
Association of genetic variants of TMEM135 and PEX5 in the peroxisome pathway with cutaneous melanoma-specific survival
Introduction
Skin cancers, including cutaneous melanoma (CM), are among the most common cancers worldwide. In the United States, their incidence continues to rise dramatically (1), having doubled in the last 4 decades. In 2019, approximately 96,480 patients were diagnosed with CM in the United States, accounting for 5.5% of all new cancer cases. As the most lethal skin cancer, deaths from CM account for 1.2% of cancer-related mortality (2). Although Breslow thickness, ulceration, and the mitotic rate are considered to be important prognostic indicators, many primary CM cases unexpectedly metastasize after complete surgical resection (3); consequently, CM has the highest mortality rate among all skin cancers. Therefore, it is of great urgency to identify and better understand additional prognostic biomarkers that may provide CM patients with more appropriate and individualized treatment options.
CM is a multifactorial disease originating from melanocytes. Host factors, such as fair skin pigmentation, and environmental factors, such as ultraviolet radiation, can contribute to the development of CM (4). Studies have also shown that genetic susceptibility is involved in both the development and progression of CM (5,6). In recent genome-wide association studies (GWASs), genetic variants have been found to be associated with both risk and survival of CM (7). However, few of the identified genetic variants, particularly single-nucleotide polymorphisms (SNPs), were biologically functional, nor did these studies provide support for the biological plausibility of their findings. To identify truly functional variants, more sophisticated and combined analyses in the post-GWAS era are needed, including the combination of hypothesis-based pathway analysis, meta-analysis, and functional analysis.
Peroxisomes are ubiquitous and dynamic organelles that are involved in the metabolism of reactive oxygen species (ROS) and lipids (8,9). They also act as intracellular signaling platforms in inflammatory, redox, lipid, and innate immune signaling (10-13). Emerging studies suggest that peroxisomes play roles in aging, nonalcoholic fatty liver disease, and neurodegenerative disorders (14,15). Other studies have suggested that peroxisomes play an important role in the progression of several cancers, and abnormal peroxisome metabolism is one of the established characteristics of cancer cells (16-19). However, few studies have investigated the role of peroxisomes in disease progression of CM, and little is known about the effect of genetic variation in the peroxisome pathway genes on the clinical outcomes of CM patients.
Given that some genetic variants are known to be associated with CM prognosis, identifying additional genetic variants in some specific signaling pathways may provide novel prognostic biomarkers of CM. Considering the importance of the peroxisome pathway in cancers, it is likely that genetic variants in peroxisome pathway genes are associated with survival in CM patients. Therefore, we tested this hypothesis by using genotyping datasets from 2 previously published GWASs.
We present the following article in accordance with the TRIPOD reporting checklist (available at http://dx.doi.org/10.21037/atm-20-2117).
Methods
Study populations
In the present study, we used the data of 858 patients from a CM GWAS conducted at The University of Texas MD Anderson Cancer Center (MDACC) (20) as a discovery dataset. Another GWAS of 409 CM patients from 2 Harvard University cohort studies, the Nurses Health Study (NHS) and the Health Professionals Follow-up Study (HPFS) (21), was used as a replication dataset. All the CM patients were non-Hispanic Whites. Informed consent was obtained from all patients. The access to the datasets used in the present study was approved by the Institutional Review Boards of the MDACC (No. LAB03-0048), Brigham and Women’s Hospital (No. 2007-P-000616), and Harvard T.H. Chan School of Public Health (No. 2007-P-000616), and conducted in accordance with the Declaration of Helsinki (as revised in 2013).
The discovery dataset was requested from the Database of Genotypes and Phenotypes (dbGaP: http://www.ncbi.nlm.nih.gov/gap), with an accession number of phs000187.v1.p1 (6). Genotyping was performed with the Illumina HumanOmni-Quad_v1_0_B array (Illumina Inc, San Diego, CA) and subsequent genome-wide imputation (imputation quality r2≥0.8) was performed with the MaCH software using sequencing data from the 1000 Genomes Project, phase I v2 CEU data (March 2010 release) (22). The replication dataset was generated with the Illumina HumanHap610 array, and genome-wide imputation (imputation quality r2≥0.8) was also performed with the MaCH software using sequencing data from the 1000 Genomes Project CEU data (March 2012 release) (23). Compared with the NHS/HPFS dataset, patients in the MDACC dataset had relatively complete clinical information, including age, sex, Breslow thickness, metastasis, mitotic rate, ulceration, and survival outcome, in addition to individual genotyping data, while patients in the NHS/HPFS replication dataset had information only for age, sex, and survival outcome, in addition to genotyping data.
