Molecular mechanism of Wutou Decoction in the treatment of osteoarthritis: a bioinformatics and molecular docking study
Introduction
Osteoarthritis (OA) is a common chronic disease characterized by a series of degenerative changes of the joint including articular cartilage degradation, reduction of chondrocyte count, and periarticular osteophyte formation (1). Its exact etiology and pathogenesis remain unclear. With the combined effects of ageing and increasing obesity in the global population, along with increasing numbers of joint injuries, this already burdensome syndrome is becoming more prevalent, with worldwide estimates suggesting that 250 million people are currently affected (2). Clinically, the knee is the most common site of OA, followed by the hand and hip. Therefore, taking knee osteoarthritis (KOA) as an example, according to a Chinese epidemiological survey data, the current prevalence rate of symptomatic KOA in China is 8.1%, which means that there are approximately 110 million knee osteoarthritis in China. The number of patients is increasing year by year (3). Modern treatments, including nonpharmacological, pharmacological, and surgical therapies have been widely applied, but are not ideal, with shortcomings such as indefinite efficacy, adverse effects, and high costs being common (4). According to traditional Chinese medicine (TCM), arthritis belongs to the category of bi-syndrome. In China, TCM therapies are a popular remedy for the prevention and treatment of OA, with the concept of care being a holistic approach centered on the principle of “syndrome differentiation”. As safe, convenient, effective, and affordable options, TCM therapies have garnered intense interest in the fields clinical treatment and new drug development (5). The TCM formula, Wutou Decoction (WTD) was first described in Essentials from the Golden Cabinet, authored by Zhang Zhongjing (Zhang Ji; 150-219 CE), an eminent Chinese physician in the Eastern Han dynasty. According to Zhang, “if a person is suffering from pain in the joints that cannot be flexed or extended, Wutou Decoction is the main remedy.” The formula is composed of 5 herbs, namely, Radix Aconiti, Herba Ephedrae, Radix Astragali seu Hedysari, Paeonia lactiflora Pall, and Radix glycyrrhizae, and has a long history of use in the prevention and treatment of OA (6). Its effectiveness has also been confirmed in our clinical practice. However, due to the complexity of the compounds and mechanisms of action of the TCM formula, existing detection techniques, which are often lacking in sensitivity, expensive, and non-standardized, cannot efficiently identify the core active compounds and therefore cannot accurately and reliably reveal the mechanisms of action of the herbal formula.
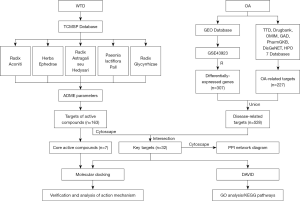
With the rapid advances in bioinformatics, along with computer science, network, and big data technologies, new tools have been extensively applied in TCM research and have borne fruit (7). Among these new techniques, molecular docking is a research method based on the interaction between drug molecules and target protein receptors. It is able to search for the optimal conformation of a ligand binding with a receptor protein and to predict the binding mode and affinity for drug design (8,9). Molecular docking has been widely used in the screening of important and complex active compounds and in efficacy prediction, opening new avenues in the research into TCM drug efficacy. In our study, both bioinformatic and molecular docking methods were innovative applied, that is better than previous pure network pharmacology analysis: data mining techniques with numerous databases [e.g., Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform (TCMSP), Therapeutic Target Database (TTD), Gene Expression Omnibus (GEO), Database for Annotation, Visualization, and Integrated Discovery (DAVID), STRING Database] were used to screen and predict the potential targets and signaling pathways of the active compounds of WTD for the treatment of OA; while molecular docking was used to select and evaluate the core drug compounds in WTD, verify that the compound matches the target. It is explained that the mechanism of TCM formulae from the perspectives of key compounds, targets and signal pathway will shed new light on the modernization of TCM formulae and the research and development of new drugs (Figure 1). We present the following article in accordance with the MDAR reporting checklist (available at https://dx.doi.org/10.21037/apm-21-1691).
Methods
Materials
The databases, software, and related analytical platforms used in this study are detailed in Table 1.

Full table
Collection and screening of chemical compounds of five Chinese herbs in WTD
When Chinese herbs enter the human digestive tract after oral administration, they must undergo four physiological processes including absorption, distribution, metabolism, and excretion (ADME), which represent the typical pharmacokinetic properties (10,11). The ADME parameters of the relevant compounds of five herbal medicines in WTD were searched for in the Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform (TCMSP) (12), and two key qualitative indicators [oral bioavailability (OB) ≥30% and drug likeness (DL) ≥0.18] were used to screen for the active compounds in the formula (13,14).
