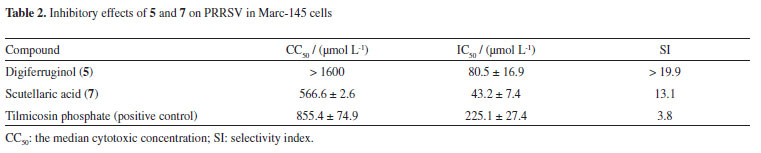Abstracts
Two new quinonoids chiritalone A and B, and a new neolignan 7'E-4,9-dihydroxy-3,3',5'-trimethoxy-8,4'-oxyneolign-7'-en-9'-al, along with known (-)-8-hydroxy-α-dunnione, digiferruginol, 2,5-dimethoxy-1,4-benzoquinone and hederagenin, were isolated from the stems of Chirita longgangensis var. hongyao. The structures of the new compounds were elucidated by detailed analysis from NMR (nuclear magnetic resonance) and MS (mass spectrometry) data, and the absolute configuration of chiritalone A was determined by single crystal X-ray diffraction analysis using the Flack parameter. The inhibitory activity of compounds against porcine respiratory and reproductive syndrome virus (PRRSV) was measured by the cytopathic effect (CPE) method. Digiferruginol and hederagenin showed weak effect on PRRSV with an IC50 value of 80.5 ± 16.9 µmol L-1 (SI = 19.9) and 43.2 ± 7.4 µmol L-1 (SI = 13.1), respectively.
Gesneriaceae; Chirita longgangensis var. hongyao; quinines; porcine reproductive and respiratory syndrome virus
Dois novos quinenóides chiritalona A and B, e uma nova neolignina 7'E-4,9-dihidroxi-3,3',5'-trimethoxi-8,4'-oxineolign-7'-en-9'-al, além dos conhecidos (-)-8-hidroxi-α-dunniona, digiferruginol, 2,5-dimetoxi-1,4-benzoquinona e hederagenina, foram isolados do caule de Chirita longgangensis var. hongyao. As estruturas dos novos compostos foram elucidadas por análise detalhada de dados obtidos pela técnicas de NMR (ressonância magnética nuclear) e MS (espectrometria de massas), e a configuração absoluta de chiritalona A foi determinada por análise de difração de raios X de monocristal utilizando o parâmetro Flack. A atividade inibitória dos compostos com relação ao vírus da síndrome respiratória e reprodutiva suína (PRRSV) foi medida pelo método do efeito citopático (CPE). Digiferruginol e hederagenina apresentaram efeito fraco com relação ao PRRSV com valor IC50 de 80,5 ± 16,9 µmol L-1 (SI = 19,9) e 43,2 ± 7,4 µmol L-1 (SI = 13,1), respectivamente.
Chemical constituents from Chirita longgangensis var. hongyao with inhibitory activity against porcine respiratory and reproductive syndrome virus
Yao SuI,II; Jun-Long BiIII; Yue-Hu WangI, Ying TanI; Jun YangI,II; Hong-Xin LiuI; Wei GuI; Ge-Fen YinIII,** e-mail: long@mail.kib.ac.cn; yingefen383@sohu.com; Chun-Lin LongI,IV,** e-mail: long@mail.kib.ac.cn; yingefen383@sohu.com
IKey Laboratory of Economic Plants and Biotechnology, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming 650201, P. R. China
IISchool of Agronomy and Biotechnology
IIICollege of Animal Science and Technology, Yunnan Agricultural University, Kunming 650201, P. R. China
IVCollege of Life and Environmental Sciences, Minzu University of China, Beijing 100081, P. R. China
ABSTRACT
Two new quinonoids chiritalone A and B, and a new neolignan 7'E-4,9-dihydroxy-3,3',5'-trimethoxy-8,4'-oxyneolign-7'-en-9'-al, along with known (-)-8-hydroxy-α-dunnione, digiferruginol, 2,5-dimethoxy-1,4-benzoquinone and hederagenin, were isolated from the stems of Chirita longgangensis var. hongyao. The structures of the new compounds were elucidated by detailed analysis from NMR (nuclear magnetic resonance) and MS (mass spectrometry) data, and the absolute configuration of chiritalone A was determined by single crystal X-ray diffraction analysis using the Flack parameter. The inhibitory activity of compounds against porcine respiratory and reproductive syndrome virus (PRRSV) was measured by the cytopathic effect (CPE) method. Digiferruginol and hederagenin showed weak effect on PRRSV with an IC50 value of 80.5 ± 16.9 µmol L-1 (SI = 19.9) and 43.2 ± 7.4 µmol L-1 (SI = 13.1), respectively.
Keywords: Gesneriaceae; Chirita longgangensis var. hongyao, quinines, porcine reproductive and respiratory syndrome virus
RESUMO
Dois novos quinenóides chiritalona A and B, e uma nova neolignina 7'E-4,9-dihidroxi-3,3',5'-trimethoxi-8,4'-oxineolign-7'-en-9'-al, além dos conhecidos (-)-8-hidroxi-α-dunniona, digiferruginol, 2,5-dimetoxi-1,4-benzoquinona e hederagenina, foram isolados do caule de Chirita longgangensis var. hongyao. As estruturas dos novos compostos foram elucidadas por análise detalhada de dados obtidos pela técnicas de NMR (ressonância magnética nuclear) e MS (espectrometria de massas), e a configuração absoluta de chiritalona A foi determinada por análise de difração de raios X de monocristal utilizando o parâmetro Flack. A atividade inibitória dos compostos com relação ao vírus da síndrome respiratória e reprodutiva suína (PRRSV) foi medida pelo método do efeito citopático (CPE). Digiferruginol e hederagenina apresentaram efeito fraco com relação ao PRRSV com valor IC50 de 80,5 ± 16,9 µmol L-1 (SI = 19,9) e 43,2 ± 7,4 µmol L-1 (SI = 13,1), respectivamente.