Gene and SNP extraction
We selected 90 peroxisome pathway genes, according to the Kyoto Encyclopedia of Genes and Genomes (KEGG), BioCarta, Reactome, and Gene Ontology (GO) databases in the Molecular Signatures Database (http://software.broadinstitute.org/gsea/msigdb/index.jsp). Because females carry 2 copies of the X chromosome (males are hemizygous), and no standard statistics have been established for sex-specific analysis, genes on the X chromosome were excluded from further analyses. Finally, after the exclusion of 2 genes (ABCD1 and ACSL4) on the X chromosome (Table S1), 88 genes located on the autosomes remained as candidate genes. SNPs within these 88 genes including 2 kb upstream and downstream were extracted from the MDACC GWAS dataset. The quality control criteria for genotyped and imputed SNPs were: (I) a minor allelic frequency (MAF) ≥0.05; (II) a genotyping success rate ≥95%; and (III) Hardy-Weinberg equilibrium (HWE) P≥1×10−5.
Statistical analysis
CM-specific survival (CMSS) was defined as the time from diagnosis with CM to CM-related death or the date of last follow-up. Multivariate Cox proportional hazards regression analysis was performed to assess associations between SNPs and CMSS with an additive model using the GenABEL package of R software. The Bayesian false-discovery probability (BFDP) approach was used, instead of the false discovery rate, for multiple testing correction with a prior probability setting of 0.1 and an upper detected hazards ratio of 3 (24). Only SNPs with a BFDP value <0.8 in both the discovery and replication datasets were chosen for further independent analysis with adjustment for clinical covariates and previously published 40 SNPs from the same MDACC GWAS dataset.
A meta-analysis was further performed using the results of the SNPs from both the discovery and replication datasets using PLINK 1.90. Cochran’s Q statistic and the I2 index were used to assess inter-study heterogeneity. A Cochran’s Q test P value >0.1 and I2<50% indicated no substantial heterogeneity between 2 studies; in such cases, a fixed-effects model was employed. Otherwise, a random-effects model was used. Potential functions of the identified SNPs and those with high linkage disequilibrium (LD) (r2≥0.8) were predicted by RegulomeDB (http://www.regulomedb.org/) and HaploReg4.1 (25). Additionally, Haploview v4.2 (26) was applied to construct a Manhattan plot, and LocusZoom (27) was used to produce regional association plots. A Kaplan-Meier curve was used to estimate survival function, and log-rank tests were performed to compare the effects of different genotypes on CMSS. To estimate the prediction accuracy of the model with demographic variables and the identified SNPs, time-dependent receiver operating characteristic (ROC) analysis was carried out with the “timeROC” package in R (28). Other statistical analyses were performed using SAS software version 9.4 (SAS Institute, Cary, NC, USA), if not specified otherwise.
Results
Basic characteristics of the study populations
The basic characteristics of the 858 CM patients from the MDACC dataset and the 409 CM patients from the NHS/HPFS dataset have been described elsewhere (6). Briefly, the MDACC CM patients were enrolled in a hospital-based case-control study at a tertiary care center; they tended to be younger with more late-stage disease than patients undergoing follow-up in the NHS/HPFS cohort studies. In the MDACC study, patient age at diagnosis ranged between 17 and 94 years, with a mean age of 52.4±14.4 years, and 57.8% (496/858) of the patients were males. The median follow-up time was 81.1 months, and at the last follow-up, 133 patients had died of various causes, including 95 (11.1%) who had died from CM. In the NHS/HPFS studies, patient age at diagnosis ranged between 34 and 87 years, with a mean age of 61.1±10.8 years, and 33.7% (138/409) of the patients were males. The median follow-up time was 179.0 months, which was considerably longer than that of the MDACC study. However, the mortality rate of CM in the NHS/HPFS studies was 11.7% (48/409), which was similar to that in the MDACC patients.
