
SOX2 amplification and chromosome 3 gain significantly impact prognosis in esophageal squamous cell carcinoma
Introduction
Esophageal cancer (EC) is the leading cause of cancer mortality in China, where it was responsible for over 193,000 deaths in 2014 (1). Approximately 90% of the new EC cases diagnosed each year are esophageal squamous cell carcinoma (ESCC). Despite the optimization of surgery, radiotherapy, and cytotoxic chemotherapy, the survival of patients with advanced ESCC remains poor, with an age-standardized 5-year relative survival rate of only 30% (2). Recent advances in genetic profiling have led to more refined molecular classifications of specific tumor entities (3-5). To determine the optimal therapeutic approach for a given patient with ESCC, prognostic biomarkers that would predict outcomes more accurately than existing ones are urgently needed. Gene copy number alterations are frequently found in human epithelial malignancies and often play an essential role in tumor development (5-7). The detection of these alterations may allow the development of novel diagnostic, prognostic, and predictive biomarkers, and promote the use of effective therapeutic regimens.
Sex determining region Y-box 2 (SOX2) is a highly conserved, single exon gene that is located at 3q26.33 and encodes for a 317-amino acid protein. SOX2 contains a high mobility group (HMG) DNA-binding domain and belongs to the SOX family of transcription factors, which are essential for preserving the pluripotency of embryonic stem cells and self-renewal capacity of tissue-specific adult stem cells (8). SOX2 has been found to be recurrently amplified in squamous cell carcinomas (SCC) of different organ sites, such as the head and neck (9), lung (10) and anus (11), and it is considered as a lineage-survival oncogene. In lung SCC (LSCC), SOX2 amplification is frequently observed and associated with favorable prognosis (10). In head and neck SCC (HNSCC), SOX2 activation is associated with improved prognosis (9). In sinonasal SCC (SSCC), SOX2 amplification is an independent indicator of disease recurrence (12).
Recently, with the availability of single-nucleotide polymorphism array and array comparative genomic hybridization, SOX2 was found to be amplified in 10% ESCC cases (13), which was a little less than 15–18% in other reports on ESCC (14,15). Few studies have explored the association between SOX2 amplification and patients’ prognosis. Some studies, wherein the clinical significance of SOX2 expression in ESCC was evaluated, had controversial conclusions. A study from Norway showed that SOX2 expression correlated with higher histological grade and poorer clinical survival (16). A later Japanese study showed the negative expression of SOX2 appeared to be an independently poor prognostic factor (17). Therefore, further studies are required to validate the prognostic role of SOX2 in patients with ESCC.
In the present study, we aimed to investigate the prevalence of SOX2 gene copy number changes in a large cohort of surgically resected ESCC patients using fluorescence in situ hybridization (FISH) analysis, and determine whether copy number alterations in selected genes affect patient outcomes. Furthermore, we examined whether gene amplification is associated with increased SOX2 protein expression in ESCC.
We present the following article in accordance with the REMARK reporting checklist (available at http://dx.doi.org/10.21037/atm-20-1290).
Methods
Patients and samples
This retrospective study consisted of 450 ESCC patients who had undergone esophagectomy without neoadjuvant treatment at the Department of Thoracic Surgery, Zhongshan Hospital, Fudan University, China, between 2007 and 2010. The patients who exhibited disease progression within three months after surgery were excluded from further analysis. Clinical information including patient prognoses was obtained from stored medical and imaging records. Patients were followed up with a clinical examination every 3 months for the first year, every 6 months for the second year, and every 6–12 months thereafter. Histology slides were reviewed in terms of differentiation, depth of invasion, vessel and nerve invasion, and lymph node metastasis. Tumor stages were defined based on the 8th edition of the American Joint Committee on Cancer/Union for International Cancer Control (AJCC/UICC) pTNM-staging system for ESCC.
Informed consent forms were obtained from all patients, and the study was conducted in accordance with the approved ethical standards of Zhongshan Hospital (B2016-135), which conforms to the provisions of the Helsinki Declaration (as revised in 2013).
