modules) were defined as the modules’ hub mRNAs with a significant association with prenatal CPF exposure. The top 10 lncRNAs with the largest MM values (i.e., green4, darkolivegreen2, and lavenderblush modules) were defined as the modules’ hub lncRNAs related to different development time points.
LncRNA-mRNA network construction
LncRNA-mRNA interaction networks were generated of the selected modules’ hub genes from the “CytoscapeInpute-edges-module” files with weight values between genes. Cytoscape 3.7.1 was used to visualize the networks.
qRT-PCR
RNA was extracted using the TRIzol (Life Technologies, CA, USA) manual extraction method for E18.5 kidneys. Total RNA (1 µg per sample) was reversed into complementary deoxyribonucleic acid (cDNA) using the PrimerScriptTM RT reagent Kit (Takara, Dalian, China). Quantitative polymerase chain reactions (PCR) were performed on a CFX96 TouchTM System (Bio-Rad, CA, USA) using iQTM SYBR Green Supermix (Bio-Rad, CA, USA). The following primers were used:
Gapdh-F 5'TGCCCCCATGTTTGTGATG;
Gapdh-R 5'TGTGGTCATGAGCCCTTCC;
Slc34a1-F 5'GGCTCCAACATTGGCACTACCA;
Slc34a1-R 5'ACCACAGTAGGATGCCCGAGAT;
Slc6a19-F 5'CGTGGTCTACTCCATCATTGGC;
Slc6a19-R 5'GTGGCATTGCACCACTGTTGGT;
Aqp1-F 5'CTTGCCATTGGCTTGTCTGTGG;
Aqp1-R 5'CCAGTGGTTTGAGAAGTTGCGG.
Immunohistochemistry
E18.5 kidneys were fixed in formalin and embedded in paraffin following a standard protocol. Sections were deparaffinized in xylene and ethanol gradients and then rehydrated in water. Antigen unmasking was performed with a 10 mM sodium citrate buffer at a sub-boiling temperature. Hydrogen peroxide (3%) was used to quench endogenous peroxidase enzyme activity. Tissues were blocked with 5% BSA for 1 h at room temperature, and the tissues were then incubated with a primary antibody overnight at 4 °C. Secondary antibody incubation and Diaminobenzidine (DAB) color-developing solution were performed with a carboxyfluorescein diacetate (CFDA) immunohistochemistry detection kit (Genetec, Shanghai, China) following the manufacturer’s recommendations. Images were taken using a Leica DM4000 microscope (Leica, Frankfurt, Germany).
Statistical analyses
The results are displayed as means ± standard deviation (SD). A statistical analysis using the two-tailed t-test was performed with GraphPad Prism 6. A P<0.05 was considered statistically significant.
Results
Construction of a WGCNA of prenatal CPF exposure mouse embryonic kidneys
The prenatal CPF exposure mouse model was established as previously reported (Figure 1A) (26). RNA-sequencing was performed on 4 prenatal CPF-exposed embryonic kidney samples and 4 control samples. The samples were obtained at different developmental time points; 1 sample for each group was obtained at E12.5, E14.5, E16.5, and E18.5 (Figure 1A). Sample clustering was performed to detect outliers, and all samples were retained (Figure S1A, B). A principal component analysis (PCA) was also conducted to detect Soft thresholding power β was automatically calculated using a “pickSoftThreshold” function in WGCNA; this value was chosen to raise co-expression similarity to calculate adjacency (27). In the present study, when β =14, the scale R2 was 0.91 (Figure S1D). Gene network clustering and module identification were performed using the “blockwiseModules” function. Modules with a correlation coefficient >0.6 were merged (Figure S2A), and each module was assigned a unique color (Figure 1B). The analysis generated 13 modules, and an eigengene dendrogram and adjacency heatmap was generated (Figure 1C). A network heatmap plot of the selected genes reflected topological overlap; the light color indicates low topological overlap, and the darker red indicates high topological overlap (Figure S2B).
