
DNA sequence analysis and Jk blood group genotype-phenotype assessment
Introduction
Kidd (JK) blood group is one of the most critical blood groups in transfusion medicine. Antibodies that target the Jk antigens may cause severe delayed hemolytic transfusion reactions and even the hemolytic disease of the newborn (HDN) (1). More seriously, the titrations of Jk antibodies usually remain at deficient levels for routine serology tests, whereas the corresponding antigens will immediately trigger an abundant production immediately (2,3). This characteristic gives a high undetectable rate and causes the Jk blood group to be much more concerned with clinical transfusion.
To date, three blood group antigens, Jka, Jkb, and Jk3, have been described in this blood group system (4). These clinically important antigens are combined into four different phenotypes: Jk (a+b-), Jk (a+b+), Jk (a-b+), and Jk (a-b-). The frequency of the latter is less than 0.02% in Chinese Han individuals (5). The frequencies of Jka and Jkb were 67.99% and 75.57%, respectively, in mainland China. These findings were like those in the Chinese population in Taiwan (6,7). However, the frequency of Jka (92.00%) was statistically higher than Jkb (49.00%) in the Black population (8). Recently, weakened expression of Jka antigen in RBCs, termed as Jk (aw+), has been reported by us and others (9,10). The Jk antigens are carriers of the glycoprotein, the urea transporter (UT-B) in red blood cells (RBCs) (11). The Jk gene, also known as SLC14A1, located in the long arm of chromosome 18 and showing 11 exons distributed over 30 kb of DNA, encodes the Kidd glycoprotein (12,13). The translation initiation codon has been found in exon 4, and the three 5' upstream exons are not translated (14). Through lacking the functional UT-B, the Jk (a-b-) RBCs own the resistant ability from lysing by 2M urea (15). Despite this, the Jk(a-b-) RBCs are healthy and own a normal shape and lifespan. This characteristic is commonly used in screening for Jk(a-b-) individuals in blood centers worldwide. However, because of the similar performance with Jk(a-b-) samples in the urea hemolysis test, the Jk(aw+b-) samples could not be identified easily in this way (9,10). Still, the standard serology test is indispensable for the identification of Jk(aw+) with Jk(a-) phenotypes.
To supplement serological genotyping, several molecular techniques, including sequence-specific primer (SSP) PCR (16,17), restriction fragment length polymorphism (RFLP) (18), single-strand conformation polymorphism (SSCP) analysis (19), and DNA microarray hybridization (20), have been developed for identification of the Jk gene. Using molecular techniques to investigate unusual blood group phenotypes has led to the clarification of many serologic anomalies. Most molecular typing techniques target exons 4-11, the coding sequence of the Jk gene, and focus on single-nucleotide polymorphisms (SNPs) in these sequences. For the limitation of primers and protein structure, these methods could only detect the common phenotypes and known mutations. Novel mutations are unlikely to be discovered.
DNA sequencing is an advanced technique that gives more exact information about the DNA sequence. The method for blood group genotyping by DNA sequencing has been established from 1997 (21,22). Several studies have discussed the application of this method in the typing of blood groups, including ABO, Rh, and Diego, together with allele frequency determination at population-scales (23,24). Our center has been applying DNA sequencing for HLA genotyping for the direct sequencing of amplicons on the ABI 3730 platforms for decades. This method is being routinely used in our stem cell donor center, affiliated to China Marrow Donor Program (CMDP), to characterize newly registered potential stem cell donors and selection of matched donors for bone marrow transplantation (25,26). Since the improving requirement for finding the molecular variations, we extended the existing Sanger-based genotyping workflow to additionally Jk genotyping. While this technique satisfies clinical needs, it is a time-consuming and high cost to apply them into routine Jk genotyping of large scales of blood donors.
Here, we describe the workflow for sequencing the Jk exons 4-11 and intron-exon borders for finding three novels, Jk variants, and reporting frequencies of SNPs in distinct antigen groups. It is intriguing for us to find that a Jk-antigens preferred approach is exhibiting the specific polymorphisms, and the genetic association and linkage of the two SNPs offer a potential sequencing target for rapid Jk typing.
We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/atm-20-6504).