Tissue microarray (TMA) construction
TMA construction was performed as previously described (18). Hematoxylin and eosin (HE) sections were evaluated and confirmed by two pathologists (DJ and YH), who validated diagnosis and marked normal tissue and carcinoma tissue as target areas for TMA construction. The regions of interest (2 mm wide and 6 mm long) were obtained from the corresponding donor blocks. Two to three representative cores of viable tissue from each tumor were included. Additional control cores of normal esophageal tissue were incorporated. Then donor tissues were then manually planted into the recipient block one by one according to the corresponding locations indicated by letters and numbers. The planting surface was aggregated on the aggregation instrument. Subsequently, the recipient block with a transparent box was incubated at 4 °C for 10 min until the paraffin could be easily separated from the transparent box. TMA recipient blocks were collected and sectioned on a routine microtome machine for further IHC and FISH staining.
Fluorescence in-situ hybridization assay (FISH)
To assess the SOX2 amplification status at the chromosomal level in TMA slides, we applied the same 2-color FISH assay as previously described by Wilbertz et al. (10). Briefly, a SOX2 target probe (red fluorescent signal) spanning the locus 3q26.33 and a green centromeric probe on chromosome 3 (Empire Genomics, New York, USA) were selected for hybridization. Only the nuclei that displayed green reference signals were included for determination of the SOX2 copy number status. All TMA slides were analyzed by two independent evaluators (J.H. and Q.S.) under an oil immersion objective using a fluorescence microscope (BX43; Olympus, Tokyo, Japan) equipped with a microscope digital camera (DP73; Olympus). In each case, we assessed at least 50 tumor cell nuclei.
Two red and two green signals in a cell were observed in a wild-type nucleus. A sample was considered amplified if SOX2 amplification was observed in at least 30% of the nuclei. Amplification status was defined according to the criteria established by Maier et al. (19). SOX2 amplification was defined as four red target signals exceeding the number of green signals. Three or more green reference signals were defined as chromosome 3 gain.
Immunohistochemistry (IHC)
The IHC was performed on a Bond Max autostainer (Leica Microsystems, Wetzlar, Germany) according to the manufacturers’ instructions using SOX2 rabbit monoclonal antibody (clone SP76, dilution 1:300, Genetech, China). Normal IgG from the same species of primary antibody diluted to match the concentration of the primary antibody was used as the negative control.
For SOX2, only nuclear staining was considered specific. A Histo-score (H-score; range, 0–300) was calculated by multiplying the intensity score (0= negative; 1= weak; 2= intermediate; 3= strong) and the fraction score (percentage of positive tumor cells; range, 0–100) (20). All stained slides were scored independently by two investigators (HW and YH), who were blinded for clinical outcome. In cases of disagreement between the two investigators, slides were simultaneously reviewed by both investigators until a consensus was reached.
Statistical analysis
The results of FISH and IHC were analyzed in subgroups and compared with clinical parameters, histologic subtypes, and grade using χ2 or Fisher exact test, whichever was appropriate. Disease-free survival (DFS) was defined as the time between the date of surgery and the date of recurrence, death, or latest follow-up. Overall survival (OS) was defined as the interval between the date of surgery and the documented date of ESCC-associated death or latest follow-up. Survival curves were constructed according to the Kaplan-Meier method and compared using the log-rank test. Associations between clinicopathological factors and DFS/OS were estimated according to odds ratios (ORs) and 95% CIs. The factors with a P value <0.05 for either OS or DFS that met the proportional hazards criteria in the univariate analysis, were included in stepwise multivariate Cox regression.
For all tests, statistical significance was defined as P<0.05. Statistical analyses were performed using IBM SPSS (SPSS Inc., Chicago, IL), version 20.0.