Multivariate analyses of the associations between SNPs in the peroxisome pathway genes and CMSS
As summarized in the study flowchart shown in Figure 1, we extracted 1,215 genotyped and 7,182 imputed SNPs in the peroxisome pathway genes from the MDACC dataset. Associations between these SNPs and CMSS were evaluated through multivariate Cox proportional hazards regression analysis with adjustment for age, sex, regional/distant metastasis, Breslow thickness, ulceration, and the mitotic rate. In the single-locus analysis of these 8,397 SNPs, 332 were found to be significantly associated with CMSS at P<0.05 in an additive genetic model, and after multiple testing correction, 227 SNPs, with BFDP <0.8, remained statistically noteworthy; these 227 SNPs were then replicated in the NHS/HPFS dataset (Figure S1A). After multivariate Cox regression analysis with adjustment for age and sex in the NHS/HPFS dataset (Figure S1B and Table S2), 27 SNPs in 4 genes, with P<0.05 and BFDP <0.8, remained significantly associated with CMSS. Among the 27 SNPs, there were 16 in TMEM135 (transmembrane protein 135), 7 in ACOX2 (acyl-CoA oxidase 2), 3 in PEX5 (peroxisomal biogenesis factor 5), and 1 in RAB8B (member RAS oncogene family). In the subsequent meta-analysis, these 27 SNPs remained significantly associated with CMSS, with no heterogeneity observed across the 2 datasets (Table 1).
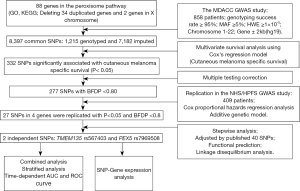
Full table
Genetic variants in the peroxisome pathway genes as independent predictors of survival
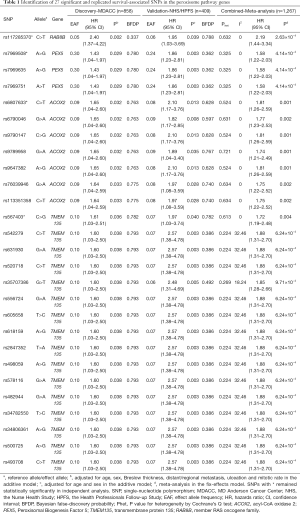
To further identify the independent predictors for CMSS survival, initial stepwise multivariate Cox regression analyses were performed to assess the effects of the 27 replicated SNPs on the survival of CM patients. These analyses were performed in the MDACC dataset only, because the NHS/HPFS dataset did not have the same detailed genotyping data and clinical covariates. Four SNPs in 4 genes remained statistically significantly associated with CMSS in the presence of covariates (i.e., age, sex, Breslow tumor thickness, regional/distant metastasis, tumor cell mitotic rate, and ulceration of tumors). These SNPs were ACOX2 rs6807633 (imputed), TMEM135 rs567403 (imputed), PEX5 rs7969508 (genotyped), and RAB8B rs117285370 (imputed). Finally, following adjustment for the 40 additional previously published survival-associated SNPs with the same MDACC GWAS dataset, we found that 2 SNPs (TMEM135 rs567403 C>G and PEX5 rs7969508 A>G) remained significant (P=0.003 and 0.031, respectively) for further analysis (Table 2).

Full table
As shown in Table 3, in the MDACC dataset, the effects of the TMEM135 rs567403 G and PEX5 rs7969508 G alleles on the survival of CM patients were statistically significant (trend test: P=0.036 and 0.030, respectively), and similar results were observed in the NHS/HPFS studies (trend test: P=0.040 and 0.004, respectively) as well as in their combined dataset (trend test: P=0.023 and 0.001, respectively). We then depicted these associations by using Kaplan-Meier survival curves (Figure 2A,B,C,D,E,F). Regional association plots were also generated to display the LD between the identified SNPs and those in TMEM135 and PEX5, including the 50 kb regions flanking these 2 genes (Figure S2A,B).