Methods
Blood samples
Shenzhen Blood Center collected EDTA-anticoagulated blood samples between January 2015 and December 2017 from voluntary blood donors. All samples were taken after informed consent at the referring institutions, and the donors were not known to be related. This cohort (n=174) consisted of blood donors with random JK phenotypes. Also, the Blood Group Reference Laboratory collected three Jk(aw+b-) samples in Shenzhen Blood Center during the routine clinical work conducted at the laboratory. Moreover, Huizhou Blood Center kindly provided six Jk(a-b-) blood samples in Guangdong Province, China. All these 183 blood samples were collected from the Chinese Han individuals. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The Ethics Committee approved the study. Patients who took part in this research signed informed consent and had complete clinical data.
Serology test
Initial serology was performed on RBCs from the samples by the referring laboratory using standard techniques with monoclonal anti-Jka and anti-Jkb test reagents (human immunoglobulin [Ig]M, Sanquin, Holland) respectively. One drop of the cell suspension was mixed with one drop of anti-Jka or anti-Jkb^ respectively, centrifuging the mixture at 1,000 rpm for 1 minute at room temperature. Clumping occurs in RBCs carrying the Jka or Jkb antigen while lacking agglutination shows the absence of the Jka or Jkb antigen.
Urea hemolysis test
A urea hemolysis test was performed for the mass screen of the Jk(a-b-) phenotype from donors, with the resistant ability of Jk(a-b-) RBCs from lysing by 2M urea. Add 100 µL 2M urea-0.4% PBS solution into a U-shaped 96-well plate, and then 20 µL 2% RBCs suspension was pipetted into each well and thoroughly mixed with urea solution. Centrifuge the plate and detect whether the hemolysis occurred or not.
DNA isolation, quantification, PCR amplification, and purification
Genomic DNA was isolated from 300 µL whole blood using the MagNA Pure LC DNA Isolation Kit I (Roche, Basel, Switzerland). DNA’s concentrations were measured using the NanoDrop Microvolume Spectrophotometers and Fluorometer (ThermoFisher, USA). Genomic DNA was diluted in ddH2O to 100 ng/µL. Samples with concentrations less than 2 ng/µL were excluded from the following procedures.
Sanger-sequencing decided the nucleotide sequences of the 4-11 JK exons and intron-exon borders. PCR amplification is performed according to in-house routine procedures. All primers (Table 1) are referred and synthesized (TAKARA, BIO INC., China) according to the SLC14A1 sequence (GenBank Accession No.: NG_011775.3) (13,27). Amplification was performed in 96-well plates with 2 µL template DNA, 1 µL 10× PCR buffer without MgCl2 (Applied Biosystems, USA), 3 µL 5 mM MgCl2, 0.8 µL 2.5 mM dNTPs, 0.1 µL Taq Polymerase (5 U/µL), 1.9 µL ddH2O, and 1.2 µL of target-specific primer mix. We used a thermal profile of 96 °C for 7 minutes followed by 35 cycles at 96 °C for 30 seconds, 58 °C for 30 seconds and 72 °C for 60 seconds, and a finishing step at 72 °C for 7 minutes. PCR products were subjected to DNA gel electrophoresis using 2% agarose gel. DNA Ladder (TaKaRa, China) was loaded for band size standardization. A gel documentation system was applied to capture the images of the DNA gel under UV radiation. Estimate the quality of products by detecting the brightness and size of bands. Then, add the exonuclease I and alkaline phosphatase (ThermoFisher, USA) to purify the PCR products and prepare for sequencing.

Full table
Sequencing
The template was prepared as described above. Nucleotide sequencing was performed with a DNA sequencing unit prism dye terminator cycle sequencing kit with AmpliTaq FS DNA polymerase (Applied Biosystems, USA). Nucleotide sequencing of genomic DNA stretches representative for 4-11 JK exons, and intron-exon borders were carried out using primers (Table 1) and 2nd PCR amplification procedures. The 2nd PCR was performed using the BigDye Terminator V3.1 kit (Applied Biosystems, USA) according to the manufacture’s recommendation. The amplification system has 5 µL of puried products in a total volume of 10 µL, with 0.25 µL Bigdye 3.1 and 0.32 µL 10 µM specific primer in the buffer supplied. Amplification was performed in the GeneAmp PCR system: 1 minute at 95 °C, followed by 25 cycles of 10 seconds at 95 °C, 5 seconds at 50 °C, and 4 minutes at 60 °C.