Results
Patient characteristics
The clinical and pathological characteristics of the 450 ESCC patients are summarized in Table 1. The mean age was 61.4 years (range, 34-83 years). Among the patients, 364 (80.9%) were male and 171 (38.0%) were smokers (ever or now smoked). The mean tumor size was 3.4 cm (range, 0.3–10 cm). In terms of anatomic site, 4.7% of tumors were located in the upper esophagus, whereas 46.0% and 44.9% of tumors were located in the middle and lower esophagus, respectively. Among these tumors, 18 (4.0%) were histologically graded as well differentiated (Grade I), 267 (59.3%) were moderately differentiated (Grade II), and 165 (36.7%) were poorly differentiated (Grade III). Vessel and nerve invasions were identified in 97 (21.6%) and 146 (32.4%) tumors, respectively. Lymph node metastasis was observed in 43.3% (195/450) of patients. Invasive depths were also evaluated, 13 (2.9%) cases were confined to the mucosal layer, 34 (7.6%) in submucosal layer, 100 (22.2%) in muscular layer, and 303 (67.3%) beyond muscular layer. Pathological stages of all the tumors were evaluated according to the 8th edition pTNM classification; stage IB was identified in 9.3% of tumors, stage IIA in 44.2%, stage IIB in 4.7%, stage IIIA in 5.3%, and stage IIIB in 36.4%.

Full table
Association between gene copy number variation and clinicopathological characteristics
Among 450 patients, 20 (4.4%) met the inclusion criteria (≥4 red target signals in no less than 30% tumor cells and less than three green signals), and were classified as cases with SOX2 amplification. Fifty-eight (12.9%) cases were defined as chromosome 3 gain with three or more green signals, among which 28 (6.2%) cases also had ≥4 red SOX2 signals in no less than 30% tumor cells. Low polysomy or disomy (82.7%) was observed in other specimens (Figure 1).
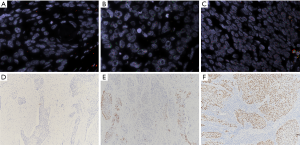
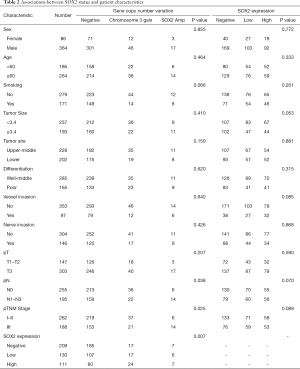
Table 2 reveals that SOX2 amplification and chromosome 3 gain were not significantly associated with sex (P=0.855), age (P=0.464), smoking (P=0.066), tumor size (P=0.410), tumor site (P=0.150), differentiation (P=0.620), vessel (P=0.640) and nerve invasions (P=0.426), and pT (P=0.207). SOX2 amplification was associated with later clinical stage, but chromosome 3 gain was associated with earlier clinical stage (P=0.025). Similarly, SOX2 amplification was associated with lymph node metastasis, however chromosome 3 gain tended to be associated with no lymph node metastasis (P=0.038).
Full table
SOX2 expression and the clinicopathological characteristics
SOX2 protein expression was highly heterogeneous among the patients, with 53.6% tumor samples having 1–80% positive stained cells, and no staining was detected in 46.4% of the samples. The H-scores of the SOX2 positive tumors ranged from 1 to 160, with a median of 30. Therefore, an H-score higher than 30 was considered indicative of high SOX2 expression (n=111), an H-score of 30 or lower was considered indicative of low expression (n=130) and an H-score of 0 was considered indicative of negative expression (n=209) (Figure 1).
The clinicopathological characteristics of patients grouped according to SOX2 expression are listed in Table 2. SOX2 expression was significantly associated with copy number variation (P=0.007). We found no correlation of SOX2 expression with sex (P=0.772), age (P=0.333), smoking (P=0.261), tumor site (P=0.881), differentiation (P=0.315), vessel or nerve invasion (P=0.085 or 0.868), pT stage (P=0.590), pN stage (P=0.070), and pTNM stage (P=0.089).