Prediction of the action targets and construction of the “TCM drugs-Active compounds-Targets” network
A target prediction model in TCMSP was used to predict the action targets of the active compounds in WTD. Cytoscape 3.6.1 (15), a biological network visualization and analysis software, was used to construct a visual network diagram of the active compounds and targets, which revealed the interactions among herbs, compounds, and targets in a more vivid manner. The relevant targets of the active compounds in WTD were obtained through literature review. The network was analyzed using the “NetworkAnalyzer” plug-in in Cytoscape. Since the active compounds with a higher degree may play a more critical role in the treatment of OA, the core compounds with a degree 2 times higher than the median in the network were selected for molecular docking verification.
Screening of OA disease targets
Using “Osteoarthritis” as the keyword, we screened Therapeutic Target Database (TTD) (16), DrugBank (17), Online Mendelian Inheritance in Man (OMIM) (18), Genetic Association Database (GAD) (19), Pharmacogenomics Knowledgebase (PharmGKB) (20), DisGeNET (21) (a database of gene-disease associations), and Human Phenotype Ontology (HPO) (22) for targets related to the pathogenesis of OA.
The microarray was searched in the National Center for Biotechnology Information (NCBI) NCBI Gene Expression Omnibus (GEO) (23) database with the keyword “Osteoarthritis”. The original microarray data file numbered GSE43923 and the microarray gene annotation file GPL570 were obtained, which contained 6 articular cartilage samples, 3 of which were from OA patients and 3 of which were from healthy controls. The raw microarray data were analyzed using R language software (The R Project for Statistical Computing), and the background correction and quartile data normalization were conducted using the robust multi-array average (RMA) algorithm. The “limma” package was used to analyze the differentially expressed genes (DEGs) in the microarray data. In this study, genes with a P value <0.05 and [log fold change (FC)] >2 were defined as DEGs. Thus, the DEGs of OA were obtained. Finally, the common OA targets obtained from the above 7 disease databases and the DEGs obtained in R language were used to identify the OA targets. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
The Database for Annotation, Visualization and Integrated Discovery enrichment analyses
The targets related to the active compounds of WTD for OA were intersected with the OA targets to obtain the key therapeutic targets of WTD for OA. The Database for Annotation, Visualization and Integrated Discovery (DAVID) (24) was used for the batch processing and annotation of gene expression data and for the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses. Functional enrichment analysis of the above key genes was performed using the DAVID 6.8 online analysis tool, and scatter plots were created using “ggplot2” in R.
Construction of a protein-protein interaction (PPI) network for key targets
STRING (25) is a web-based database for online retrieval of interacting genes and proteins. We entered the above key targets into the search field and integrated the results to construct a PPI network diagram of key targets of WTD for OA using Cytoscape software. Subsequently, the “NetworkAnalyzer” plug-in was used to analyze the network, and the core targets with a network median degree >20 were selected for molecular docking to verify the key molecular mechanisms of WTD intervention in OA.
Molecular docking validation
First, based on the results of network analysis, the core active pharmaceutical compounds of WTD were selected using the criterion of greater than 2 times the median degree value of all the pharmaceutical compounds, and the three-dimensional (3D) structure files of all core active compounds were downloaded (in .mol2 format) from the ZINC database. Meanwhile, the RCSB Protein Data Bank (PDB) was used to search and download the 3D structure files (in .pdb format) of the core targets screened in above results. The protein preparation wizard module in Maestro software version 11.5 (Schrödinger) is used for protein preparation, hydrogenation, charging, and deletion of water molecules; protein receptor grid point creation and active pocket coordinate determination, and grid point files are generated with the original ligand in the crystal structure as the center. The box size is set to 1 nm (10 Å), and the system default values are selected for other parameters. The Lig Prep module is used for ligand processing, the Epik method is selected to generate all the conformations of pH 7.0±2.0, and other system defaults are selected; the Ligand Docking module is used for ligand docking, and the receptor grid module that has been created is selected on the Receptor grid module, Select the created ligand on the File name module. A flexible docking method was adopted. The validation was based on the matching of results between the core active compounds and the key target proteins according to the docking score.
Statistical analysis
The data were analyzed with R Package, online tool DAVID and Cytoscape 3.6.1 software. P<0.05 was considered as a statistically significant value.
Results
Chemical compounds of 5 Chinese herbs in WTD
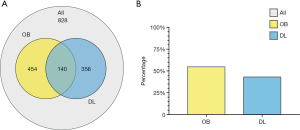
According to the ADME parameters, a total of 828 compounds of Radix Aconiti, Herba Ephedrae, Radix Astragali seu Hedysari, Paeonia lactiflora Pall, and Radix glycyrrhizae were identified in the TCMSP database, 454 of which met the criterion of OB ≥30% and 356 of which met the criterion of DL ≥0.18. According to the screening criteria of ADME parameters, there were 140 compounds meeting both OB ≥30% and DL ≥0.18. Thus, 140 active compounds in WTD were initially obtained (Figure 2).