Introduction
Porcine reproductive and respiratory syndrome (PRRS), caused by porcine reproductive and respiratory syndrome virus (PRRSV), is characterized by respiratory disorders in young pigs and reproductive failure in sows. It is widespread in most major pig-producing areas throughout the world and is one of the most important causes of economic loss to the swine industry. In China, in 2006 only, PRRS spread to more than 10 provinces (or autonomous regions) and affected over 2,000,000 pigs with about 400,000 fatal cases.1 Although previous studies provided a basis for development of pharmacological agents to inhibit PRRSV replication, so far there are no effective drugs to overcome this problem,2 and many vaccine strategies developed to control the disease are not yet completely successful.3,4
Quinones are a widespread group of oxygen-substituted aromatic compounds, and some of them are considered as inhibitory agents against both RNA and DNA virus.5 Plants of Gesneriaceae such as Streptocarpus dunnii Mast., Chirita eburnea Hance, Sinningia aggregata (Ker-Gawl.) Wiehler, and Didymocarpus hedyotideus Chun. are rich in quinones.6-10 Chirita longgangensis var. hongyao, a species of Gesneriaceae, is distributed in Guangxi of China,11 and is used in the treatment of bone fractures, wounds and pains.12 The chemical constituents of Chirita are mainly phenylethanoid glycosides, flavonoids and quinones.13-17 Our group conducted a phytochemical research on the stems of the plant, which led to the isolation of seven compounds (1-7) including two new quinones (1 and 2) and the new neolignan (3) (Figure 1). All of the isolates were evaluated for their inhibitory activity against PRRSV. The structure elucidation of the new constituents and the bioassay results are reported.
Results and Discussion
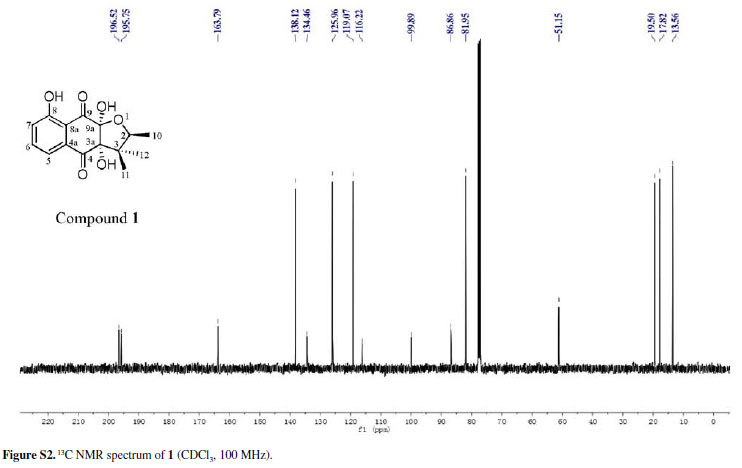
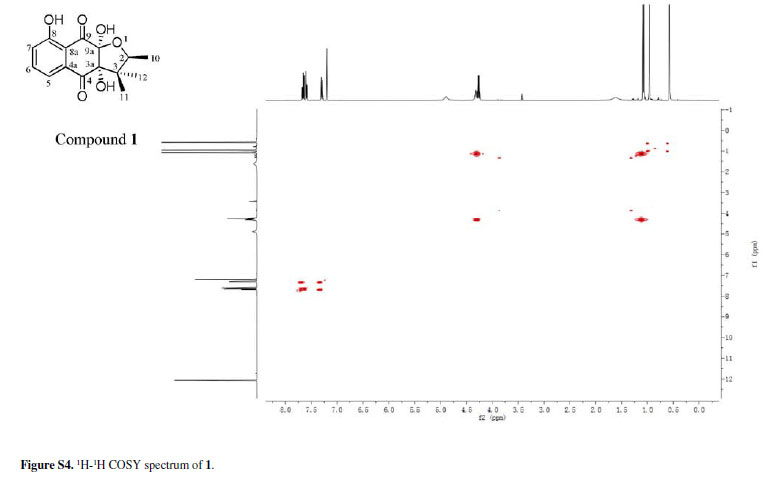
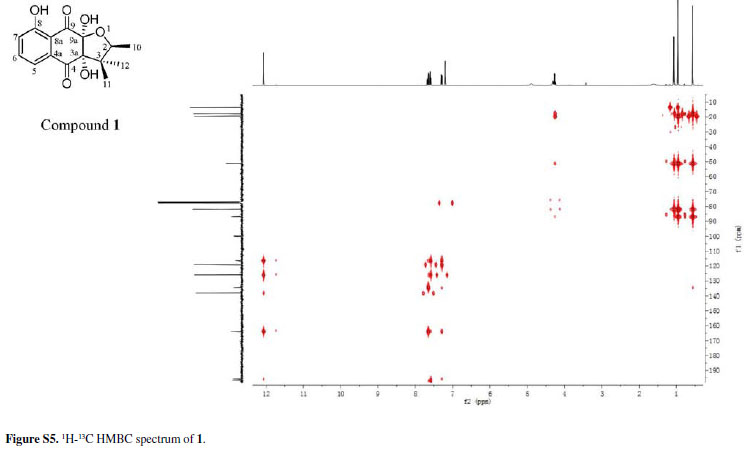
Chiritalone A (1) was obtained as colorless crystals. The molecular formula C15H16O6, indicating eight degrees of unsaturation, was determined from the HRESIMS (highresolution electrospray ionization mass spectrometry) quasi-molecular ion [M -H]-at m/z 291.0865 (calcd. 291.0868). The IR spectrum revealed absorption bands for free hydroxy (3521 cm-1), chelated OH (3381 cm-1) and CO groups (1685 cm-1). The 1H and 13C nuclear magnetic resonance (NMR) spectra of 1 (Table 1) showed signals for two conjugated CO groups (dC 196.5 and 195.8 ppm), a trisubstituted Ph ring [δH 7.66 ppm (dd, J 8.3, 7.6 Hz), 7.59 ppm (dd, J 7.6, 1.2 Hz) and 7.30 ppm (d, J 8.3, 1.2 Hz)], three sp3 quaternary C-atoms including two O-bearing ones (δC 99.9, 86.9 and 51.2 ppm), one O-bearing CH group (δC 82.0 ppm), and three Me groups [δH 1.08 ppm (d, J 6.4 Hz), 0.96 ppm (s), and 0.57 ppm (s)]. Comparison of NMR data of 1 with those of the known quinone (-)-8-hydroxy-α-dunnione (4)8 revealed that compound 1 possessed signals for the enone in 4, thus suggesting that the C-3a and C-9a in 1 might be both hydroxylated. Most of the 13C NMR signals for 1 were assigned by heteronuclear multiple bond correlation (HMBCs) (Figure 2).
To elucidate the absolute configuration, the crystals of 1 was obtained from a mixture solvent of petroleum ether/CHCl3 (2:1), and its integrated structure and absolute configuration were finally determined by the single-crystal X-ray diffraction study using graphite-monochromated Cu Kα radiation in light of the Flack parameter of 0.1(2) (Figure 3). Thus, 1 was elucidated as (2S,3aR,9aS)3a,8,9a-trihydroxy-2,3,3-trimethyl-2,3,3a,9atetrahydronaphtho[2,3-b]furan-4,9-dione, and named as chiritalone A.