Statistical analysis
The genotyping results were analyzed using computer software (uTYPE, Chromas, Graphpad Software). A database search in NCBI (https://www.ncbi.nlm.nih.gov/ncbisearch), on the SLC14A1 sequence (GenBank Accession No.: NG_011775.3) was performed to compare results of the sequencing in different serological phenotypes, including Jk(a+b+), Jk(a+b-), Jk(a-b+), Jk(aw+b-) and Jk(a-b-).
Terminology
We use the officially accepted blood group terminology together with the guidelines currently under consideration by the ISBT Working Party on Red Cell Immunogenetics and Blood Group Terminology. The letter w is used to show a weakened antigen in RBCs.
Results
This study proves the biochemical, serologic, and genetic findings retrieved in 183 independent samples in the Blood Group Reference Laboratory in Shenzhen Blood Center. By the Sanger sequencing approach, we found the molecular alterations in exon 4–11, including adjacent intronic sequences of the Jk locus. We detected up to six mutations per sample, forming homo- and heterozygous nucleotide exchanges.
Urea hemolytic and serologic test
The urea hemolysis test has been originally conducted to discriminate Jk(a-b-) samples from Jk(a+) and/or Jk(b+) samples. Jk(a-b-) RBCs take at least 30 minutes to lyse, while RBCs with Jk(a+) and/or Jk(b+) phenotypes lyse within 2 minutes. The 174 independent blood samples collected from voluntary blood donors gave 100% urea-induced lysis compared to water after 2 minutes incubation, demonstrating the Jk phenotypes. Then, we confirmed our workflow by finding the samples by known serologic blood group tests, using both the human monoclonal anti-Jka and anti-Jkb. The results showed that the Jk phenotypes of 174 samples were 80 Jk(a+b+), 40 Jk (a+b-), and 54 Jk (a-b+), all of which were common Jk phenotypes.
However, in the urea hemolysis test, the Jk(aw+b-) RBCs exhibit similarity with Jk(a-b-) samples, showing no hemolysis. Thus, the urea hemolysis test designed to screen for Jk(a-b-) samples was unable to find the Jk(aw+b-) samples. However, Jk(aw+b-) RBCs reacted weakly with human monoclonal anti-Jka, from which the serologic test owns the ability to distinguish it from Jk(a-b-) RBCs. Despite this low expression of the Jka-antigen, Jk(aw+b-) RBCs has been reported to induce anti-Jka immunization. Thus, these individuals should be regarded as Jka-positive donors, Jka-negative recipients. Furthermore, the RBCs from the Jk(a-b-) samples were resisted from lysing by urea, and this phenotype was inconsistent with the serologic test.
Evaluation of amplification and sequencing reactions in blood samples
Evaluation of the amplification reactions for Jk exons 4-11 and intron-exon borders gave strong positive results in all samples, showing the Jk specificity for all reactions (Figure 1). The Jk(aw+b-) and Jk(a-b-) samples also showed positive results in all reactions, and sequencing procedures of them showed no deviation from the control samples. Then, procedures were conducted to read the complete sequence from the coding region with one sequencing primer for each exon.
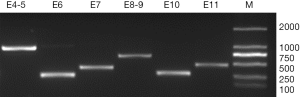
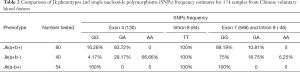
Several SNPs were noted through the alignment of our data with published Jk sequences (GenBank Accession No.: NG_011775.3). Distinct frequencies of single-nucleotide mutations of exon 4 (130), exon 7 (588), and intron 9 (-46) have been revealed in samples of each Jk phenotype (Table 2). And the computer simulation of amino acid mutations according to Jk polymorphism exon 4 (130G > A) showed variants of Glu44Lys (Figure S1A,B). Furthermore, a homozygous T/T mutation in intron 8 (-84) was found in all 174 samples in Table 2. The SNPs frequencies in the Shenzhen voluntary blood donor cohort, as well as results retrieved from the database, are summarized in Table 2. Although it differs further, the frequency of these SNPs in Jk(a+b+), Jk(a+b-), and Jk(a-b+) samples demonstrates the characteristic in Chinese Han individuals.