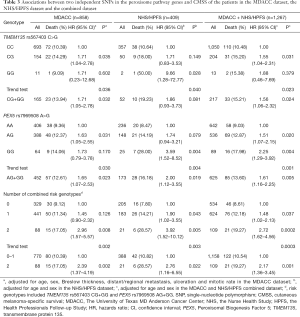
Full table
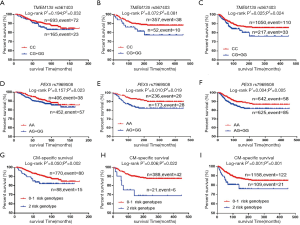
Combined genotype analyses of the 2 independent SNPs
To assess the joint effect of the 2 independent SNPs on CMSS, we further combined the risk genotypes of TMEM135 rs567403 CG+GG and PEX5 rs7969508 AG+GG into a genetic score as the number of risk genotypes (NRG), and categorized all the patients into 3 groups with 0, 1, and 2 risk genotypes. The trend test showed that a higher NRG was associated with a progressively worse survival in the MDACC dataset in a dose-response manner (P=0.002), as well as in the NHS/HPFS dataset (P=0.003) and the combined dataset (P=0.0003), with adjustments for the available covariables (Table 3). Next, we further dichotomized the NRG score into 2 groups: low-risk (0-1 risk genotype) and high-risk (2 risk genotypes). We found that patients in the high-risk group had a higher CM-death risk in the MDACC dataset (HR =2.39, 95% CI: 1.37–4.19, P=0.002) and the NHS/HPFS dataset (HR =1.97, 95% CI: 1.16–6.55, P=0.022), as well as in the combined dataset (HR =2.17, 95% CI: 1.36–3.45, P=0.001), compared to those in the low-risk group (Table 3). These associations were also depicted by Kaplan-Meier curves (Figure 2G,H,I).
Stratified analyses for the effect of combined risk genotypes on CMSS
In the previous analysis we found that patients in the high NRG score group showed a substantially greater risk of CM-related death in the presence of the available clinical variables. The risk was more evident in the subgroup aged ≤50, male patients, patients with ulceration or late-stage disease, and those with a tumor cell mitotic rate >1/mm2 in the MDACC dataset, as well as in the subgroup aged >50 and female patients in the NHS/HPFS dataset. However, no interactions were found (Table S3).
ROC curve and area under the curve (AUC) for prediction of CMSS
We further assessed the predictive effect of the 2 independent SNPs on CMSS using the time-dependent AUC of the ROC curve. The ROC of the 5-year CMSS showed that the inclusion of the 2 identified SNPs only improved the predictive performance of the models including clinical variables without and with risk genotypes by 1.6% in the MDACC dataset (AUC =65.88% vs. 67.48%, P=0.336) (Figure S3A,B). However, we observed that the model with age, sex, and genetic variants had a better performance than those only including demographic variables in the NHS/HPFS dataset (54.05% vs. 70.88%, P=0.003) (Figure S3C,D) and in the combined dataset (AUC =63.6% vs. 67.61%, P=0.073) (Figure S3E,F).
Functional predictions of the 2 independent SNPs
Functional prediction by RegulomeDB showed that TMEM135 rs567403 C>G and PEX5 rs7969508 A>G had RegulomeDB scores of 5 and 1f, respectively, and also indicated that these SNPs might be located at DNase I regulating sites or transcription factor binding sites. We further searched for SNPs with high LD (r2≥0.8) with these 2 independent SNPs and made functional predictions by using HaploReg. We found that TMEM135 rs567403 C>G might disrupt or change the motifs of Dobox4 and Irf, whereas PEX5 rs7969508 A>G, located in DNase I hypersensitive sites, may be a marker of promoter histone and enhancer histone in embryonic stem cell-derived tissue (ESDR), and may have a high linear correlation with mRNA expression of its corresponding gene PEX5 (Table S4). We further assessed the potential functions of these 2 independent SNPs by using data from the ENCODE Project. TMEM135 rs567403 C>G has no significant finding (Figure S4A), while PEX5 rs7969508 A>G is located in the H3K4Me1 region, which may have potential enhancer activity (Figure S4B).