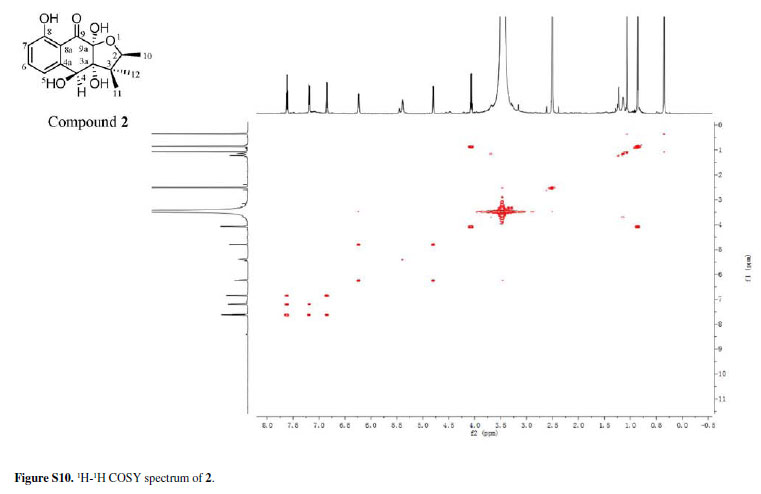
Compound 2 was isolated as a colorless amorphous solid. Its molecular formula was assigned as C15H18O6 from the HRESIMS quasimolecular ion [M -H]-at m/z 293.1024 space (calcd. 293.1025), which indicated seven degrees of unsaturation. The IR spectrum of 2 showed absorption bands for OH (3441 cm-1) and CO (1631 cm-1) groups. The 1H and 13C NMR spectra of 2 (Table 1) showed signals for one conjugated CO group (δC 198.2 ppm), a trisubstituted Ph ring [δH 7.62 ppm (t, J 8.0 Hz), 7.20 ppm (d, J 8.0 Hz) and 6.85 ppm (d, J 8.0 Hz)], three sp3 quaternary C-atoms including two O-bearing ones (δC 99.7, 83.1 and 48.6 ppm), two O-bearing CH groups (δC 80.4 and 72.4 ppm) and three Me groups [δH 1.06 ppm (s), 0.86 ppm (d, J 6.4 Hz), and 0.35 ppm (s)]. These data were very similar to those of 1 except that the signal for one ketone group in 1 was replaced by the signal for the O-bearing CH group (dC 72.4 ppm) in 2. This implied that one of the ketone groups was reduced to an O-bearing CH group in 2. The group was located at C-4 by the HMBCs correlations from H- 4 to C-3 and C-5, and 4-OH to C-4a (Figure 2). Accordingly, the planar structure of 2 was elucidated as 3a,4,8,9a-tetrahydroxy-2,3,3-trimethyl-2,3,3a,4,9apentahydronaphtho[2,3-b]furan-9-one.
The relative configuration of 2 was determined by the rotating-frame Overhauser spectroscopy (ROESY) spectrum of 2 (Figure 4). Key ROESY correlations H-2/3a-OH, H-2/12-Me and H-4/3a-OH suggested these protons to be cofacial and were arbitrarily assigned in α-orientation. Therefore, 2 was determined as rel-(2R,3aS,4R,9aR)-3a,4,8,9a-tetrahydroxy-2,3,3trimethyl-2,3, 3a,4,9a-pentahydronaphtho[2,3-b]furan9-one and given the trivial name chiritalone B.
Compound 3 was isolated as an oil, and its molecular formula established as C21H24O8 from the HRESIMS quasi-molecular ion [M -H]-at m/z 403.1390 (calcd. from H- 4 to C-3 and C-5, and 4-OH to C-4a (Figure 2). 403.1392). The IR absorption bands of 3 were indicative Accordingly, the planar structure of 2 was elucidated of OH presence of OH (3440 cm-1), CHO (1659 cm-1) and Ph (1583, 1502 and 1425 cm-1) groups. The 1H and 13C NMR data of 3 showed high similarity with those of the known neolignan 8 isolated from Eucommia ulmoides Oliver (Figure S18 in the Supplementary Information (SI) section),18 which displays a negative optical rotation [[α] -15.5 (c 0.04, MeOH)]. Since the optical rotation of 3 was positive, its 7R,8S configuration was excluded. Nevertheless, due to the lack of available sample material, the CD spectrum of 3 could not be recorded, and the absolute configuration could not be assigned. The structure of 3 was further supported by HMBCs as shown in Figure 2. The coupling constant of 5.0 Hz between H-7 and H-8 suggested the two H-atoms were in erythro-configuration.19 Accordingly, 3 was determined as 7'E-4,9-dihydroxy3,3',5'-trimethoxy-8,4'-oxyneolign-7'-en-9'-al.
-15.5 (c 0.04, MeOH)]. Since the optical rotation of 3 was positive, its 7R,8S configuration was excluded. Nevertheless, due to the lack of available sample material, the CD spectrum of 3 could not be recorded, and the absolute configuration could not be assigned. The structure of 3 was further supported by HMBCs as shown in Figure 2. The coupling constant of 5.0 Hz between H-7 and H-8 suggested the two H-atoms were in erythro-configuration.19 Accordingly, 3 was determined as 7'E-4,9-dihydroxy3,3',5'-trimethoxy-8,4'-oxyneolign-7'-en-9'-al.
Compounds 4-7 (Figure 1) were identified as (-)-8-hydroxy-α-dunnione (4),8 digiferruginol (5),20 2,5-dimethoxy-1,4-benzoquinone (6) 21 and 3α,24dihydroxy-olean-12-en-28-oic acid (scutellaric acid, 7),22,23 respectively, by comparison of their spectroscopic and physical data with those reported in the literature.
The inhibitory activity of compounds 1-7 against PRRSV was measured by the cytopathic effect (CPE) method.24 Digiferruginol (5) and scutellaric acid (7) showed weak effect on PRRSV with an IC50 (concentration of compound required for 50% inhibition) value of 80.5 ± 16.9 µmol L-1 (selectivity index, SI > 19.9) and 43.2 ± 7.4 µmol L-1 (SI = 13.1), respectively, compared with tilmicosin phosphate (IC50 = 225.1 ± 27.4 µmol L-1, SI = 3.8) (Table 2). No significant inhibitory effects were observed for other compounds at a concentration of 200 µmol L-1. Among the quinones (1, 2 and 4-6), only the active compound (5) belongs to the anthraquinone type. These results suggest that the carbon skeleton of anthraquinone might be necessary for the PRRSV-inhibitory activity. Previous studies find that digiferruginol (5) is also cytotoxic against human epidermoid carcinoma KB (IC50 = 9 × 10-8 g mL-1), Chinese hamster V-79 (IC50 = 7.8 × 10-6 g mL-1), human hepatoma Hepa-3B (IC50 = 3.85 × 10-6 g mL-1) and human cervical carcinoma HeLa (IC50 = 2.45 × 10-5 g mL-1) cell lines.25-27 The inhibitory activity of the oleanane-type triterpene 7 against protein tyrosine phosphatase 1B (PTP1B) has been evaluated, but it was found to be inactive.28 The antiviral activity of compounds 5 and 7 against PRRSV is here reported for the first time.