Survival analyses
The 5-year DFS and disease-specific OS rates of patients were 46.7% and 47.2%, respectively, with a median follow-up period of 36 months (range, 4–102 months). There were 232 disease progression documented. The mean and median DFS were 58.0 and 40.0 months, respectively. A total of 231 patients (51.3%) died during the follow-up period, and 224 (49.8%) patients died of EC. The mean and median disease-specific OS were 61.4 and 47.0 months, respectively.
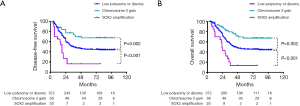
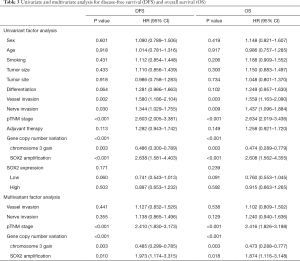
A significantly shorter DFS or OS in the SOX2 amplification group, whereas a significantly longer DFS or OS in the chromosome 3 gain group, than those in the group with low polysomy or disomy were observed (Figure 2). In detail, a significantly poorer prognosis was observed in 20 patients with SOX2 amplification, whose median DFS or OS were 18.0 or 23.0 months, respectively, whereas a significantly better prognosis was observed in 59 patients with chromosome 3 gain, whose median DFS or OS were no-reached, compared with 36.0 or 43.0 months in the group with low polysomy or disomy (P<0.001). The difference in terms of patient 5-year survival between SOX2 amplification and normal groups was deceased 28.8% for DFS and 32.3% for OS, while the difference between chromosome 3 gain and normal groups was increased 22.4% for DFS and 21.6% for OS. SOX2 expression was not associated with DFS or OS (P=0.159 and 0.230). In addition, SOX2 amplification or chromosome 3 gain, vessel invasion, nerve invasion, and pTNM stage were associated with DFS and OS in univariate analysis. With variables that were found to be significant in the univariate analyses (P<0.05), a multivariate analysis using the cox proportional hazard model was performed. Multivariate analysis indicated that chromosome 3 gain was an independently better prognostic factor (DFS, P=0.003, HR 0.486, 95% CI, 0.300–0.789; OS, P=0.003, HR 0.474, 95% CI, 0.289–0.779), while SOX2 amplification was an independently poorer prognostic factor (DFS, P<0.001, HR 2.638, 95% CI, 1.581–4.403; OS, P<0.001, HR 2.608, 95% CI, 1.562–4.355), along with pTNM stage (Table 3).
Full table
Discussion
Identifying the copy number variation of target genes represents an important step toward understanding and potentially preventing the complex changes a cell undergoes during ESCC tumor development, which also facilitates the further outcome stratification and treatment decisions of these patients (5-7). The aim of our study was to elucidate whether SOX2 amplification is a common event in ESCC and to determine whether SOX2 amplification is correlated with prognosis. FISH analysis was performed to detect SOX2 copy number gains, followed by IHC SOX2 protein expression analysis. At present, there are no data available in literature on both SOX2 amplification and SOX2 expression in large scale of ESCC patients.
In our study, SOX2 was amplified in 4.4% ESCC, less than 10–18% in other ESCC reports (13,14,19). These variations in frequency are likely due to (I) the molecular method applied for detection of SOX2 copy number status, (II) the threshold set to distinguish gene amplification, (III) the differences between ESCC cohorts, and (IV) tumor heterogeneity (21). A key finding of this study was the observation that green signals (Chromosome 3 gain) were found in 12.9% ESCC, among which 6.2% tumors with SOX2 copies were ≥4 in more than 30% cells. If 6.2% patients were also regarded as SOX2 amplification, the percentage of 10.6% (6.2%+4.4%) was consistent with other reports in ESCC. However, it is possible that there are multiple targets for chromosome 3 gain, and co-amplification of adjacent oncogenes can have a synergistic effect. Other genes reported to be putative targets in chromosome 3 include PIK3CA (22,23), SKIL (24), TERC (25), DCUN1D1 (26), TP63 (27), and EVI1 (28). For example, PIK3CA, which is located at the 2.6 Mb centromeric side of SOX2 and DCUN1D1, which is located at the 1.2 Mb telomeric side of SOX2, are reported to be molecular biomarkers in many tumors (14,29). Therefore, the division of SOX2 amplification and chromosome 3 gain should be necessary. In clinicopathological analysis, SOX2 amplification was higher in later pTNM stage, however, chromosome 3 gain was higher in earlier pTNM stage (P=0.039).