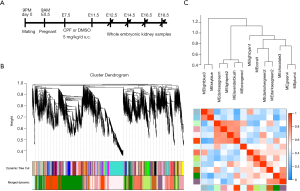
Co-expression modules corresponding to prenatal CPF exposure
Next, an association analysis was performed to examine the gene expression patterns in the modules and the particular trait. The r and P values are set out in the module-trait relationships heatmap (Figure 2A). The results showed that the lightpink2 (r =0.64, P=0.09) and the plum4 (r =–0.66, P=0.08) modules were significantly associated with prenatal CPF exposure. After screening with an absolute GS value >0.2 and an absolute MM value >0.8, 108 lncRNAs and 735 mRNAs in the lightpink2 module and 137 lncRNAs and 121 mRNAs in the plum4 module remained (Tables 1, https://cdn.amegroups.cn/static/public/atm-20-6632-1.pdf). A scatterplot between MM and GS revealed that MM in the lightpink2 and plum4 modules was significantly correlated with their GS (r =0.4, P=1.1E–90; r =0.37, P=5.8E–32) (Figure 2B). A higher MM value indicated a higher GS value in module-trait relationships, which suggested that hub genes in the lightpink2 and plum4 modules were highly associated with prenatal CPF exposure.
Full table
A LncRNA-mRNA network and a functional enrichment analysis of modules corresponding to prenatal CPF exposure
The selected mRNAs (absolute value of GS >0.2, the absolute value of MM >0.8) from the lightpink2 and plum4 modules were examined using a GSEA. The mRNAs with the top 10 frequencies in the GSEA results were defined as the hub mRNAs. The top 10 differentially expressed lncRNAs with the smallest P values of selected lncRNAs (absolute value of GS >0.2, the absolute value of MM >0.8) from the lightpink2 and plum4 modules were defined as the hub lncRNAs. Finally, there were 9 hub lncRNAs and 12 hub mRNAs in the lightpink2 module, and 6 hub lncRNAs and 0 hub mRNAs in the plum4 module (Table 1). The differentially expressed lncRNAs are set out in https://cdn.amegroups.cn/static/public/atm-20-6632-2.pdf.
Next, a lncRNA-mRNA network was constructed of hub genes in the lightpink2 module using Cytoscape. LncRNAs are displayed as circles and mRNAs as diamonds. The blue color indicates the downregulation, and the red color indicates the upregulation of a gene in the prenatal CPF exposure group compared to those for the DMSO control group (Figure 3A). The color’s intensity is proportional to the logarithm of the fold change of CPF/DMSO (logFC) in expression. The connection of genes reflects Topological Overlap Matrix (TOM) similarity, which represents the co-expression similarity based on the gene’s topological overlap (thicker edges indicate stronger connections).
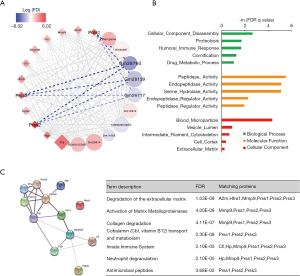
3 mRNAs from the trypsin family of serine proteases, Prss1, Prss2, and Prss3, were significantly upregulated in the CPF group. These genes encode trypsinogen and are members of the protein digestion and absorption pathway (28). They all showed strong TOM similarity with lncRNAs Gm28760, Gm28139, and Gm26717 (Figure 3A, Table 2). A GO enrichment analysis of selected mRNAs in the lightpink2 module was performed using GSEA. The top 5 items with the largest −ln (FDR q value) values of biological process (BP), molecular function (MF), and cellular component (CC) were shown (Figure 3B). The results indicated that the selected mRNAs of the lightpink2 module were mainly involved in cellular component disassembly and proteolysis. The protein-protein interactions (PPI) of hub mRNAs was also constructed in the lightpink2 module using STRING. The PPI results showed that the Prss1-3 interactions were closely connected. A STRING pathway analysis of the hub mRNAs in this module showed that Prss1-3 plays an important role in extracellular matrix and collagen degradation (Figure 3C).