Furthermore, by the real-time fluorescent quantitative reverse transcription-polymerase chain reaction (FQ RT-PCR),29-31 the relative expression ratio of non-strach polysaccharides NSP9 and open reading frame ORF7 genes of PRRSV was tested. As shown in Table 3 and Figure 5, NSP9 mRNA relative expression level was significantly reduced by compounds 5 and 7 at the concentrations of 50 µmol L-1 or more (P < 0.001) in a dose-dependent manner. Also, ORF7 mRNA relative expression level was significantly reduced by compounds 5 and 7 at the concentration of 200 µmol L-1 (P < 0.001). The viral RNA-dependent RNA polymerase (RdRp; nsp9), a key enzyme for the RNA synthesis of PRRSV, is enc oded by the NSP9 gene.32 ORF7 encodes the nucleocapsid protein N which is the most abundant viral protein in virus-infected cells and essential during the assembly and disassembly of virions.33-35 The down regulation of NSP9 and ORF7 mRNA expression implied that the replication of PRRSV RNA and the assembly of the virons might be inhibited by the compounds.
Experimental
General experimental procedures
Melting points were determined using an X-4 melting point apparatus (Yingyu Yuhua Apparatus Factory, Gongyi, P. R. China), and were not corrected. Bruker SMART APEX-II and Bruker APEX DUO diffractometers using graphite-monochromated Cu Kα radiations were employed for the intensity data collection, and the structures of compounds were solved by direct methods (SHELXS97).36 Optical rotations were determined on a JASCO DIP-370 automatic digital polarimeter. UV spectra were recorded on a Shimadzu double-beam 210A spectrometer. IR spectra were recorded on a Bio-Rad FTS-135 infrared spectrophotometer. Circular dichroism (CD) spectra were obtained using a JASCO J-715 spectropolarimeter (Japan Spectroscopic, Tokyo, Japan) in CD3OD solution. NMR spectra: Bruker AM-400, DRX-500 and Avance III-600 spectrometers with chemical shifts in ppm relative to tetramethylsilane (TMS) as an internal standard. ESIMS and HRESIMS were measured using an API QSTAR Pulsar 1 spectrometer. Column chromatography (CC): MCI gel (70-150 µm; Mitsubishi Chemical Corporation), C18 silica gel (40-75 µm; Fuji Silysia Chemical Ltd.), Sephadex LH-20 gel (40-70 µm, GE Healthcare Bio-Xciences AB), silica gel G (SiO2; 80-100 and 300-400 mesh; Qingdao Meigao Chemical Co.) and silica gel H (10-40 µm). High performance liquid chromatographic (HPLC) separations were performed using an Agilent 1200 series pump equipped with a diode array detector and a semi-preparative Zorbax SB-C18 (5 µm, 9.4 × 250 mm) column. Thin layer chromatography (TLC) was conducted on precoated silica gel plates GF254 (Qingdao).
Plant material
The stems of Chirita longgangensis var. hongyao were collected from Jingxi County of Guangxi Zhuang Autonomous Region, P. R. China, in June 2010. The plant material was identified by Prof. Chun-Lin Long, and a voucher specimen (No. JX1001) was deposited at the Key Laboratory of Economic Plants and Biotechnology, Kunming Institute of Botany, Chinese Academy of Sciences.
Extraction and isolation
The air-dried stems of Chirita longgangensis var. hongyao (4.5 kg) were exhaustively extracted with MeOH (4, 3 and 3 h, successively) at 70 ºC, and the solvent evaporated under reduced pressure. The MeOH extracts were evaporated under reduced pressure. The residue was suspended in H2O and extracted with petroleum ether and AcOEt. The AcOEt-soluble portion (28 g) was chromatographed by MCI gel (MeOH/H2O, 90%) and C18 MeOH/H2O (10 to 100%, v/v) to give Fr. A(10% MeOH/H2O), Fr. B (40% MeOH/H2O), Fr. C (50% MeOH/H2O), Fr. D (70% MeOH/H2O) and Fr. E (80% MeOH/H2O). Fr. A was separated by Sephadex LH-20 (MeOH) to yield 6 (5.3 mg). Fr. B was fractionated by Sephadex LH-20 (MeOH) and silica gel [CHCl3 (v)] to give 3 (3.9 mg). Fr. C was fractionated by Sephadex LH-20 (MeOH) and silica gel [CHCl3/MeOH, 50:1 to 40:1( v/v)] to give 1 (21.6 mg) and a mixture. The later was purified by semi-prep. HPLC (MeOH/H2O, 40%; tR = 25.09 min) to yield 2 (17.5 mg). Fr. D was separated by silica gel [CHCl3/AcOEt, 50:1(v/v)] to give 4 (3.9 mg) and 5 (9.3 mg). Fr. E was purified by recrystallisation (MeOH) to give 7 (3.3 mg).
Chiritalone A (1)
Colorless crystals (from petroleum ether/CHCl3, 2:1); mp. 175-178 ºC; [α] -3.1º (c 0.28, MeOH); UV (MeOH) λmax/nm (log ε) 359 (2.81) and 240 (3.29); IR (KBr) νmax/cm-1 3521, 3381, 1685, 1649, 1576, 1456, 1225, 1170, 984 and 856; CD (c 0.0125, MeOH) 359 (+0.37), 279 (-1.05), 251 (-0.16), 235 (+2.22) and 211 (+0.07) nm; 1H and 13C NMR data, see in Table 1; ESIMS m/z 291 [M -H]-, 327 [M + Cl]-; HRESIMS m/z (%) 291.0865 [M]-(C15H16O6, calcd. 291.0868).
-3.1º (c 0.28, MeOH); UV (MeOH) λmax/nm (log ε) 359 (2.81) and 240 (3.29); IR (KBr) νmax/cm-1 3521, 3381, 1685, 1649, 1576, 1456, 1225, 1170, 984 and 856; CD (c 0.0125, MeOH) 359 (+0.37), 279 (-1.05), 251 (-0.16), 235 (+2.22) and 211 (+0.07) nm; 1H and 13C NMR data, see in Table 1; ESIMS m/z 291 [M -H]-, 327 [M + Cl]-; HRESIMS m/z (%) 291.0865 [M]-(C15H16O6, calcd. 291.0868).