Full table

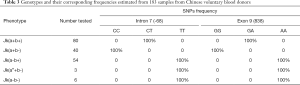
Screening of all 183 samples containing the 174 voluntary blood donors, 3 Jk(aw+b-), and 6 Jk(a-b-) by Sanger sequencing revealed a fantastic regularity between genotype and phenotype in all samples tested. Results showed that Jk(a+b+) samples were heterozygous for intron 7(-68C/T) and exon 9 (838G/A) (Table 3). Moreover, homozygous intron 7 (-68C/C) and exon 9 (838G/G) was demonstrated in Jk(a+b-) samples, whereas intron 7 (-68T/T) and exon 9 (838A/A) in Jk(a-b+) retrieved from the sequencing results are also summarized in Table 3. The determination of the SNPs was then performed on the Jk(aw+b-) and Jk(a-b-) samples and showed homo- or heterozygosity, respectively. Intriguingly, not only in Jk(a-b+) samples, the homozygote intron 7 (-68T/T) and exon 9 (838A/A) but also demonstrated in Jk(aw+b-) and Jk(a-b-) groups, suggesting the co-inheritance of these adjacent DNA variants in all Jk(aw/-) samples. Assuming that these are genetic association and linkage in these two sites of intron 7 (-68) and exon 9 (838), described above, offers a promising marker for rapid typing of the Jk blood group. Moreover, we also simulated the Jk polymorphism on exon 9 (838G>A) and confirmed its variation on Asp280Asn amino acid substitution (Figure S1A,C).
Full table
Discussion
From the perspective of a patient immunized against a blood group antigen, it is vital to make sure that compatible blood is transfused and that the antibody response is not boosted. Thus, it is vital to detect the correct blood type before clinical transfusion.
We have developed a sequencing strategy for the Jk exons that can be applied to unravel the Jk status even under unclear situations. Because the translation is started from the 5' upstream of exon 4 (28), we lately sequenced the Jk exons 4-11 and intron-exon borders in this study. It is an applicable method for the molecular characterization of Jk-variants that are caused by single nucleotide exchanges restricted in these sequences. The sequencing of these Jk exons of 183 independent blood samples of Chinese individuals proved the feasibility of the method. All primers were perfectly matched to the procedure and gave undoubted amplification and sequencing results. Primers were also previously applied successfully in Jk(aw+b-) and Jk(a-b-) samples.
Urea hemolytic and serologic test showed there were 80 Jk(a+b+), 40 Jk(a+b-), and 54 Jk(a-b+) samples in 174 samples from individual donors. In this study, six SNPs in a total of 183 samples, holding Jk(aw+b-) and Jk(a-b-) samples, were identified, as shown in Tables 2 and 3. Distinct frequency of polymorphisms in exon 4 (-130), exon 7 (-588), and intron 9 (-46) were shown in each Jk phenotype. However, our computer simulation of Jk antigens on exon 4 (130G > A) showed variants of Glu44Lys, and the structure of Jk protein proved this amino acid located inside the membrane. All the samples sequenced, containing the 174 voluntary blood donors, 3 Jk(aw+b-), and 6 Jk(a-b-), exhibited homozygous T/T in intron 8 (-84), suggesting a common genotype in the Chinese Han population.
In addition, the intron 7 (-68C/T) and exon 9 (838G/A) SNPs were found in 80 DNA samples from blood donors with Jk(a+b+) phenotype. In the present study, homozygous mutations of -68C/C in intron 7 and 838G/G in exon 9 were also found in Jk(a+b-) RBCs. Intron 7 (-68T/T) and exon 9 (838A/A) has been showed in Jk(a-b+). Such a polymorphism was intriguingly found in 3 (100%) Jk(aw+b-) and 6 (100%) Jk(a-b-) blood samples. Besides, we also simulated the Jk antigens on exon 9 (838G > A) and confirmed its variation on Asp280Asn. We assume the co-inheritance of adjacent DNA sites, intron 7 (-68) and exon 9 (838), allows us to predict the exact Jk phenotype. This assumption is after the pattern of SNPs shown in Table 3. This is the first report of a novel mutation intron 7 (-68), which is associated with the Jka/Jkb polymorphism, beside exon 9 (-838). When we retrieved the sequencing result of a blood sample, revealing the mutant pattern of these two sites, intron 7 (-68) and exon 9 (-838), which exhibited the astonishing regularity with phenotypes, we could predict the Jk type of the sample.