Full table
Co-expression modules corresponding to effects of prenatal CPF exposure at different developmental times
The line plot of r values of the module-trait relationship at different developmental time points showed that the r values of the darkolivegreen2 and green4 modules increased as the development time increased, while the r values of lavenderblush decreased as the development time increased (Figure 4A). MM and GS were also plotted in a scatterplot, and the results showed that MM in the darkolivegreen2 and green4 modules was significant with GS at E12.5 development time (r =0.73, P<1E–200; r =0.57, P=1E–139), while in the lavenderblush module, MM was significantly correlated with its GS at E18.5 development time (r =0.72, P<1E−200) (Figure 4B). Hub mRNAs were confirmed by a GSEA among selected mRNAs (https://cdn.amegroups.cn/static/public/atm-20-6632-3.pdf). The top 10 lncRNAs with the largest MM values were defined as the hub lncRNAs. Finally, there are 10 hub lncRNAs and 16 hub mRNAs in the green4 module, 10 hub lncRNAs and 12 hub mRNAs in the darkolivegreen2 module (Table 1).
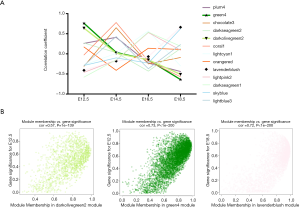
E12.5 kidney development was hindered in the prenatal CPF exposure mouse model
LncRNA-mRNA networks of hub genes in the green4 and darkolivegreen2 modules were constructed separately. The lncRNAs are shown as circles, while the mRNAs are shown as diamonds. The blue color indicates the downregulation, and the red color indicates the upregulation of a gene in E12.5 under prenatal CPF exposure (Figure 5A). GO analyses of the selected mRNAs of the green4, and darkolivegreen2 modules were performed using a GSEA. The top 10 items with the largest −ln (FDR q value) values are shown of BP for the darkolivegreen2 module (Figure 5B). The results showed that the selected mRNAs were mainly involved in urogenital system development, especially kidney development. The hub mRNAs of the darkolivegreen2 module were all downregulated, as reflected in the E12.5 kidney development, which was hindered. The top 5 items with the largest −ln (FDR q value) values were shown of BP, MF and CC for the green4 module (Figure 5C). The results indicated that the selected mRNAs of the green4 module were mainly involved in the muscle regulation process. PPI were also examined using the STRING of the hub mRNAs for these two modules. Using the cluster function in STRING, the hub mRNAs were divided into two clusters in accordance with the two modules. The red cluster comprised hub mRNAs from the darkolivegreen2 module, and the green cluster comprised hub mRNAs from the green4 module. The GO analysis results of the hub mRNAs using STRING were similar to those of the GO analysis of selected mRNAs using a GSEA. Thus, hub mRNAs play an important role in the enriched items (Figure 5C).
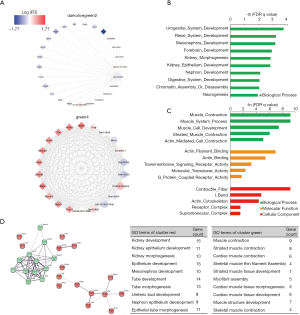
Abnormal proximal tubules development in E18.5 kidney from the prenatal CPF exposure mouse model
As the MM in the lavenderblush module was significantly correlated with its GS at the E18.5 development time point, a lncRNA-mRNA network was constructed of the hub genes in the lavenderblush module. Hub mRNAs were confirmed using a GSEA among the selected mRNAs (https://cdn.amegroups.cn/static/public/atm-20-6632-4.pdf; https://cdn.amegroups.cn/static/public/atm-20-6632-5.pdf). The top 10 lncRNAs with the largest MM values were defined as the hub lncRNAs. There were 10 hub lncRNAs and 10 hub mRNAs in this module. LncRNAs are shown as circles, and mRNAs are shown as diamonds. The blue color indicates the downregulation, and the red color indicates the upregulation of a gene in E18.5 under prenatal CPF exposure (Figure 6A). GO analysis of hub mRNAs for the lavenderblush module was performed by STRING. The results showed that the hub mRNAs were mainly involved in ion transport processes (Figure 6B). We searched the single-cell sequencing database and found that these hub mRNAs are mainly proximal tubule markers (Table 3). According to all the databases, Sla34a1, Sla6a19, and Aqp1 were confirmed proximal tubule markers. Thus, a quantitative real-time polymerase chain reaction (qRT-PCR) was performed for Sla34a1, Sla6a19, and Aqp1, and the results indicated that the mRNA levels of these genes were significantly increased in the E18.5 kidney of the CPF group compared to that of the DMSO control group (Figure 6C). Immunohistochemistry staining against Aqp1 of E18.5 kidneys showed that the ratio of Aqp1+ tubules/all tubules was significantly increased in the CPF group than that of the DMSO control group (Figure 6D).