Crystal data for chiritalone A (1)
C15H16O6, MW = 292.28, colorless prisms, size 0.41 × 0.48 × 0.60 mm3, monoclinic system, space group P2(1); a = 7.7816 (2) Å, b = 6.33270 (10) Å, c = 13.4381 (3)Å, α = γ = 90º, b = 93.4560 (10)º, V = 661.01(2) Å3, T = 296 (2) K, Z = 2, ρcalcd = 1.468 Mg m-3, µ(Cu Kα) = 0.963 mm-1, F(000) = 308, 5415 reflections in h (-9/7), k (-7/6), l (-16/16), measured in the range 3.93º < θ < 68.13º, completeness θmax = 92.5%, 1844 independent reflections, Rint = 0.0325, 1840 reflections with |F|2> 2s|F|2, 196 parameters, 1 restrains, GOF = 1.244. Final R indices [I > 2sigma(I)]: R1 = 0.0522, wR2 = 0.1467. R indices (all data): R1 = 0.0523, wR2 = 0.1471. Flack parameter 0.1 (2), largest difference peak and hole = 0.332 and -0.437 e Å-3. The intensity data for 1 were collected on a Bruker APEX DUO diffractometer using graphite-monochromated Cu Kα radiation. The structure of 1 was solved by direct methods (SHELXS97),36 expanded using difference Fourier techniques, and refined by the program and full-matrix least-squares calculations. The nonhydrogen atoms were refined anisotropically, and hydrogen atoms were fixed at calculated positions. Crystallographic data for the structure of 1 were deposited in the Cambridge Crystallographic Data Centre (deposition No. CCDC 852437).
Chiritalone B (2)
Colorless amorphous solid; [α] +27.4º (c 0.08, MeOH); UV (MeOH) λmax/nm (log ε) 342.6 (2.84), 270.6 (3.17) and 215.6 (3.45); IR (KBr) νmax/cm-1 3441, 1631, 1511, 1459, 1127, 1091, 899 and 660; CD (c 0.0116, MeOH) 337 (+1.17), 270 (+3.05) and 217 (-3.25) nm; 1H and 13C NMR data, see in Table 1; ESIMS m/z 293 [M -H]-, 329 [M + Cl]-; HRESIMS m/z (%) 293.1024 [M]-(C15H18O6, calcd. 293.1025).
+27.4º (c 0.08, MeOH); UV (MeOH) λmax/nm (log ε) 342.6 (2.84), 270.6 (3.17) and 215.6 (3.45); IR (KBr) νmax/cm-1 3441, 1631, 1511, 1459, 1127, 1091, 899 and 660; CD (c 0.0116, MeOH) 337 (+1.17), 270 (+3.05) and 217 (-3.25) nm; 1H and 13C NMR data, see in Table 1; ESIMS m/z 293 [M -H]-, 329 [M + Cl]-; HRESIMS m/z (%) 293.1024 [M]-(C15H18O6, calcd. 293.1025).
7'E-4,9-Dihydroxy-3,3',5'-trimethoxy-8,4'-oxyneolign7'-en-9'-al (3)
Pale yellow oil; [α] +6.1º (c 0.49, MeOH); UV (MeOH) λmax/nm (log ε) 320.0 (3.37), 229.0 (3.56) and 202.5 (3.94); IR (KBr) νmax/cm-1 3441, 1659, 1624, 1583, 1502, 1425, 1243, 1224, 1033, 976 and 817; 1H NMR (acetone) 9.56 (d, J 7.5 Hz, H-9'); 7.50 (d, J 15.7 Hz, H-7'); 7.03 (br. s, H-2', 6'); 6.93 (br. s, H-2); 6.73 (d, J 7.6 Hz, H-6); 6.68 (dd, J 15.7, 7.5 Hz, H-8'); 6.66 (d, J 7.6 Hz, H-5); 4.88 (d, J 5.0 Hz, H-7); 4.17 (br. s, H-8); 3.81 (s, MeO-3', 5'); 3.74-3.79 (m, Ha-9); 3.37 (br. d, J 11.8 Hz, Hb-9); 13C NMR 194.1 (C-9'); 154.6 (C-3', 5'); 153.6 (C-7'); 148.0 (C-3); 146.6 (C-4); 139.3 (C-4'); 133.8(C-1); 131.1 (C-1'); 129.2 (C-8'); 120.1 (C-6); 115.3 (C-5); 110.9 (C-2); 107.0 (C-2', 6'); 88.1(C-8); 73.5 (C-7); 61.0 (C-9); 56.8 (C-3', 5'-OMe); 56.2 (C-3-OMe); ESIMS m/z 403 [M -H]-, 439 [M + Cl]-; HRESIMS m/z (%) 403.1390 [M]-(C21H24O8, calcd. 403.1392).
+6.1º (c 0.49, MeOH); UV (MeOH) λmax/nm (log ε) 320.0 (3.37), 229.0 (3.56) and 202.5 (3.94); IR (KBr) νmax/cm-1 3441, 1659, 1624, 1583, 1502, 1425, 1243, 1224, 1033, 976 and 817; 1H NMR (acetone) 9.56 (d, J 7.5 Hz, H-9'); 7.50 (d, J 15.7 Hz, H-7'); 7.03 (br. s, H-2', 6'); 6.93 (br. s, H-2); 6.73 (d, J 7.6 Hz, H-6); 6.68 (dd, J 15.7, 7.5 Hz, H-8'); 6.66 (d, J 7.6 Hz, H-5); 4.88 (d, J 5.0 Hz, H-7); 4.17 (br. s, H-8); 3.81 (s, MeO-3', 5'); 3.74-3.79 (m, Ha-9); 3.37 (br. d, J 11.8 Hz, Hb-9); 13C NMR 194.1 (C-9'); 154.6 (C-3', 5'); 153.6 (C-7'); 148.0 (C-3); 146.6 (C-4); 139.3 (C-4'); 133.8(C-1); 131.1 (C-1'); 129.2 (C-8'); 120.1 (C-6); 115.3 (C-5); 110.9 (C-2); 107.0 (C-2', 6'); 88.1(C-8); 73.5 (C-7); 61.0 (C-9); 56.8 (C-3', 5'-OMe); 56.2 (C-3-OMe); ESIMS m/z 403 [M -H]-, 439 [M + Cl]-; HRESIMS m/z (%) 403.1390 [M]-(C21H24O8, calcd. 403.1392).