Prediction of the action targets and construction of the “TCM drugs-Active compounds-Targets” network
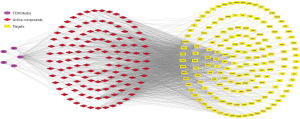
After the matching analysis based on the target prediction model in the TCMSP database was completed, 123 active compounds were identified to have corresponding targets. A visual network of “TCM Drugs-Active Compounds-Targets” was drawn using the Cytoscape software version 3.7.2 (Figure 3). Within the network, there were 2,051 interrelations between 123 active compounds of WTD and 163 targets, and thus 163 action targets of WTD were obtained (Figure 3). Each node was then analyzed for relevant network parameters using the “NetworkAnalyzer” plug-in of Cytoscape software: the more components a target corresponds to than other targets, the greater its degree value, which suggests that the importance of the target may be relatively higher. The network was analyzed using the “NetworkAnalyzer” plug-in in Cytoscape, which yielded 7 core compounds with a degree 2 times higher than the median 34 in the network (Table 2). These core compounds were used for molecular docking verification.

Full table
Screening of OA-related targets
In 7 databases (including TTD), 35, 1, 61, 97, 2, 80, and 33 OA-related targets were obtained. After obtaining common targets and removing duplicates, 227 OA-related targets were obtained.
The raw data file of microarray GSE43923 contains 6 articular cartilage samples, and we compared the osteochondral tissues of 3 OA patients with the cartilage tissues of 3 healthy individuals. Analysis with the help of R language yielded a total of 331 significantly altered and affected genes. After removing duplicates, 307 genes were left, of which 256 were upregulated and 75 were downregulated.
The common OA targets from the above 7 databases and the DEGs in R language were obtained, and the intersection with the action targets of active compounds of the 5 herbs in WTD was determined. In this way, 32 key targets of WTD for OA were obtained (Table 3).

Full table
DAVID enrichment analyses
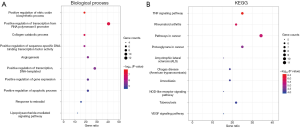
The 32 key targets of WTD for OA were uploaded to the online software DAVID for GO and KEGG pathway analyses, and 186 biological functions and 60 signaling pathways were identified. Based on our professional knowledge and literature review, the top 10 biological processes (ranked in ascending order by P value) and 10 relevant signaling pathways are presented in the form of bubble diagrams (Figure 4). It was found that the biological processes of WTD for OA mainly involved the positive regulation of nitric oxide (NO) biosynthesis, positive regulation of RNA polymerase II promoter transcription, positive regulation of sequence-specific DNA-binding transcription factor activity, angiogenesis, apoptosis, estradiol response, and lipopolysaccharide-mediated signaling pathway; in addition, the signaling pathways involved mainly included tumor necrosis factor (TNF) signaling pathway, rheumatoid arthritis signaling pathway, cancer-related signaling pathway, amyotrophic lateral sclerosis signaling pathway, nucleotide oligomerization domain-like receptors, tuberculosis signaling pathway, and vascular endothelial growth factor (VEGF).
Construction of a PPI network for key targets
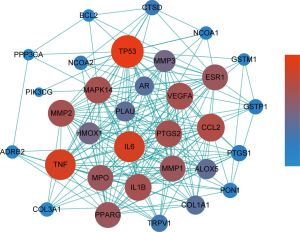
The above 32 key targets of WTD for OA (Table 3) were entered into the search function of the STRING database to obtain the target interaction networks. Cytoscape software was used to visually integrate the data and draw the PPI network diagram of key targets of WTD for OA (Figure 5). The results showed that there were 218 interactions among these 32 key targets. The network parameters were analyzed using the “NetworkAnalyzer” plug-in, in which the values of degree and closeness centrality were positively correlated with their importance in the network.
Molecular docking validation
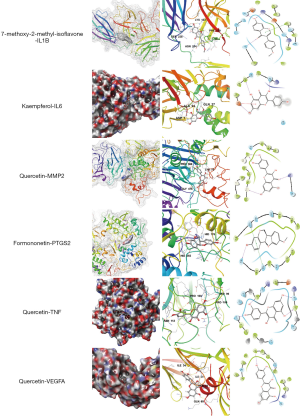
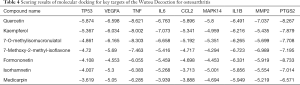
After analysis of the network diagram (described in PPI network for key targets), 9 core targets with network neutrality values of >20 were selected for molecular docking with the 7 core drug compounds screened (described in the “TCM drugs-Active compounds-Targets” network). It is generally believed that a smaller molecular docking value indicates a stronger binding ability and affinity of a compound to the target and a more stable binding conformation. The binding of a compound to its target is regarded as good if the value is <–5.0 and extremely strong if it is <–7.0. The results of molecular docking are shown in Table 4. The mean scores of the binding of quercetin to cellular tumor antigen p53 (TP53), vascular endothelial growth factor A (VEGFA), TNF, C-C motif chemokine 2 (CCL2), mitogen-activated protein kinase 14 (MAPK14), and 72 kDa type IV collagenase (MMP2) were all <–5.0, and its binding capacities were higher than those of the other 6 compounds. Kaempferol had the optimum binding ability with interleukin-6 (IL6), 7-methoxy-2-methylisoflavone had the strongest binding ability with IL-1β; and formononetin had the strongest affinity with prostaglandin G/H synthase 2 (PTGS2) (Figure 6). Notably, quercetin, kaempferol, 7-O-methylisoxylitol, and 7-methoxy-2-methylisoflavone had docking scores of <–4.25 with the core key target of WTD for OA and therefore may be the active compounds of WTD for OA.