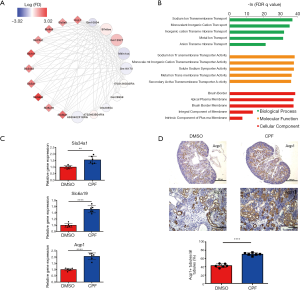
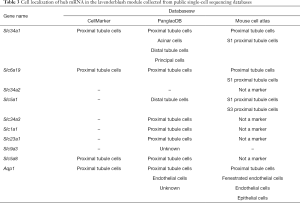
Full table
Discussion
Prenatal CPF exposure has been found to affect neurodevelopment (8-10), and lncRNAs appear to be involved in biological development processes (19-21). To date, research on prenatal CPF exposure and lncRNAs in kidney development has been limited. Our study investigated the lncRNA-mRNA regulation network by undertaking a WGCNA of kidney development in a prenatal CPF exposure mouse model.
A WGCNA is based on a systems biology method for constructing correlation networks; it describes the correlation patterns among genes across microarray samples (29). Researchers found that WGCNA had a higher validation rate in a study of glioblastoma than a differential expression analysis in biomarker identification (30). Also, WGCNA is widely applied in molecular targets and disease-associated pathway screenings of many diseases and biological processes (31-33).
In our study, high-throughput RNA-sequencing was performed on embryonic kidneys obtained at E12.5, E14.5, E16.5, and E18.5 from prenatal CPF exposure mice and DMSO control mice. The WGCNA produced 13 module clusters based on the sequenced data, and each module was assigned a unique color. A module-trait relationships analysis indicated that the lightpink2 and plum4 modules were related to prenatal CPF exposure. A scatterplot between MM and GS also showed that MM in the lightpink2 and plum4 modules correlated significantly with GS, suggesting that the hub genes in the lightpink2 and plum4 modules were highly associated with prenatal CPF exposure. Hub genes were selected (seen the methods section); however, it should be noted that there were no hub mRNAs in the plum4 module. Thus, we only constructed the hub lncRNA-mRNA network of the lightpink2 module using Cytoscape. The result showed that 3 hub mRNAs of the trypsin family of serine proteases, Prss1, Prss2, and Prss3, were significantly upregulated in the CPF group.
Further, they all had strong TOM similarity with 3 significantly downregulated hub lncRNAs, Gm28760, Gm28139, and Gm26717. GO analysis revealed that Prss1-3 plays an important role in the extracellular matrix and collagen degradation. This lncRNA-mRNA network showed that prenatal CPF exposure mainly affected the degradation of extracellular matrix and collagen, and Prss1-3 under the regulation of Gm28760, Gm28139, and Gm26717 were the core genes.
We also investigated the effect of prenatal CPF exposure on kidney development at different embryonic days. The darkolivegreen2 and green4 modules were found to be related to E12.5 kidney development with prenatal CPF exposure, while the lavenderblush module was found to be related to E18.5 kidney development. GO analysis of selected mRNAs from the darkolivegreen2 module revealed that kidney development was hindered at E12.5 after prenatal CPF exposure. All the hub mRNAs were downregulated in the darkolivegreen2 module. Osr1, Wt1, and foxd1 were known nephron progenitor cell (NPC) markers, the low expression of NPC markers reflects the NPC self-renewal process’s inhibition, which leads to abnormal nephron development (34-36). The LncRNA-mRNA network showed that all of the hub mRNAs were connected with lncRNA Gm13446. GO analysis of the green4 module revealed the lncRNA-mRNA network mainly involved in muscle regulation. Notably, hub mRNAs in the lavenderblush module were all membrane transporter genes, and most of them were proteins expressed on proximal tubules. These results showed that ectopic proximal tubules formed in E18.5 kidneys in the CPF group. We verified this finding through qRT-PCR and immunohistochemistry staining.