Cell culture and cytotoxicity assays
Marc-145 cells (Cell Bank of the Chinese Academy of Sciences, Shanghai) were grown in monolayer in a 5% carbon dioxide and 95% atmosphere at 37 ºC, with Dulbecco's Modified Eagle Medium (DMEM; HyClone) containing 10% fetalbovineserum (HyClone), 100 U mL-1 of penicillin, and 1 × 10-4 g mL-1 of streptomycin. Cytotoxicity assays were performed by the WST-8 method,37,38 using Cell Counting Kit-8 (CCK-8; Beyotime Co.) according to the supplier recommendations. Briefly, cells were incubated in a 96-well microculture plate (Corning) in the absence or presence of twofold serial dilutions of compounds 1-7, and tilmicosin phosphate (Hubei Hengshuo Chemical Co. Ltd., China). After 3 days of culture, 10 µL of CCK-8 solution were added, and the cells were incubated for 1.5 h. The number of surviving cells was measured with a Bio-Tek ELx 800. ELISAmicroplate reader could show the detection wavelength of 450 nm (L1) and the reference wavelength of 650 nm (L2). The 50% cytotoxic concentration (CC50) was obtained by nonlinear regression analysis of logistic curves (the value of L1 -L2 to different concentrations of compounds).
Cytopathic effect inhibition assay
YN-1 strain of PRRSV was isolated from local pigs in Yunnan Province, P. R. China.39 The antiviral activity of tested compounds against viruses was measured by the CPE inhibition assay.24 Tissue culture medium infective dose (TCID50) of 500 viral particles with twofold serial dilutions of the compounds were added to each test well, and the plates were reincubated for 4 days to allow development of a CPE if any. A non-infection control was made in the absence of natural products and tilmicosin phosphate was used for drug control. The concentration reducing CPE by 50% with respect to virus control was estimated from graphic plots and was defined IC50. The selectivity index was calculated from the ratio CC50/IC50.
PRRSV mRNA expression inhibition assay
The mRNA expression of PRRSV ORF7 and NSP9 genes was determined by real-time RT-PCR.29-31 Briefly, after 4 days of incubation, total virus RNA of both administration and control groups was isolated using RNAisoTM Plus (TaKaRa Biotechnology, Dalian, China), dissolved in 30 µL of RNase-free water (TaKaRa), and then stored at -80 ºC. According to the GenBank data base Accession No. PRU87392, primers were selected and designed from conserved regions based on the ORF7 and NSP9 sequences using Primer5.0 and Oligo6.0 software. A 330 bp fragment of PRRSV ORF7 gene was amplified using the following primers: forward primer was 5'-AATGGCCAGCCAGTCAATCA-3' and reverse primer was 5'-TCATGCTGAGGGTGATGCTG-3'. A 162 bp fragment of PRRSV NSP9 gene was amplified using the following primers: forward primer was 5'-CACTAAAGAGGAAGTCGCACTCA-3' and reverse primer was 5'-GGTATGTCTCCAAACCTTGTATTCTG-3'. A 130 bp fragment of beta-actin gene was amplified using the following primers: forward primer was 5'-ATCCAGGCTGTGCTGTCC-3' and reverse primer was 5'-GAGGATCTTCATGAGGTAGTCG-3'.
cDNAs were synthesized using PrimeScript RT® Reagent Kit (TaKaRa) with 10 µL of reaction mixtures containing 4.5 µL of RNase Free dH2O, 2 µL of 5 × PrimeScript Buffer, 0.5 µL of PrimeScript RT Enzyme Mix I, 0.5 µL of Random 6 mers (100 µmol L-1), 0.5 µL of Oligo dT primer (50 µmol L-1) and 2 µL of total RNA. The reaction programme was as follows: 37 ºC for 15 min, 85 ºC for 5 s. The PCR mixture (25 µL) contained 12.5 µL of SYBR® Primix Ex TaqTMII (TaKaRa), 0.5 µL of PCR forward primer (10 µmol L-1), 0.5 µL of PCR reverse primer (10 µmol L-1), 9.5 µL of dH2O and 2 µL of cDNA. The reactions were carried out in an iQ5 real time PCR (Bio-Rad. Co. Ltd.). The reaction programme was as follows: one cycle at 95 ºC for 30 s, followed by 40 cycles at 95 ºC for 5 s, 60 ºC for 30 s.
Statistical analyses
All experiments were performed in three replications. Continuous variables, expressed as mean ± standard deviation, were compared using one-way ANOVA (analysis of variance). Statistical analyses were conducted with SPSS 17.0 Statistics software.
Conclusions
In this research, seven compounds were isolated from the stems of C. longgangensis var. hongyao. Compounds 1 and 2 are new quinonoids, and 3 is a new neolignan. The inhibitory activity of all compounds against PRRSV was measured by the CPE method. Among the quinones (1, 2 and 4-6), only the active compound 5 belongs to the anthraquinone type. The results suggest that the carbon skeleton of anthraquinone might be necessary for the PRRSV-inhibitory activity.
Supplementary Information
Supplementary data (Figures S1-S18) are available free of charge at http://jbcs.sbq.org.br as PDF file.
Acknowledgements
This work was funded by the National Natural Science Foundation of China (Nos. 31070288, 20972166, 31160509 and 31161140345), Yunnan Provincial
Program for Excellent Scientists (No. 2009CI125), the Key Research Direction Project of Knowledge Innovation Program for Chinese Academy of Sciences (No. KSCX2-EW-J-24), and the Ministry of Education of China through its 111 and 985 projects (Nos. B08044, MUC985-9 and MUC98506-01000101). We thank Dr. Xiao-Nian Li at the Kunming Institute of Botany who measured and elucidated the crystal structure.
References
1. Tian, K. G.; Yu, X. L.; Zhao, T. Z.; Feng, Y. J.; Cao, Z.; Wang, C. B.; Hu, Y.; Chen, X. Z.; Hu, D. M.; Tian, X. S.; Liu, D.; Zhang, S.; Deng, X. Y.; Ding, Y. Q.; Yang, L.; Zhang, Y. X.; Xiao, H. X.; Qiao, M. M.; Wang, B.; Hou, L. L.; Wang, X. Y.; Yang, X. Y.; Kang, L. P.; Sun, M.; Jin, P.; Wang, S. J.; Kitamura, Y.; Yan, J. H.; Gao, G. F.; PLoS One 2007,6,1.