Full table
Discussion
OA is a multifactorial degenerative joint disease characterized by cartilage damage and destruction, inflammatory hyperplasia at the joint edges, and increased joint wear and tear, which leads to pain. With a high disability rate, OA seriously affects joint function and quality of life. Although OA has some similarities with rheumatoid arthritis (RA) in clinical symptoms such as joint swelling and pain, deformity, and inflammatory changes. In fact, RA is an autoimmune syndrome, and its chronic inflammation and the occurrence of hyperplasia often starts in the synovium of the joints and gradually invades the articular cartilage, subchondral bone, ligaments and other tissues. But the origin of OA is often cartilage. Traditionally, it is believed that OA is caused by the disruption of the delicate balance between the synthesis and catabolism of chondrocytes, subchondral bone, and extracellular matrix (ECM), and a combination of human factors, mechanics, and biomechanics. The lesions can affect all joint tissues, including cartilage, subchondral bones, and synovial tissue (26,27). Unfortunately, the pathogenic mechanism is not yet clear. Compared with Western medicine, TCM therapy has been increasingly used in the prevention and treatment of OA because of its long history, rich clinical experience, and lower toxicity. OA is categorized as “bi-syndrome” in TCM. TCM believes that the core pathogenesis of OA is the “deficiency in origin and excess in superficiality”. Wind, cold and dampness arthralgia troubles the meridians, resulting in poor circulation of qi and blood, mixed with phlegm, dampness, and blood stasis to form a “excess in superficiality”, which causes OA on the basis of the “deficiency in origin” of the patient’s liver and kidney deficiency and muscle and bone dystrophy. In the WTD, Radix Aconiti warms the meridians to disperse cold and remove dampness to relieve pain, serving as the sovereign drug, while Herba Ephedrae disperses wind-cold and dispels cold and dampness, serving as the minister drug. These two drugs, with the similar and synergistic effects, combine to reinforce yang, relieve exterior syndrome, and disperse cold and dampness; Radix Astragali seu Hedysari invigorates qi for consolidating immune system, assists the sovereign and minister drugs to warm the meridians and relieve pain, and suppresses the excessive effect of Herba Ephedrae in dispersing superficies. Paeonia lactiflora Pall dredges channel blockade and moves blood, and can alleviate pain when used in combination with Radix glycyrrhizae. The combination of all these 5 herbs can dispel cold, promote circulation, eliminate dampness, and relieve pain, especially for cold and painful joints that are unable to flex or extend (6,28). This is also the mechanism of WTD in treating OA. In fact, searching for effective, active monomers from classic TCM formulae has become an area of intense research focus in recent years.