In our univariate and multivariate analyses, SOX2 amplification was an independently poorer prognostic factor (DFS, P<0.001, HR 2.638, 95% CI, 1.581–4.403; OS, P<0.001, HR 2.608, 95% CI, 1.562–4.355), and chromosome 3 gain was an independently better prognostic factor (DFS, P=0.003, HR 0.486, 95% CI, 0.300–0.789; OS, P=0.003, HR 0.474, 95% CI, 0.289–0.779). This is the first study simultaneously explored the prognostic significance of SOX2 amplification and chromosome 3 gain in a relatively large-scale ESCC cohort study using the FISH method. SOX2 amplification was detected more frequently in SCC than in adenocarcinomas (19). Therefore, SOX2 amplification was believed to be an important mechanism of tumor progression in a considerable subset of SCC (30). In LSCC, SOX2 amplification is associated with indicators of favorable prognosis (10). In HNSCC, SOX2 copy number gain is associated with improved patients’ prognosis (9). In SSCC, patients with SOX2 amplification experienced a significantly higher incidence of recurrence (12). In vulvar SCC, SOX2 amplification was not associated with OS (31). None of the above studies analyzed the prognostic difference between chromosome 3 gain and SOX2 amplification. Unlike other SCCs, gene focus within 3q26-3q33 containing candidate oncogenes, including TP63 (6.5–20%) (13,32) and PIK3CA (7–40.7%) (13,33), were amplified to higher levels in ESCC. We speculated why chromosome 3 gain is associated with a better prognosis might due to chromosome 3 gain includes multiple oncogenes. The prognostic significance of these gene amplifications is not well known in ESCC, and needs to be explored in the future.
SOX2 expression was also evaluated in our ESCC patients with a negative rate of 46.4%, low expression rate of 28.9% and high expression rate of 24.7%. Consistent with our results, reports from the literature demonstrated that SOX2 expression rate was 22.8–93% (11,16,17). SOX2 expression occurred more frequently than SOX2 amplification. SOX2 expression was significantly associated with SOX2 amplification or chromosome 3 gain (P=0.007). Among patients with SOX2 amplification or chromosome 3 gain, high SOX2 expression was higher than low SOX2 expression and negative. However, SOX2 expression was not associated with DFS and OS (P=0.159 and 0.230), which was inconsistent with SOX2 amplification or chromosome 3 gain. This is because SOX2 amplification is only one of the mechanisms capable of up-regulating gene expression. Namely, SOX2 expression can be up-regulated not only by amplification, but also at the transcription and translation levels. To date, SOX2 expression has been documented in other studies about ESCC. In Wang’ s study, high SOX2 expression was significantly associated with higher histological grade and poorer clinical survival (16), however, Maehara et al. demonstrated that the negative expression of SOX2 appeared to be an independently poor prognostic factor (OR 7.05, 95% CI, 1.27–39.0) (17). In terms of the above results, more detailed analyses are needed to elucidate the prognostic significance of SOX2 expression.
To our knowledge, there is limited study correlating SOX2 amplification and chromosome 3 gain with its clinical significance in patients with ESCC. In the present study, SOX2 amplification is an independently poorer prognostic factor, but chromosome 3 gain is an independently favorable prognostic factor. SOX2 expression was significantly associated with copy number variation (P=0.007), but not associated with DFS and OS. In summary, these results suggested that SOX2 amplification and chromosome 3 gain might play important roles in tumor development and could serve as an independent predictor of poorer and better prognosis for ESCC.