In summary, our study revealed molecular phenotype changes in the kidney development of a prenatal CPF exposure mouse model via RNA-sequencing and a WGCNA. The novel findings that a lncRNA-mRNA network regulated the kidney development process in our mouse model may provide a new treatment strategy for early kidney disease intervention. We will use small interfering RNA and Gabmer to confirm mRNA-lncRNA relationships in kidney cell lines in our future research. Additionally, an in vitro culture of the embryonic kidney will be performed in a functional study to confirm the direct role of hub lncRNAs as found in our research.
Acknowledgments
We would like to express our thanks to all the staff at the Kidney Disease Center of The First Affiliated Hospital of Zhejiang University for their sincere support.
Funding: This work was supported by the National Natural Science Foundation of China (No. 81470938 and No. 81770697) to HJ, and National Basic Research Program of China (973 Program) (No. 2011CB944002) to JC.
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-6632
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-6632
Peer Review File: Available at http://dx.doi.org/10.21037/atm-20-6632
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-6632). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work, including ensuring that questions related to the accuracy or integrity of any part of the work have been appropriately investigated and resolved. The Research Ethics Committee of the First Affiliated Hospital, College of Medicine, Zhejiang University approved the experimental protocols (No. 2018361). All the experiments were carried out in accordance with the NIH Guide for the Care and Use of Laboratory Animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Clevers H. Modeling Development and Disease with Organoids. Cell 2016;165:1586-97. [Crossref] [PubMed]
- Buser MC, Ingber SZ, Raines N, et al. Urinary and blood cadmium and lead and kidney function: NHANES 2007-2012. Int J Hyg Environ Health 2016;219:261-7. [Crossref] [PubMed]
- Kipling RM, Cruickshank AN. Organophosphate insecticide poisoning. Anaesthesia 1985;40:281-4. [Crossref] [PubMed]
- Furlong MA, Herring A, Buckley JP, et al. Prenatal exposure to organophosphorus pesticides and childhood neurodevelopmental phenotypes. Environ Res 2017;158:737-747. [Crossref] [PubMed]
- Bellinger DC. Prenatal Exposures to Environmental Chemicals and Children's Neurodevelopment: An Update. Saf Health Work 2013;4:1-11. [Crossref] [PubMed]
- Engel SM, Bradman A, Wolff MS, et al. Prenatal Organophosphorus Pesticide Exposure and Child Neurodevelopment at 24 Months: An Analysis of Four Birth Cohorts. Environ Health Perspect 2016;124:822-30. [Crossref] [PubMed]
- Mie A, Rudén C, Grandjean P. Safety of Safety Evaluation of Pesticides: developmental neurotoxicity of chlorpyrifos and chlorpyrifos-methyl. Environ Health 2018;17:77. [Crossref] [PubMed]
- Lan A, Kalimian M, Amram B, et al. Prenatal chlorpyrifos leads to autism-like deficits in C57Bl6/J mice. Environ Health 2017;16:43. [Crossref] [PubMed]
- Guo J, Zhang J, Wu C, et al. Associations of prenatal and childhood chlorpyrifos exposure with Neurodevelopment of 3-year-old children. Environ Pollut 2019;251:538-546. [Crossref] [PubMed]
- von Ehrenstein OS, Ling C, Cui X, et al. Prenatal and infant exposure to ambient pesticides and autism spectrum disorder in children: population based case-control study. BMJ 2019;364:l962. [Crossref] [PubMed]
- Tanvir EM, Afroz R, Chowdhury M, et al. A model of chlorpyrifos distribution and its biochemical effects on the liver and kidneys of rats. Hum Exp Toxicol 2016;35:991-1004. [Crossref] [PubMed]