Therapeutic effects of the core compounds in WTD on OA
In our current study, we used bioinformatics and network pharmacology to construct the “TCM Drugs-Active Compounds-Targets” network of WTD and obtained 123 active compounds and 163 action targets in WTD. Seven core compounds, mostly flavonoids, were screened by network analysis. Modern pharmacological studies have shown that flavonoids are a class of naturally occurring polyphenolic compounds. They exhibit a 3-ring structure base (C6-C3-C6) and are abundant in Chinese herbal medicines, such as malaytea scurfpea fruit, epimedium herb, baical skullcap root, tree peony root bark, chrysanthemum flower, kudzuvine root, and purple common perilla. They mainly interfere with the apoptosis, proliferation, and metabolism of chondrocytes, regulate inflammatory cells and cytokines, and affect the differentiation from bone marrow stromal stem cells to chondrocytes, thus exerting their therapeutic effects against OA (29-32). Quercetin, a flavonoid with various biological activities, is widely found in the flowers, leaves, fruits, and seeds of many medicinal plants and pharmacological properties of antioxidant, anti-inflammatory, free radical-scavenging, and immune-boosting activity (33,34). Ex vivo experiments have demonstrated that quercetin can effectively reduce the expressions and secretion of inflammatory cytokines, such as tumor necrosis factor-α (TNFα), IL-1β, and IL-6 (35-37), and its potential role in treating OA warrants further exploration. Kaempferol, a natural flavonol component, has a variety of pharmacological activities. Ex vivo experiments have demonstrated that it can significantly lower the expression levels of IL-1β and other inflammatory markers in rat chondrocytes, thereby suppressing the degradation of type II collagen; also, it exerts its antiarthritic effect by regulating intestinal flora and microbial metabolism (38,39). A study using liquid chromatography-electrospray ionization-mass spectrometry (LC-ESI/MS) revealed that Pergularia daemia extract can effectively treat joint pain and arthritis, with its major active compounds being flavonoids, such as formononetin and quercetin (40). Isorhamnetin, a natural 3'-O-methylated flavonoid, has been shown to exert strong anti-inflammatory and antioxidant activity. It can inhibit oxidative stress in mice with collagen-induced arthritis by regulating the expressions of cytokines. Research has demonstrated its effect on IL-6, and thus isorhamnetin can be applied in the treatment of rheumatoid arthritis (41). In an animal study, medicarpin, an active component with anti-osteoclast properties, significantly downregulated the destruction of protective cartilage by downregulating proinflammatory cytokines TNF-α, IL-6, and IL-17A and upregulating the anti-inflammatory cytokine IL-10 (42). Thus far, no literature has described the roles of 7-O-methylisoxylitol and 7-methoxy-2 methyl isoflavones in the prevention and treatment of OA, but it is speculated that they may function similarly to other flavonoids, which should be confirmed by further research.
Association of key targets, biological processes, and signaling pathways with OA
Molecular docking showed that all of the 7 core drug compounds had the strongest binding ability to TNF and PTGS2 and also had strong affinity to VEGFA, IL-6, IL-1β, and MMP2. TNF belongs to a class of cytokines that can trigger tumor bleeding and necrosis, and is mainly involved in immune response, antitumor activity, inflammatory response, and other physiopathological processes. It can induce the secretion of IL-1, IL-6, IL-8, cerebrospinal fluid (CSF), matrix metalloproteinases (MMPs), and other cytokines and chemokines, accelerating the development of inflammatory response. Among them, TNF-α plays an important role in the pathogenesis of OA: the inflammatory cells can activate trypsinogen via TNF-α, and TNF-α, when abundant, can accelerate the death of chondrocytes, which in turn further aggravates the systemic and local inflammatory responses (43). Quercetin, kaempferol, 7-O-methylisoxylitol, and 7-methoxy-2-methyl isoflavone have a strong affinity to TNF and may inhibit its expression, thus exerting a protective effect on cartilage. The activity of PTGS2, which encodes cyclooxygenase-2 (COX-2), directly affects the synthesis of prostaglandin (PG), which mainly exists in synovial tissue and is thought to be closely related to vasodilation, fluid exudation, and local pain. Certain cyclooxygenase-2 (COX-2) inhibitors can exert similar pharmacological effects as nonsteroidal anti-inflammatory drugs (NSAIDs) (44). MMPs can catalyze the metabolism of collagen and proteoglycan in the soft ECM mainly through hydrolytic enzymes. MMP2 is a gelatinase A enzyme that can directly cause the degradation of ECM, the excess of which can result in the loss of the structural integrity of the articular cartilage fibers and collagen network, leading to the decreased elasticity and protective capability of these structures, ultimately resulting in OA (45). TP53 is thought to be closely associated with fibroblast like synovial cell proliferation and joint destruction in rheumatoid arthritis (46). Articular cartilage is devoid of blood vessels, and thus the expression of VEGFA is extremely low in normal chondrocytes. Studies (47-49) have revealed the presence of large amounts of VEGFA in articular cartilage, synovium, and synovial fluid of OA patients, and VEGFA expression has been positively correlated with OA severity and pain intensity. Thus, VEGFA is considered to be a key contributor to the pathogenesis of OA (50). CCL2 is a chemokine that activates monocytes, macrophages, and lymphocytes in inflammatory and immune responses. CCL2 expression is elevated in the peripheral blood of OA patients and in bone and joint tissues of OA rats, suggesting its potential application as a biomarker for the diagnosis, prognosis, and treatment of OA (51,52). TNF-α in the TNF signaling pathway can induce the activation of NF-κB signaling pathway in synovial cells (53). The dysregulation of NF-κB signaling plays a vital role in the regulation of OA inflammation and chondrocyte proliferation, differentiation, and apoptosis, which ultimately leads to cartilage degradation and joint damage (54). The MAPK pathway is involved in regulating cell proliferation, autophagy, and apoptosis; inflammation; and other physiopathological processes. In particular, MAPK pathway plays a key role in balancing the level and metabolism of chondrocytes by regulating articular cartilage autophagy (55). Additionally, by degrading the cartilage ECM, it can directly induce chondrocyte hypertrophy and apoptosis, resulting in degenerative joint changes (56,57). It has been shown that IL-1β is a key proinflammatory cytokine in OA, as it can lead to disruption of the normal metabolic and synthetic processes of chondrocytes; also, it can promote the secretion of TNF-Α, which causes chondrocyte death and aggravates the pathogenesis of OA (58). IL-6 is one of the key acute response mediators in cartilage injury and repair. It is positively correlated with TNF-α expression and trauma severity. It has been found IL-6 can stimulate the production of other inflammatory mediators after trauma and contribute to the occurrence of systemic inflammatory response syndrome (59,60).