In summary, our results show that the molecular analysis of the Jk gene by sequencing is required in some cases to provide the appropriate recommendations for anti-Jk prophylaxis in transfusions. Confirmation of blood type is the necessary and essential procedure before clinical transfusion. For blood genotyping, a medium-throughput technique like DNA sequencing is time-and cost-consuming (29). In DNA sequencing of all exons of a blood gene, extensive data are generated that are difficult to store, and analysis of the data takes days to weeks (30). According to the regularity, Jk genotyping of far more blood samples than present would be possible within one sequencing cycle. However, our knowledge of the co-inheritance of the Jk base substitution is from the results retrieved from 183 Chinese individuals. Differences among different racial populations can be assumed. Still, more data need to be collected for the confirmation of this assumption.
Conclusions
Through finding the genotype of intron 7 (-68) and exon 9 (-838), we would estimate the Jk antigens and narrow down the possible phenotype. Phenotypes of Jk(a+) and Jk(aw+/-) can be found by detecting these two SNPs. Under that circumstance, sequencing displays a higher cost-effective technique for future routine testing on the Jk blood system in blood centers.
Acknowledgments
Funding: This work was supported by the Wei Gao Project of the Chinese Society of Blood Transfusion (CSBT-WG-2018-09), Sanming Project of Medicine in Shenzhen (No.: SZSM201811092), Shenzhen Key Medical Discipline (No. SZXK070), Shenzhen Science and Technology Plan Project (No.: JCYJ20180302153621300).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-6504
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-6504
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-6504). SL reports grants from The Chinese Society of Blood Transfusion, during the conduct of the study; HZ reports grants from Shenzhen Science and technology innovation Commission, during the conduct of the study; WXH and YPX report grants from Shenzhen Municipal Health Commission, during the conduct of the study. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The Ethics Committee approved the study. Patients who took part in this research signed informed consent and had complete clinical data.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Daniels G. Human Blood Groups. First edition. Blackwell Science Limited 1995:Chapter 9.
- Velasco Rodríguez D, Pérez-Segura G, Jiménez-Ubieto A, et al. Hemolytic disease of the newborn due to anti-jkb: case report and review of the literature. Indian J Hematol Blood Transfus 2014;30:135-8. [Crossref] [PubMed]
- Dean L. Blood Groups and Red Cell Antigens. Bethesda (MD): National Center for Biotechnology Information (US) 2005:Chapter 10.
- Issitt P, Anstee DJ. Applied Blood Group Serology. 4th edition. Montgomery Scienti®c Publications (USA) 1998.
- Wen Z, Defa L, Hongmei W, et al. Genomic characterization of the Jk(a-b-) phenotype for children in Shenzhen,China. Chin J Blood Transfusion 2015;28:24-5.
- Lin-Chu M, Broadberry RE, Chang FJ. The distribution of blood group antigens and alloantibodies among Chinese in Taiwan region. Transfusion 1988;28:350-2. [Crossref] [PubMed]
- Yung CH, Chow MP, Hu HY, et al. Blood group phenotypes in Taiwan. Transfusion 1989;29:233-5. [Crossref] [PubMed]
- Yu Y, Ma C, Sun X, et al. Frequencies of red blood cell major blood group antigens and phenotypes in the Chinese Han population from Mainland China. Int J Immunogenet 2016;43:226-35. [Crossref] [PubMed]
- Wester ES, Storry JR, Olsson ML. Characterization of Jk(a+(weak)): a new blood group phenotype associated with an altered JK*01 allele. Transfusion 2011;51:380-92. [Crossref] [PubMed]
- Su YQ, Li WB, Liang YL, et al. The correlation analysis of the weakly Jka antigen with 130A in exon 4 of the JK gene. Chinese Journal of Blood Transfusion 2018;31:228-30.