- Zahran E, Risha E, Awadin W, et al. Acute exposure to chlorpyrifos induces reversible changes in health parameters of Nile tilapia (Oreochromis niloticus). Aquat Toxicol 2018;197:47-59. [Crossref] [PubMed]
- Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem 2012;81:145-66. [Crossref] [PubMed]
- Esteller M. Non-coding RNAs in human disease. Nat Rev Genet 2011;12:861-74. [Crossref] [PubMed]
- Huarte M. The emerging role of lncRNAs in cancer. Nat Med 2015;21:1253-61. [Crossref] [PubMed]
- Hirata H, Hinoda Y, Shahryari V, et al. Long Noncoding RNA MALAT1 Promotes Aggressive Renal Cell Carcinoma through Ezh2 and Interacts with miR-205. Cancer Res 2015;75:1322-31. [Crossref] [PubMed]
- Wang M, Yao D, Wang S, et al. Long non-coding RNA ENSMUST00000147869 protects mesangial cells from proliferation and fibrosis induced by diabetic nephropathy. Endocrine 2016;54:81-92. [Crossref] [PubMed]
- Zhou Q, Huang XR, Yu J, et al. Long Noncoding RNA Arid2-IR Is a Novel Therapeutic Target for Renal Inflammation. Mol Ther 2015;23:1034-1043. [Crossref] [PubMed]
- Lorenzen JM, Thum T. Long noncoding RNAs in kidney and cardiovascular diseases. Nat Rev Nephrol 2016;12:360-73. [Crossref] [PubMed]
- Bae BI, Jayaraman D, Walsh CA. Genetic changes shaping the human brain. Dev Cell 2015;32:423-34. [Crossref] [PubMed]
- Goff LA, Groff AF, Sauvageau M, et al. Spatiotemporal expression and transcriptional perturbations by long noncoding RNAs in the mouse brain. Proc Natl Acad Sci U S A 2015;112:6855-62. [Crossref] [PubMed]
- Barak H, Boyle SC. Organ culture and immunostaining of mouse embryonic kidneys. Cold Spring Harb Protoc 2011;2011:pdb.prot 5558.
- Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15-21. [Crossref] [PubMed]
- Trapnell C, Hendrickson DG, Sauvageau M, et al. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol 2013;31:46-53. [Crossref] [PubMed]
- Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545-15550. [Crossref] [PubMed]
- Chen W, Jiang H, Wang M, et al. Effects of chlorpyrifos exposure on kidney Notch2-Jagged1 pathway of early prenatal embryo. Birth Defects Res B Dev Reprod Toxicol 2011;92:97-101. [Crossref] [PubMed]
- Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol 2005;4:Article17.
- Emi M, Nakamura Y, Ogawa M, et al. Cloning, characterization and nucleotide sequences of two cDNAs encoding human pancreatic trypsinogens. Gene 1986;41:305-10. [Crossref] [PubMed]
- Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 2008;9:559. [Crossref] [PubMed]
- Horvath S, Zhang B, Carlson M, et al. Analysis of oncogenic signaling networks in glioblastoma identifies ASPM as a molecular target. Proc Natl Acad Sci U S A 2006;103:17402-7. [Crossref] [PubMed]
- Niemira M, Collin F, Szalkowska A, et al. Molecular Signature of Subtypes of Non-Small-Cell Lung Cancer by Large-Scale Transcriptional Profiling: Identification of Key Modules and Genes by Weighted Gene Co-Expression Network Analysis (WGCNA). Cancers (Basel) 2019;12:E37 [Crossref] [PubMed]
- Maertens A, Tran V, Kleensang A, et al. Weighted Gene Correlation Network Analysis (WGCNA) Reveals Novel Transcription Factors Associated With Bisphenol A Dose-Response. Front Genet 2018;9:508. [Crossref] [PubMed]
- Ding M, Li F, Wang B, et al. A comprehensive analysis of WGCNA and serum metabolomics manifests the lung cancer-associated disordered glucose metabolism. J Cell Biochem 2019;120:10855-10863. [Crossref] [PubMed]
- Li Z, Araoka T, Wu J, et al. 3D Culture Supports Long-Term Expansion of Mouse and Human Nephrogenic Progenitors. Cell Stem Cell 2016;19:516-529. [Crossref] [PubMed]
- Wanner N, Vornweg J, Combes A, et al. DNA Methyltransferase 1 Controls Nephron Progenitor Cell Renewal and Differentiation. J Am Soc Nephrol 2019;30:63-78. [Crossref] [PubMed]
- O'Brien LL. Nephron progenitor cell commitment: Striking the right bala