In summary, the core drug compounds, key targets of action, and major signaling pathways of WTD for OA were identified through bioinformatics and molecular docking analyses. The 7 core compounds found regulate inflammation, immunity, autophagy, apoptosis, metabolism, and endocrine signaling pathways mainly through the targets of TNF, PTGS2, MMP2, IL-6, and IL-1β, which may be the key regulatory mechanism of WTD in the treatment of OA. However, they are still in the early stage of the study and exist some limitations. For example, bioinformatics and molecular docking analyses uses computer screening to achieve target selection, and some results do not have enough experimental data support. In the same way, more corresponding experiments are still needed to support our results further. We will continue to conduct in vivo and ex vivo experiments to investigate the core mechanisms of WTD in the prevention and treatment of OA, so as to further promote the application of classic TCM formulae in clinical settings and to gather new insights for the development of new drug therapies.
Acknowledgments
Funding: This work was supported by the National Natural Science Foundation of China (No. 81774333 and 82074453).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://dx.doi.org/10.21037/apm-21-1691
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/apm-21-1691). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Zhang Z, Huang C, Jiang Q, et al. Guidelines for the diagnosis and treatment of osteoarthritis in China (2019 edition). Ann Transl Med 2020;8:1213. [Crossref] [PubMed]
- Hunter DJ, Bierma-Zeinstra S. Osteoarthritis. Lancet 2019;393:1745-59. [PubMed]
- Tang X, Wang S, Zhan S, et al. The Prevalence of Symptomatic Knee Osteoarthritis in China: Results From the China Health and Retirement Longitudinal Study. Arthritis Rheumatol 2016;68:648-53. [Crossref] [PubMed]
- DeRogatis M, Anis HK, Sodhi N, et al. Non-operative treatment options for knee osteoarthritis. Ann Transl Med 2019;7:S245. [Crossref] [PubMed]
- Sun Q, Zhang K, Chen J, et al. Traditional Chinese medicine classification of knee osteoarthritis with proteomics analysis. Ann Palliat Med 2020;9:3750-6. [Crossref] [PubMed]
- Mei YY, Fu CL, Pang SQ, et al. Study on the pharmacological effects of Wutou Decoction and its mechanism of intervention in osteoarthritis. Rheumatism and Arthritis 2016;5:47-50.
- Zhang YQ, Li S. Progress in network pharmacology for modern research of traditional Chinese medicine. Chinese Journal of Pharmacology and Toxicology 2015;29:883-92.
- Duan AX, Chen J, Liu HD, et al. Applications and Developments of Molecular Docking Method. Journal of Analytical Science 2009;25:473-7.
- Yu YN, Chen XX, Liu Y, et al. Virtual screening of anti-angiogenesis quinolizidine alkaloids from Leguminosae plant. Chinese Traditional and Herbal Drugs 2018;49:1640-6.
- Xu X, Zhang W, Huang C, et al. A Novel Chemometric Method for the Prediction of Human Oral Bioavailability. International Journal of Molecular Sciences 2012;13:6964-82. [Crossref] [PubMed]
- Liu H, Wang J, Zhou W, et al. Systems approaches and polypharmacology for drug discovery from herbal medicines: An example using licorice. Journal of Ethnopharmacology 2013;146:773-93. [Crossref] [PubMed]
- Ru J, Li P, Wang J, et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J Cheminform 2014;6:13. [Crossref] [PubMed]
- Li J, Zhao P, Li Y, et al. Systems pharmacology-based dissection of mechanisms of Chinese medicinal formula Bufei Yishen as an effective treatment for chronic obstructive pulmonary disease. Sci Rep 2015;5:15290. [Crossref] [PubMed]
- Wang S, Wang H, Lu Y. Tianfoshen oral liquid: a CFDA approved clinical traditional Chinese medicine, normalizes major cellular pathways disordered during colorectal carcinogenesis. Oncotarget 2017;8:14549-69. [Crossref] [PubMed]
- Shannon P, Markiel A, Ozier O, et al. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Cold Spring Harbor Laboratory Press 2003;13:2498-504.