Acknowledgments
Funding: This work was supported by National Natural Science Foundation of China (No. 81702372 to DJ), Shanghai Natural Science Foundation of China (No. 18ZR1406800 to YH), Xiamen Science and Technology Project of Fujian Province, China (No. 3502Z20184003 to YH), Shanghai Municipal Commission of Science and Technology (No. 19441904000 to YH), Shanghai Municipal Key Clinical Specialty (No. shslczdzk01302 to YH), and Shanghai Science and Technology Development Fund (No. 19MC1911000 to YH).
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-1290
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-1290
Peer Review File: Available at http://dx.doi.org/10.21037/atm-20-1290
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-1290). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All patient provided their informed consent and the study was conducted in accordance with the approved ethical standards of Zhongshan Hospital (B2016-135), which conforms to the provisions of the Helsinki Declaration (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Chen W, Sun K, Zheng R, et al. Cancer incidence and mortality in china, 2014. Chin J Cancer Res 2018;30:1-12. [Crossref] [PubMed]
- Zeng H, Chen W, Zheng R, et al. Changing cancer survival in china during 2003-15: A pooled analysis of 17 population-based cancer registries. Lancet Glob health 2018;6:e555-67. [Crossref] [PubMed]
- Levy SE, Myers RM. Advancements in next-generation sequencing. Annu Rev Genomics Hum Genet 2016;17:95-115. [Crossref] [PubMed]
- Dinan MA, Mi X, Reed SD, et al. Association between use of the 21-gene recurrence score assay and receipt of chemotherapy among medicare beneficiaries with early-stage breast cancer, 2005-2009. JAMA oncol 2015;1:1098-109. [Crossref] [PubMed]
- Liu X, Zhang M, Ying S, et al. Genetic alterations in esophageal tissues from squamous dysplasia to carcinoma. Gastroenterology 2017;153:166-77. [Crossref] [PubMed]
- Integrated genomic characterization of oesophageal carcinoma. Nature 2017;541:169-75. [Crossref] [PubMed]
- Beroukhim R, Mermel CH, Porter D, et al. The landscape of somatic copy-number alteration across human cancers. Nature 2010;463:899-905. [Crossref] [PubMed]
- Feng R, Wen J. Overview of the roles of sox2 in stem cell and development. Biol Chem 2015;396:883-91. [Crossref] [PubMed]
- Chung JH, Jung HR, Jung AR, et al. Sox2 activation predicts prognosis in patients with head and neck squamous cell carcinoma. Sci Rep 2018;8:1677. [Crossref] [PubMed]
- Wilbertz T, Wagner P, Petersen K, et al. Sox2 gene amplification and protein overexpression are associated with better outcome in squamous cell lung cancer. Mod Pathol 2011;24:944-53. [Crossref] [PubMed]
- Long KB, Hornick JL. Sox2 is highly expressed in squamous cell carcinomas of the gastrointestinal tract. Hum Pathol 2009;40:1768-73. [Crossref] [PubMed]
- Schröck A, Goke F, Wagner P, et al. Sex determining region y-box 2 (sox2) amplification is an independent indicator of disease recurrence in sinonasal cancer. PloS One 2013;8:e59201. [Crossref] [PubMed]
- Lin DC, Hao JJ, Nagata Y, et al. Genomic and molecular characterization of esophageal squamous cell carcinoma. Nat Genet 2014;46:467-73. [Crossref] [PubMed]
- Gen Y, Yasui K, Zen Y, et al. Sox2 identified as a target gene for the amplification at 3q26 that is frequently detected in esophageal squamous cell carcinoma. Cancer Genet Cytogenet 2010;202:82-93. [Crossref] [PubMed]
- Wang K, Johnson A, Ali SM, et al. Comprehensive genomic profiling of advanced esophageal squamous cell carcinomas and esophageal adenocarcinomas reveals similarities and differences. Oncologist 2015;20:1132-39. [Crossref] [PubMed]