Two independent SNPs and mRNA expression levels of the associated genes
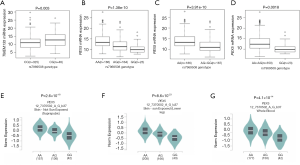
To provide molecular support for the observed survival associations with, and predictions by, the genotypes, we further performed expression quantitative trait locus (eQTL) analysis to explore correlations between genotypes of the 2 independent SNPs and their associated mRNA expression levels in the publicly available RNA sequencing data of lymphoblastoid cell lines generated from 373 European descendants in the 1000 Genomes Project. The eQTL analysis revealed that rs567403 C>G was significantly correlated with elevated mRNA expression levels of TMEM135 in an additive model (P=0.003, Figure 3A), while rs7969508 A>G demonstrated a significant correlation with decreased mRNA expression levels of PEX5 in all genetic models (P=1.38×10–10, P=3.91×10–10, and P=0.0018 for the additive, dominant, and recessive models, respectively; Figure 3B,C,D). We also performed an eQTL analysis using data from the Genotype-Tissue Expression (GTEx) Project V7.p2 (http://www.gtexportal.org/home). The results showed that rs7969508 A>G was correlated with significantly decreased PEX5 mRNA expression in normal tissues from unexposed suprapubic skin (P=2.6×10–18), sun-exposed lower leg skin (P=8.6×10–23), and whole blood cells (P=4.1×10–14) (Figure 3E,F,G). These results were consistent with the findings of the 1000 Genomes Project. However, there were no significant correlations between rs567403 genotypes and mRNA expression levels of TMEM135 in normal skin tissues from unexposed suprapubic skin (P=0.260), sun-exposed lower leg skin (P=0.480), or whole blood cells (P=0.730) (Figure S5) from the GTEx project.
Discussion
In the present study, we assessed the associations between SNPs in the peroxisome pathway-related genes and CM survival. We found that 2 independent and potentially functional SNPs (TMEM135 rs567403 C>G and PEX5 rs7969508 A>G) were independently or jointly associated with survival in CM patients. Patients with a higher NRG score of these 2 genetic variants had a progressively worse survival. Importantly, the effect was consistent across most subgroups. Moreover, the TMEM135 rs567403 risk G allele was found to be significantly correlated with increased mRNA expression levels of TMEM135, whereas the PEX5 rs7969508 risk G allele was found to be significantly correlated with decreased mRNA expression levels of PEX5. These findings suggest that SNPs in peroxisomal pathway genes may play biological roles in the prognosis of CM, suggesting some biological plausibility of the survival-associated SNPs.
Peroxisome homeostasis is achieved by mediating the responses of peroxisome biogenesis and degradation to environmental conditions. By maintaining the integrity and number of organelles under different environmental stresses, pexophagy, as the process of selective autophagy, is essential for maintaining the stability of cellular homeostasis, which is one of the mechanisms underlying peroxisome degradation (29). Peroxisomes are important sites of ROS production and degradation, and thus, play crucial roles in the development and progression of cancer (30). High levels of intracellular ROS can trigger cancer development by promoting pro-oncogenic mutations, and excessive ROS may contribute to progression of cancer cells (31). These may be the mechanisms through which peroxisomes affect cancer development and prognosis. However, no published studies have investigated the molecular mechanisms of the peroxisomes involved in CM survival.
TMEM135, located on chromosome 11q14.2, encodes transmembrane protein 135, which is one of peroxisomal proteins belonging to the Tim17 protein family. To date, the role of TMEM135 in the biological processes of energy expenditure and the related fatty acid metabolism have remained unclear (32,33). Few studies have investigated the roles of TMEM135 in cancer. One study that used sequencing analysis of independent cohorts for breast cancer found that TMEM135 was a potential novel gene (34), and another study reported that rs11235127 near TMEM135 had a statistically significant association with breast cancer risk (35). In prostate cancer patients, a novel fusion transcript in TMEM135-CCDC67 was found to be associated with prostate cancer metastasis, recurrence, and prostate cancer-specific death after operation (36). A genome-wide linkage analysis of Spanish melanoma-prone families found a locus at 11q14.1-q14.3 with significant linkage evidence from multiple pedigrees, and the subregion contained TMEM135 (37). In the present study, we found that the rs567403 G allele might up-regulate the expression of TMEM135, suggesting that TMEM135 may play an oncogenic role in CM biology. However, additional experimental investigations are required to determine how TMEM135 rs567403 C>G influences CMSS.