- Chen X, Ji ZL, Chen YZ. TTD: Therapeutic Target Database. Nucleic Acids Res 2002;30:412-5. [Crossref] [PubMed]
- Wishart DS, Knox C, Guo AC, et al. DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res 2008;36:D901-D906. [Crossref] [PubMed]
- Hamosh A, Scott AF, Amberger JS, et al. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res 2005;33:D514-7. [Crossref] [PubMed]
- Becker KG, Barnes KC, Bright TJ, et al. The Genetic Association Database. Nat Genet 2004;36:431-2. [Crossref] [PubMed]
- Hewett M. PharmGKB: the Pharmacogenetics Knowledge Base. Nucleic Acids Res 2002;30:163-5. [Crossref] [PubMed]
- Piñero J, Queralt-Rosinach N, Bravo À, et al. DisGeNET: a discovery platform for the dynamical exploration of human diseases and their genes. Database (Oxford) 2015;2015:bav028 [Crossref] [PubMed]
- Robinson PN, Köhler S, Bauer S, et al. The Human Phenotype Ontology: A Tool for Annotating and Analyzing Human Hereditary Disease. Am J Hum Genet 2008;83:610-5. [Crossref] [PubMed]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 2002;30:207-10. [Crossref] [PubMed]
- Dennis G, Sherman BT, Hosack DA, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol 2003;4:3. [Crossref] [PubMed]
- Szklarczyk D, Franceschini A, Kuhn M, et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res 2011;39:D561-D568. [Crossref] [PubMed]
- Fraysse F, Arnold J, Thewlis D. A method for concise reporting of joint reaction forces orientation during gait. J Biomech 2016;49:3538-42. [Crossref] [PubMed]
- Ding CB, Zhou Y. Pathogenesis of synovitis in knee osteoarthritis patients. Chinese Journal of Tissue Engineering Research 2015;19:8327-32.
- Chen D, Chen HH, Shao X, et al. Study on the mechanism of Wutou decoction in inhibiting inflammation of osteoarthritis. Rheumatism and Arthritis 2016;5:62-6.
- Li Y, Wang J, Song X, et al. Effects of baicalein on IL-1β-induced inflammation and apoptosis in rat articular chondrocytes. Oncotarget 2017;8:90781-95. [Crossref] [PubMed]
- Yang Y, Liu Y. Study on Effect of Puerarin on Nitric Oxide and Nitric Oxide Synthase of Articular Chondrocytes in IL-1β Injury Rats. Yunnan Journal of Traditional Chinese Medicine and Materia Medica 2017;38:83-5.
- Yuan XL, Li LF, Shi WM, et al. Effects of quercetin on the expression of MMP-13, TIMP-1 in joint cartilage. Lishizhen Medicine and Materia Medica Research 2016;27:283-5.
- Zhao Y, Li Z, Wang W, et al. Naringin Protects Against Cartilage Destruction in Osteoarthritis Through Repression of NF-κB Signaling Pathway. Springer US 2016;39:385-92.
- Wiczkowski W, Romaszko J, Bucinski A, et al. Quercetin from shallots (Allium cepa L. var. aggregatum) is more bioavailable than its glucosides. J Nutr 2008;138:885-8. [Crossref] [PubMed]
- Liu SW, Liu JY. Advances in the pharmacological effects of quercetin. Chinese Journal of Lung Diseases 2020;13:104-6. (Electronic Edition).
- Saberi Hosnijeh F, Bierma-Zeinstra SM, Bay-Jensen AC. Osteoarthritis year in review 2018: biomarkers (biochemical markers). Osteoarthritis Cartilage 2019;27:412-23. [Crossref] [PubMed]
- Lee YM, Son E, Kim S, et al. Anti-inflammatory and anti-osteoarthritis effect of Mollugo pentaphylla extract. Pharm Biol 2019;57:74-81. [Crossref] [PubMed]
- Haleagrahara N, Hodgson K, Miranda-Hernandez S, et al. Flavonoid quercetin–methotrexate combination inhibits inflammatory mediators and matrix metalloproteinase expression, providing protection to joints in collagen-induced arthritis. Inflammopharmacology 2018;26:1219-32. [Crossref] [PubMed]
- Huang X, Pan Q, Mao Z, et al. Kaempferol inhibits interleukin-1β stimulated matrix metalloproteinasesby suppressing the MAPK-associated ERK and P38 signaling pathways. Mol Med Rep 2018;18:2697-704. [PubMed]
- Aa LX, Fei F, Qi Q, et al. Rebalancing of the gut flora and microbial metabolism is responsible for the anti-arthritis effect of kaempferol. Acta Pharmacol Sin 2020;41:73-81. [Crossref] [PubMed]
- Ananth DA, Rameshkumar A, Jeyadevi R, et al. Amelioratory effect of flavonoids rich Pergularia daemia extract against CFA induced arthritic rats. Biomed Pharmacother 2016;80:244-52. [Crossref] [PubMed]
- Wang X, Zhong W. Isorhamnetin attenuates collagen-induced arthritis via modulating cytokines and oxidative stress in mice. Int J Clin Exp Med 2015;8:16536-42. [PubMed]
- Mansoori MN, Raghuvanshi A, Shukla P, et al. Medicarpin prevents arthritis in post-menopausal conditions by arresting the expansion of TH17 cells and pro-inflammatory cytokines. Int Immunopharmacol 2020;82:106299 [Crossref] [PubMed]
- Liu QM, Hu J, Wang WD, et al. Comparative Study of the Expression of Cartilage Related Factors in the Normal Cartilage and Osteoarthritis in the Hip Joint. Chinese Journal of Traditional Medical Traumatology & Orthopedics 2018;26:17-22.