2. Karuppannan, A. K.; Wu, K. X.; Qiang, J.; Chu, J. J. H.; Kwang, J.; Antvirial Res. 2012,94,188.
3. Meng, X. J.; Vet. Microbiol. 2000,74,309.
4. Huang, Y. W.; Meng, X. J.; Virus Res. 2010,154,141.
5. Abad, M. J.; Bermejo, P.; Stud. Nat. Prod. Chem. 2005,30,303.
6. Inoue, K.; Ueda, S.; Nayeshiro, H.; Inouye, H.; Chem. Pharm. Bull. 1982,30,2265.
7. Thomson, R. H.; Pharm. World Sci. 1991,13,70.
8. Cai, X. H.; Luo, X. D.; Zhou, J.; Hao, X. J.; J. Nat. Prod. 2005,68,797.
9. Verdan, M. H.; Barison, A.; de Sá, E. L.; Salvador, M. J.; Poliquesi, C. B.; Eberlin, M. N.; Stefanello, M. É. A.; J. Nat. Prod. 2010,73,1434.
10. Xiao, X. B.; Lin, Y. X.; Xu, G. B.; Gong, X. B.; Gu, Y.; Tong, J. F.; Yang, J.; Helv. Chim. Acta 2011,94,404.
11. Wang, W. T.; Pan, K. Y.; Li, Z. Y.; Wetzman, A. L.; Skog, L. E.; Flora of China, vol. 18; Science Press: Beijing, P. R. China, 2010, p. 334.
12. China National Group Corporation of Traditional & Herbal Medicine; The Chinese Traditional Medicine Resource Records; Science Press: Beijing, P. R. China, 1994, p. 1177.
13. Wang, M. Y.; Yang, L.; Tu, Y. Y.; China Journal of Chinese Materia Medica 2005,30,1921.
14. Wang, M. Y.; Yang, L.; Tu, Y. Y.; China Journal of Chinese Materia Medica 2006,31,307.
15. Wang, M. Y.; Fan, Y. J.; Zhang, J.; Gong, M. X.; China Journal of Chinese Materia Medica 2010,35,3188.
16. Cai, X. H.; Luo, X. D.; Zhou, J.; Hao, X. J.; J. Asian Nat. Prod. Res. 2006,8.351.
17. Wang, M. Y.; Gong M. X.; Zhang, D.; Yang, L.; Acta Pharmacol. Sin. 2011,46,179.
18. Chen, D. F.; Zhu, H. W.; Zhang, Y. Y.; Fudan University, Chin. Pat. 200710044099.5, 2007.
19. Li, S. F.; Di, Y. T.; Wang, Y. H.; Tan, C. J .; Fang, X.; Zhang, Y.; Zheng, Y. T.; Li, L.; He, H. P.; Li, S. L.; Hao, X. J.; Helv. Chim. Acta 2010,93,1795.
20. Kuo, S. C.; Chen, P. L.; Lee, S. W.; Chen, Z. T.; J. Chin. Chem. Soc. 1995,42,869.
21. Li, B.; Zhang, D. M.; Luo, Y. M.; Acta. Pharmacol. Sinica 2006,41,426.
22. Deepak, M.; Handa, S. S.; Phytochemistry 1998,49,269.
23. Morota, T.; Yang, C. X.; Sasaki, H.; Qin, W. Z.; Sugama, K.; Miao, K. L.; Yoshino, T.; Xu, L. H.; Maruno, M.; Yang, B. H.; Phytochemistry 1995,39,1153.
24. Ma, S. C.; He, Z. D.; Deng, X. L.; But, P. P. H.; Ooi, V. E. C.; Xu, H. X.; Lee, S. H. S.; Lee, S. F.; Chem. Pharm. Bull. 2001,49,1471.
25. Chang, P.; Lee, K. H.; J. Nat. Prod. 1985,48,948.
26. Itokawa, H.; Ibraheim, Z. Z.; Qiao, Y. F.; Takeya, K.; Chem. Pharm. Bull. 1993,41,1869.
27. Wu, T. S.; Lin, D. M.; Shi, L. S.; Damu, A. G.; Kuo, P. C.; Kuo, Y. H.; Chem. Pharm. Bull. 2003,51,948.
28. Thuong, P. T.; Lee, C. H.; Dao, T. T.; Nguyen, P. H.; Kim, W. G.; Lee, S. J.; Oh, W. K.; J. Nat. Prod. 2008,71,1775.
29. Pfaffl, M. W.; Nucleic Acids Res. 2001,29,2002.
30. Lurchachaiwong, W.; Payungporn, S.; Srisatidnarakul, U.; Mungkundar, C.; Theamboonlers, A.; Poovorawan, Y.; Lett. Appl. Microbiol. 2008,46,55.
31. Wang, Y. W.; Luo, R.; Fang, L. R.; Wang, D.; Bi, J.; Chen, H. C.; Xiao, S. B.; Mol. Immunol. 2011,48,586.
32. Fang, Y.; Snijder, E. J.; Virus Res. 2010,154,61.
33. Snijder, E. J.; Meulenberg, J. J. M.; J. Gen. Virol. 1998,79,961.
34. He, Y. X.; Hua, R. H.; Zhou, Y. J.; Qiu, H. J.; Tong, G. Z.; Antiviral Res. 2007,74,83.
35. Hao, X. F.; Lu, Z. J.; Kuang, W. D.; Sun, P.; Fu, Y.; Wu, L.; Zhao, Q.; Bao, H. F.; Fu, Y. F.; Cao,Y. M.; Li, P. H.; Bai, X. W.; Li, D.; Liu, Z. X.; J. Virol. 2011,8,1.
36. Sheldrick, G. M.; SHELXS-97, Program for Crystal Structure Solution; University of Göttingen: Göttingen, Germany, 1997.
37. Ishiyama, M.; Shiga, M.; Sasamoto, K.; Mizoguchi, M.; He, P. G.; Chem. Pharm. Bull. 1993,41,1118.
38. Tominaga, H.; Ishiyama, M.; Ohseto, F.; Sasamoto, K.; Hamamoto, T.; Suzuki, K.; Watanabe, M.; Anal. Commun. 1999,36,47.
39. Duan, B. F.; Shen, Y. P.; Yang, G. S.; Zhang, Y. F.; Wu, J. M.; Duan, G.; Yin, G. F.; Prog. Vet. Med. 2010,31,11.
Submitted: June 15, 2012
Published online: October 30, 2012
- 1. Tian, K. G.; Yu, X. L.; Zhao, T. Z.; Feng, Y. J.; Cao, Z.; Wang, C. B.; Hu, Y.; Chen, X. Z.; Hu, D. M.; Tian, X. S.; Liu, D.; Zhang, S.; Deng, X. Y.; Ding, Y. Q.; Yang, L.; Zhang, Y. X.; Xiao, H. X.; Qiao, M. M.; Wang, B.; Hou, L. L.; Wang, X. Y.; Yang, X. Y.; Kang, L. P.; Sun, M.; Jin, P.; Wang, S. J.; Kitamura, Y.; Yan, J. H.; Gao, G. F.; PLoS One 2007,6,1.