PEX5, located on chromosome 12p13.31, encodes the type-1 peroxisomal targeting signal (PTS1) receptor necessary for peroxisome biogenesis (38). Few studies have investigated the role of PEX5 in cancer. One study suggested that under starvation conditions, PEX5 depletion prevented the activation of autophagy (39), an important process in intracellular degradation that results in programmed cell death or the promotion of cell survival (40). Therefore, it is likely that the expression levels of PEX5 may alter the processes of autophagy or cell death, which then affects the development and prognosis of cancer. Studies have also reported that cell proliferation was inhibited after PEX5 depletion in human hepatocellular carcinoma cells (16), and PEX5 expression was significantly increased in colon carcinoma tissue compared with normal colon mucosa from 5 patients, as determined by northern blotting analysis (41). These findings suggest that PEX5 may play an oncogenic role. However, in the present study we found that the PEX5 rs7969508 G allele down-regulated the mRNA expression of PEX5, resulting in poor survival in CM patients. This suggests that PEX5 may play a suppressor role in CM biology, which appears to disagree with reports about this gene in the literature, although we did not have additional experimental data to explain this finding.
The limitations of the present study are mainly related to the 2 GWAS datasets that were available to us. The sample sizes of the 2 GWAS datasets were insufficient, and some valuable clinical information, such as treatment and response rate, were not available for adjustment in the analysis; thus, the associations between these SNPs and CM survival should be further investigated in larger studies. Furthermore, the prognosis prediction model was built in non-Hispanic white populations of the United States, which may not be generalizable to other populations. Finally, additional functional experiments for these 2 independent SNPs should be performed to identify the molecular mechanisms underlying the observed survival association in CM patients.
Conclusions
In summary, we analyzed SNPs in the peroxisome pathway genes for their associations with CM survival using data from 2 published GWAS datasets, both of which had strict quality-control standards. The 2 novel SNPs we identified were predicted to have potential biological functions. Once our findings are replicated by other investigators, these genetic variants may serve as new biomarkers for predicting survival in CM patients.
Acknowledgments
The authors would like to thank Bingrong Zhou and Lingling Zhao for their technical assistance, and all participants and staff members of the Nurses Health Study and the Health Professionals Follow-Up Study for their valuable contributions, as well as the following state cancer registries for their support: AL, AZ, AR, CA, CO, CT, DE, FL, GA, ID, IL, IN, IA, KY, LA, ME, MD, MA, MI, NE, NH, NJ, NY, NC, ND, OH, OK, OR, PA, RI, SC, TN, TX, VA, WA, and WY. The authors also thank the Channing Division of Network Medicine, Department of Medicine, Brigham and Women’s Hospital as the home of the NHS. The authors assume full responsibility for the analysis and interpretation of these data. We also thank the Johns Hopkins University Center for Inherited Disease Research for conducting high-throughput genotyping for our study. We also thank all of the investigators and funding agencies that enabled the deposition of the data in dbGaP used here (dbGaP Study Accession: phs000187.v1.p1).
Funding: This work was partly supported by NIH/NCI R01CA100264, 2P50CA093459, R01CA133996, R01CA49449, P01CA87969, UM1CA186107, UM1CA167552, The University of Texas MD Anderson Cancer Center Various Donors Melanoma and Skin Cancers Priority Program Fund, the Miriam and Jim Mulva Research Fund, the McCarthy Skin Cancer Research Fund, and the Marit Peterson Fund for Melanoma Research.