- Heaton DC, McLoughlin K. Jk (a-b-) red blood cells resist urea lysis. Transfusion 1982;22:70-1. [Crossref] [PubMed]
- Olivès B, Mattei MG, Huet M, et al. Kidd blood group and urea transport function of human erythrocytes are carried by the same protein. J Biol Chem 1995;270:15607-10. [Crossref] [PubMed]
- Irshaid NM, Henry SM, Olsson ML. Genomic characterization of the Kidd blood group gene: different molecular basis of the Jk (a-b-) phenotype in Polynesians and Finns. Transfusion 2000;40:69-74. [Crossref] [PubMed]
- Irshaid NM, Eicher NI, Hustinx H, et al. Novel alleles at the JK blood group locus explain the absence of the erythrocyte urea transporter in European families. Br J Haematol 2002;116:445-53. [Crossref] [PubMed]
- Sands JM, Gargus JJ, Fröhlich O, et al. Urinary concentrating ability in patients with Jk (a-b-) blood type who lack carrier-mediated urea transport. J Am Soc Nephrol 1992;2:1689-96. [PubMed]
- Prager M. Molecular genetic blood group typing by the use of PCR-SSP technique. Transfusion 2007;47:54S-9S. [Crossref] [PubMed]
- Liu Z, Zeng R, Chen Q, et al. Genotyping for Kidd, Kell, Duffy, Scianna, and RHCE blood group antigens polymorphisms in Jiangsu Chinese Han. Chin Med J (Engl) 2012;125:1076-81. [PubMed]
- Geitvik GA, Høyheim B, Gedde-Dahl T, et al. The Kidd (JK) blood group locus assigned to chromosome 18 by close linkage to a DNA-RFLP. Hum Genet 1987;77:205-9. [Crossref] [PubMed]
- Sell AM, Visentainer JE. Blood grouping based on PCR methods and agarose gel electrophoresis. Methods Mol Biol 2015;1310:37-49. [Crossref] [PubMed]
- Beiboer SH. Rapid genotyping of blood group antigens by multiplex polymerase chain reaction and DNA microarray hybridization. Transfusion 2005;45:667-79. [Crossref] [PubMed]
- Gorakshakar A, Gogri H, Ghosh K. Evolution of technology for molecular genotyping in blood group systems. Indian J Med Res 2017;146:305-15. [PubMed]
- Nata M, Kanetake J, Adachi N, et al. ABO genotyping by PCR-direct sequencing. Nihon Hoigaku Zasshi 1997;51:1-5. [PubMed]
- McBean RS, Hyland CA, Flower RL. Approaches to determination of a full profile of blood group genotypes: Single nucleotide variant mapping and massively parallel sequencing. Comput Struct Biotechnol J 2014;11:147-51. [Crossref] [PubMed]
- Lang K, Wagner I, Schone B, et al. ABO allele-level frequency estimation based on population-scale genotyping by next generation sequencing. BMC Genomics 2016;17:374. [Crossref] [PubMed]
- Deng Z, Wang D, Xu Y, et al. HLA-C polymorphisms and PCR dropout in exons 2 and 3 of the Cw*0706 allele in sequence-based typing for unrelated Chinese marrow donors. Hum Immunol 2010;71:577-81. [Crossref] [PubMed]
- Zhen J, Wang D, He L, et al. Genetic profile of KIR and HLA in southern Chinese Han population. Hum Immunol 2014;75:59-64. [Crossref] [PubMed]
- Wester ES, Johnson ST, Copeland T. Erythroid urea transporter deficiency due to novel JKnull alleles. Transfusion 2008;48:365-72. [PubMed]
- Lucien N, Walter FS, Olives B, et al. Characterization of the Gene Encoding the Human Kidd Blood Group/Urea Transporter Protein. J Biol Chem 1998;273:12973-80. [Crossref] [PubMed]
- Fichou Y, Audrézet MP, Guéguen P, et al. Next-generation Sequencing Is a Credible Strategy for Blood Group Genotyping. Br J Haematol 2014;167:554-62. [Crossref] [PubMed]
- Lane WJ, Vege S, Mah HH, et al. Automated typing of red blood cell and platelet antigens from whole exome sequences. Transfusion 2019;59:3253-63. [Crossref] [PubMed]