- Krasselt M, Baerwald C. Celecoxib for the treatment of musculoskeletal arthritis. Expert Opin Pharmacother 2019;20:1689-702. [Crossref] [PubMed]
- Sousa LH, Linhares EV, Alexandre JT, et al. Effects of Atorvastatin on Periodontitis of Rats Subjected to Glucocorticoid-Induced Osteoporosis. J Periodontol 2016;87:1206-16. [Crossref] [PubMed]
- Shchetynsky K, Diaz-Gallo L, Folkersen L, et al. Discovery of new candidate genes for rheumatoid arthritis through integration of genetic association data with expression pathway analysis. Arthritis Res Ther 2017;19:19. [Crossref] [PubMed]
- Hamilton JL, Nagao M, Levine BR, et al. Targeting VEGF and Its Receptors for the Treatment of Osteoarthritis and Associated Pain. J Bone Miner Res 2016;31:911-24. [Crossref] [PubMed]
- Nagao M, Hamilton JL, Kc R, et al. Vascular Endothelial Growth Factor in Cartilage Development and Osteoarthritis. Sci Rep 2017;7:13027. [Crossref] [PubMed]
- Maes C. Signaling pathways effecting crosstalk between cartilage and adjacent tissues: Seminars in cell and developmental biology: The biology and pathology of cartilage. Semin Cell Dev Biol 2017;62:16-33. [Crossref] [PubMed]
- Zhang X, Prasadam I, Fang W, et al. Chondromodulin-1 ameliorates osteoarthritis progression by inhibiting HIF-2α activity. Osteoarthritis Cartilage 2016;24:1970-80. [Crossref] [PubMed]
- Guo Q, Liu Z, Wang M, et al. Analysis on the expression and value of CCL2 and CCL3 in patients with osteoarthritis. Exp Mol Pathol 2021;118:104576 [Crossref] [PubMed]
- Bonfante HL, Almeida CS, Abramo C, et al. CCL2, CXCL8, CXCL9 and CXCL10 serum levels increase with age but are not altered by treatment with hydroxychloroquine in patients with osteoarthritis of the knees. Int J Rheum Dis 2017;20:1958-64. [Crossref] [PubMed]
- Luo XJ, Mo XR, Zhou LL. Activation of NF-κB signal pathway by TNF-α in synoviocytes of rheumatoid arthritis. Immunological Journal 2012;28:321-3, 332.
- Zhou Z, Zhang L. The research of mechanism of NF-κB signaling pathway in the initiation and development of osteoarthritis. Chinese Journal of Osteoporosis 2012;18:78-82.
- Ma JD, Zhou JJ, Zheng DH, et al. Serum matrix metalloproteinase-3 as a noninvasive biomarker of histological synovitis for diagnosis of rheumatoid arthritis. Mediators Inflamm 2014;2014:179284 [Crossref] [PubMed]
- Hu G, Zhao X, Wang C, et al. MicroRNA-145 attenuates TNF-α-driven cartilage matrix degradation in osteoarthritis via direct suppression of MKK4. Cell Death Dis 2017;8:e3140 [Crossref] [PubMed]
- Liu F, Li L, Lu W, et al. Scutellarin ameliorates cartilage degeneration in osteoarthritis by inhibiting the Wnt/β-catenin and MAPK signaling pathways. Int Immunopharmacol 2020;78:105954 [Crossref] [PubMed]
- Luo YM, Zheng WP, Wei HZ. Research Progress on the Relationship of Osteoarthritis with TNF-α,IL-6. Modern Diagnosis and Treatment 2013;24:326-7.
- Le HL, Luo GQ. Research Progress of Early Posttraumatic Inflammatory Cytokines TNF-a, IL-1, IL-6. Modern Diagnosis and Treatment 2014;25:763-5.
- Yang F, Sui FG, Zhang Y, et al. Expression of serum interleukin-6, 8, 10, C-reactive protein and deep vein thrombosis after joint replacement. Chinese Journal of Tissue Engineering Research 2011;15:8031-3.