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-2117
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-2117
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-2117). Dr. Wang reports grants from The First hospital of Jilin University, outside the submitted work. Dr. Wei reports grants from Duke Cancer Institute, outside the submitted work. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Informed consent was taken from all the patients, and the access to the datasets was approved by the Institutional Review Boards of the MDACC (No. LAB03-0048), Brigham and Women’s Hospital (No. 2007-P-000616), and Harvard T.H. Chan School of Public Health (No. 2007-P-000616) and conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Rogers HW, Weinstock MA, Feldman SR, et al. Incidence Estimate of Nonmelanoma Skin Cancer (Keratinocyte Carcinomas) in the U.S. Population, 2012. JAMA Dermatol 2015;151:1081-6. [Crossref] [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69:7-34. [Crossref] [PubMed]
- Abbas O, Miller DD, Bhawan J. Cutaneous malignant melanoma: update on diagnostic and prognostic biomarkers. Am J Dermatopathol 2014;36:363-79. [Crossref] [PubMed]
- Berwick M, Buller DB, Cust A, et al. Melanoma Epidemiology and Prevention. Cancer Treat Res 2016;167:17-49. [Crossref] [PubMed]
- Pejkova S, Dzokic G, Tudzarova-Gjorgova S, et al. Molecular Biology and Genetic Mechanisms in the Progression of the Malignant Skin Melanoma. Pril (Makedon Akad Nauk Umet Odd Med Nauki) 2016;37:89-97. [Crossref] [PubMed]
- Dai W, Liu H, Xu X, et al. Genetic variants in ELOVL2 and HSD17B12 predict melanoma-specific survival. Int J Cancer 2019;145:2619-28. [Crossref] [PubMed]
- Sud A, Kinnersley B, Houlston RS. Genome-wide association studies of cancer: current insights and future per-spectives. Nat Rev Cancer 2017;17:692-704. [Crossref] [PubMed]
- Lodhi IJ, Semenkovich CF. Peroxisomes: a nexus for lipid metabolism and cellular signaling. Cell Metab 2014;19:380-92. [Crossref] [PubMed]
- Van Veldhoven PP. Biochemistry and genetics of inherited disorders of peroxisomal fatty acid metabolism. J Lipid Res 2010;51:2863-95. [Crossref] [PubMed]
- Odendall C, Dixit E, Stavru F, et al. Diverse intracellular pathogens activate type III interferon expression from peroxisomes. Nat Immunol 2014;15:717-26. [Crossref] [PubMed]
- Dixit E, Boulant S, Zhang Y, et al. Peroxisomes are signaling platforms for antiviral innate immunity. Cell 2010;141:668-81. [Crossref] [PubMed]
- Nordgren M, Fransen M. Peroxisomal metabolism and oxidative stress. Biochimie 2014;98:56-62. [Crossref] [PubMed]
- Dorninger F, Wiesinger C, Braverman NE, et al. Ether lipid deficiency does not cause neutropenia or leukopenia in mice and men. Cell Metab 2015;21:650-1. [Crossref] [PubMed]
- Puri P, Wiest MM, Cheung O, et al. The plasma lipidomic signature of nonalcoholic steatohepatitis. Hepatology 2009;50:1827-38. [Crossref] [PubMed]
- Cipolla CM, Lodhi IJ. Peroxisomal Dysfunction in Age-Related Diseases. Trends Endocrinol Metab 2017;28:297-308. [Crossref] [PubMed]
- Cai M, Sun X, Wang W, et al. Disruption of peroxisome function leads to metabolic stress, mTOR inhibition, and lethality in liver cancer cells. Cancer Lett 2018;421:82-93. [Crossref] [PubMed]
- Shukla N, Adhya AK, Rath J. Expression of Alpha - Methylacyl - Coenzyme A Racemase (AMACR) in Colorectal Neoplasia. J Clin Diagn Res 2017;11:EC35-8. [Crossref] [PubMed]
- Zhou M, Chinnaiyan AM, Kleer CG, et al. Alpha-Methylacyl-CoA racemase: a novel tumor marker over-expressed in several human cancers and their precursor lesions. Am J Surg Pathol 2002;26:926-31. [Crossref] [PubMed]