- 2. Karuppannan, A. K.; Wu, K. X.; Qiang, J.; Chu, J. J. H.; Kwang, J.; Antvirial Res. 2012,94,188.
- 3. Meng, X. J.; Vet. Microbiol 2000,74,309.
- 4. Huang, Y. W.; Meng, X. J.; Virus Res. 2010,154,141.
- 5. Abad, M. J.; Bermejo, P.; Stud. Nat. Prod. Chem 2005,30,303.
- 6. Inoue, K.; Ueda, S.; Nayeshiro, H.; Inouye, H.; Chem. Pharm. Bull. 1982,30,2265.
- 7. Thomson, R. H.; Pharm. World Sci. 1991,13,70.
- 8. Cai, X. H.; Luo, X. D.; Zhou, J.; Hao, X. J.; J. Nat. Prod. 2005,68,797.
- 9. Verdan, M. H.; Barison, A.; de Sá, E. L.; Salvador, M. J.; Poliquesi, C. B.; Eberlin, M. N.; Stefanello, M. É. A.; J. Nat. Prod. 2010,73,1434.
- 10. Xiao, X. B.; Lin, Y. X.; Xu, G. B.; Gong, X. B.; Gu, Y.; Tong, J. F.; Yang, J.; Helv. Chim. Acta 2011,94,404.
- 11. Wang, W. T.; Pan, K. Y.; Li, Z. Y.; Wetzman, A. L.; Skog, L. E.; Flora of China, vol. 18; Science Press: Beijing, P. R. China, 2010, p. 334.
- 12. China National Group Corporation of Traditional & Herbal Medicine; The Chinese Traditional Medicine Resource Records; Science Press: Beijing, P. R. China, 1994, p. 1177.
- 13. Wang, M. Y.; Yang, L.; Tu, Y. Y.; China Journal of Chinese Materia Medica 2005,30,1921.
- 14. Wang, M. Y.; Yang, L.; Tu, Y. Y.; China Journal of Chinese Materia Medica 2006,31,307.
- 15. Wang, M. Y.; Fan, Y. J.; Zhang, J.; Gong, M. X.; China Journal of Chinese Materia Medica 2010,35,3188.
- 16. Cai, X. H.; Luo, X. D.; Zhou, J.; Hao, X. J.; J. Asian Nat. Prod. Res. 2006,8351.
- 17. Wang, M. Y.; Gong M. X.; Zhang, D.; Yang, L.; Acta Pharmacol. Sin 2011,46,179.
- 18. Chen, D. F.; Zhu, H. W.; Zhang, Y. Y.; Fudan University, Chin. Pat. 200710044099.5, 2007.
- 19. Li, S. F.; Di, Y. T.; Wang, Y. H.; Tan, C. J .; Fang, X.; Zhang, Y.; Zheng, Y. T.; Li, L.; He, H. P.; Li, S. L.; Hao, X. J.; Helv. Chim. Acta 2010,93,1795.
- 20. Kuo, S. C.; Chen, P. L.; Lee, S. W.; Chen, Z. T.; J. Chin. Chem. Soc. 1995,42,869.
- 21. Li, B.; Zhang, D. M.; Luo, Y. M.; Acta. Pharmacol. Sinica 2006,41,426.
- 22. Deepak, M.; Handa, S. S.; Phytochemistry 1998,49,269.
- 23. Morota, T.; Yang, C. X.; Sasaki, H.; Qin, W. Z.; Sugama, K.; Miao, K. L.; Yoshino, T.; Xu, L. H.; Maruno, M.; Yang, B. H.; Phytochemistry 1995,39,1153.
- 24. Ma, S. C.; He, Z. D.; Deng, X. L.; But, P. P. H.; Ooi, V. E. C.; Xu, H. X.; Lee, S. H. S.; Lee, S. F.; Chem. Pharm. Bull. 2001,49,1471.
- 25. Chang, P.; Lee, K. H.; J. Nat. Prod. 1985,48,948.
- 26. Itokawa, H.; Ibraheim, Z. Z.; Qiao, Y. F.; Takeya, K.; Chem. Pharm. Bull. 1993,41,1869.
- 27. Wu, T. S.; Lin, D. M.; Shi, L. S.; Damu, A. G.; Kuo, P. C.; Kuo, Y. H.; Chem. Pharm. Bull. 2003,51,948.
- 28. Thuong, P. T.; Lee, C. H.; Dao, T. T.; Nguyen, P. H.; Kim, W. G.; Lee, S. J.; Oh, W. K.; J. Nat. Prod 2008,71,1775.
- 29. Pfaffl, M. W.; Nucleic Acids Res. 2001,29,2002.
- 30. Lurchachaiwong, W.; Payungporn, S.; Srisatidnarakul, U.; Mungkundar, C.; Theamboonlers, A.; Poovorawan, Y.; Lett. Appl. Microbiol 2008,46,55.
- 31. Wang, Y. W.; Luo, R.; Fang, L. R.; Wang, D.; Bi, J.; Chen, H. C.; Xiao, S. B.; Mol. Immunol 2011,48,586.
- 32. Fang, Y.; Snijder, E. J.; Virus Res 2010,154,61.
- 33. Snijder, E. J.; Meulenberg, J. J. M.; J. Gen. Virol 1998,79,961.
- 34. He, Y. X.; Hua, R. H.; Zhou, Y. J.; Qiu, H. J.; Tong, G. Z.; Antiviral Res 2007,74,83.
- 35. Hao, X. F.; Lu, Z. J.; Kuang, W. D.; Sun, P.; Fu, Y.; Wu, L.; Zhao, Q.; Bao, H. F.; Fu, Y. F.; Cao,Y. M.; Li, P. H.; Bai, X. W.; Li, D.; Liu, Z. X.; J. Virol 2011,8,1.
- 36. Sheldrick, G. M.; SHELXS-97, Program for Crystal Structure Solution; University of Göttingen: Göttingen, Germany, 1997.
- 37. Ishiyama, M.; Shiga, M.; Sasamoto, K.; Mizoguchi, M.; He, P. G.; Chem. Pharm. Bull 1993,41,1118.
- 38. Tominaga, H.; Ishiyama, M.; Ohseto, F.; Sasamoto, K.; Hamamoto, T.; Suzuki, K.; Watanabe, M.; Anal. Commun 1999,36,47.
- 39. Duan, B. F.; Shen, Y. P.; Yang, G. S.; Zhang, Y. F.; Wu, J. M.; Duan, G.; Yin, G. F.; Prog. Vet. Med. 2010,31,11.
Publication Dates
-
Publication in this collection
09 Nov 2012 -
Date of issue
Oct 2012
History
-
Received
15 June 2012 -
Accepted
30 Oct 2012