- Wang Q, He W, Lu C, et al. Oct3/4 and sox2 are significantly associated with an unfavorable clinical outcome in human esophageal squamous cell carcinoma. Anticancer Res 2009;29:1233-41. [PubMed]
- Maehara R, Fujikura K, Takeuchi K, et al. Sox2-silenced squamous cell carcinoma: A highly malignant form of esophageal cancer with sox2 promoter hypermethylation. Mod Pathol 2018;31:83-92. [Crossref] [PubMed]
- Shi Y, He D, Hou Y, et al. An alternative high output tissue microarray technique. Diagn Pathol 2013;8:9. [Crossref] [PubMed]
- Maier S, Wilbertz T, Braun M, et al. Sox2 amplification is a common event in squamous cell carcinomas of different organ sites. Hum Pathol 2011;42:1078-88. [Crossref] [PubMed]
- Prat A, Cheang MC, Martin M, et al. Prognostic significance of progesterone receptor-positive tumor cells within immunohistochemically defined luminal a breast cancer. J Clin Oncol 2013;31:203-9. [Crossref] [PubMed]
- Li Q, Liu F, Zhang Y, et al. Association of sox2 and nestin DNA amplification and protein expression with clinical features and overall survival in non-small cell lung cancer: A systematic review and meta-analysis. Oncotarget 2016;7:34520-31. [Crossref] [PubMed]
- Kim HS, Lee SE, Bae YS, et al. Pik3ca amplification is associated with poor prognosis among patients with curatively resected esophageal squamous cell carcinoma. Oncotarget 2016;7:30691-701. [Crossref] [PubMed]
- Hou J, Jiang D, Zhang J, et al. Frequency, characterization, and prognostic analysis of pik3ca gene mutations in chinese esophageal squamous cell carcinoma. Hum Pathol 2014;45:352-8. [Crossref] [PubMed]
- Akagi I, Miyashita M, Makino H, et al. Snon overexpression is predictive of poor survival in patients with esophageal squamous cell carcinoma. Ann Surg Oncol 2008;15:2965-75. [Crossref] [PubMed]
- Hu Y, Teng X, Wu L, Liu W, et al. The clinicopathological correlations of hterc amplification with esophageal squamous cell precursor lesions. Dig Dis Sci 2019;64:68-75. [Crossref] [PubMed]
- Huang G, Singh B. Coamplification and cooperation: Toward identifying biologically relevant oncogenes. Clin Cancer Res 2013;19:5549-51. [Crossref] [PubMed]
- Geddert H, Kiel S, Heep HJ, et al. The role of p63 and deltanp63 (p40) protein expression and gene amplification in esophageal carcinogenesis. Hum Pathol 2003;34:850-6. [Crossref] [PubMed]
- Jomrich G, Maroske F, Stieger J, et al. Mk2 and etv1 are prognostic factors in esophageal adenocarcinomas. J Cancer 2018;9:460-8. [Crossref] [PubMed]
- McCaughan F, Pole JC, Bankier AT, et al. Progressive 3q amplification consistently targets sox2 in preinvasive squamous lung cancer. Am J Respir Crit Care Med 2010;182:83-91. [Crossref] [PubMed]
- Liu K, Jiang M, Lu Y, et al. Sox2 cooperates with inflammation-mediated Stat3 activation in the malignant transformation of foregut basal progenitor cells. Cell Stem Cell 2013;12:304-15. [Crossref] [PubMed]
- Gut A, Moch H, Choschzick M. Sox2 gene amplification and overexpression is linked to hpv-positive vulvar carcinomas. Int J Gynecol Pathol 2018;37:68-73. [Crossref] [PubMed]
- Tanière P, Martel-Planche G, Saurin JC, et al. Tp53 mutations, amplification of p63 and expression of cell cycle proteins in squamous cell carcinoma of the oesophagus from a low incidence area in western europe. Br J Cancer 2001;85:721-6. [Crossref] [PubMed]
- Song Y, Li L, Ou Y, et al. Identification of genomic alterations in oesophageal squamous cell cancer. Nature 2014;509:91-5. [Crossref] [PubMed]