- Honma I, Torigoe T, Hirohashi Y, et al. Aberrant expression and potency as a cancer immunotherapy target of alpha-methylacyl-coenzyme A racemase in prostate cancer. J Transl Med 2009;7:103. [Crossref] [PubMed]
- Amos CI, Wang LE, Lee JE, et al. Genome-wide association study identifies novel loci predisposing to cutaneous melanoma. Hum Mol Genet 2011;20:5012-23. [Crossref] [PubMed]
- Song F, Qureshi AA, Zhang J, et al. Exonuclease 1 (EXO1) gene variation and melanoma risk. DNA Repair (Amst) 2012;11:304-9. [Crossref] [PubMed]
- Li Y, Willer CJ, Ding J, et al. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol 2010;34:816-34. [Crossref] [PubMed]
- Lappalainen T, Sammeth M, Friedlander MR, et al. Transcriptome and genome sequencing uncovers functional variation in humans. Nature 2013;501:506-11. [Crossref] [PubMed]
- Wakefield J. A Bayesian measure of the probability of false discovery in genetic epidemiology studies. Am J Hum Genet 2007;81:208-27. [Crossref] [PubMed]
- Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res 2012;40:D930-4. [Crossref] [PubMed]
- Barrett JC, Fry B, Maller J, et al. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 2005;21:263-5. [Crossref] [PubMed]
- Pruim RJ, Welch RP, Sanna S, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics 2010;26:2336-7. [Crossref] [PubMed]
- Chambless LE, Diao G. Estimation of time-dependent area under the ROC curve for long-term risk prediction. Stat Med 2006;25:3474-86. [Crossref] [PubMed]
- Walker CL, Pomatto LCD, Tripathi DN, et al. Redox Regulation of Homeostasis and Proteostasis in Peroxisomes. Physiol Rev 2018;98:89-115. [Crossref] [PubMed]
- Weinberg F, Chandel NS. Reactive oxygen species-dependent signaling regulates cancer. Cell Mol Life Sci 2009;66:3663-73. [Crossref] [PubMed]
- Tubbs A, Nussenzweig A. Endogenous DNA Damage as a Source of Genomic Instability in Cancer. Cell 2017;168:644-56. [Crossref] [PubMed]
- Žárský V, Dolezal P. Evolution of the Tim17 protein family. Biol Direct 2016;11:54. [Crossref] [PubMed]
- Exil VJ, Silva Avila D, Benedetto A, et al. Stressed-induced TMEM135 protein is part of a conserved genetic network involved in fat storage and longevity regulation in Caenorhabditis elegans. PLoS One 2010;5:e14228. [Crossref] [PubMed]
- Natrajan R, Mackay A, Lambros MB, et al. A whole-genome massively parallel sequencing analysis of BRCA1 mutant oestrogen receptor-negative and -positive breast cancers. J Pathol 2012;227:29-41. [Crossref] [PubMed]
- Teraoka SN, Bernstein JL, Reiner AS, et al. Single nucleotide polymorphisms associated with risk for contralateral breast cancer in the Women's Environment, Cancer, and Radiation Epidemiology (WECARE) Study. Breast Cancer Res 2011;13:R114. [Crossref] [PubMed]
- Yu YP, Ding Y, Chen Z, et al. Novel fusion transcripts associate with progressive prostate cancer. Am J Pathol 2014;184:2840-9. [Crossref] [PubMed]
- Potrony M, Puig-Butille JA, Farnham JM, et al. Genome-wide linkage analysis in Spanish melanoma-prone families identifies a new familial melanoma susceptibility locus at 11q. Eur J Hum Genet 2018;26:1188-93. [Crossref] [PubMed]
- Dodt G, Gould SJ. Multiple PEX genes are required for proper subcellular distribution and stability of Pex5p, the PTS1 receptor: evidence that PTS1 protein import is mediated by a cycling receptor. J Cell Biol 1996;135:1763-74. [Crossref] [PubMed]
- Eun SY, Lee JN, Nam IK, et al. PEX5 regulates autophagy via the mTORC1-TFEB axis during starvation. Exp Mol Med 2018;50:1-12. [Crossref] [PubMed]
- Singletary K, Milner J. Diet, autophagy, and cancer: a review. Cancer Epidemiol Biomarkers Prev 2008;17:1596-610. [Crossref] [PubMed]
- Lauer C, Volkl A, Riedl S, et al. Impairment of peroxisomal biogenesis in human colon carcinoma. Carcinogenesis 1999;20:985-9. [Crossref] [PubMed]


