Abstract
A number of knowledge management systems have been developed to allow users to have access tolargequantity of neuroanatomical data. The advent of three-dimensional (3D) visualization techniques allows users to interact with complex 3D object. In order to better understand the structural and functional organization of the brain, we present Neuroanatomical Affiliations Visualization-Interface System (NAVIS) as the original software to see brain structures and neuroanatomical affiliations in 3D.
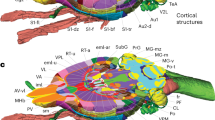
This version of NAVIS has made use of the fifth edition of “The Rat Brain in Stereotaxic coordinates” (Paxinos and Watson, 2005). The NAVIS development, environment was based on the scripting language name Python, using visualization toolkit (VTK) as 3D-library and wxPython for the graphic user interface. The following manuscript is focused on the nucleus of the solitary tract (Sol) and the set of affiliated structures in the brain to illustrate the functionality of NAVIS. The nucleus of the Sol is the primary relay center of visceral and taste information, and consists of 14 distinct subnuclei that differ in cytoarchitecture, chemoarchitecture, connections, and function. In the present study, neuroanatomical projection data of the rat Sol were collected from selected literature in PubMed since 1975. Forty-nine identified projection data of Sol were inserted in NAVIS. The standard XML format used as an input for affiliation data allows NAVIS to update data online and/or allows users to manually change or update affiliation data. NAVIS can be extended to nuclei other than Sol.
Similar content being viewed by others
References
Boldogkoi, Z., Sik, A., Denes, A., et al. (2004) Novel tracing paradigms—genetically engineered herpesviruses as tools for mapping functional circuits within the CNS: present status and future prospects. Prog. Neurobiol. 72, 417–445.
Borgefors, G. (1986) Distance transformations in digitalimages. Comput. Vision Graphics Image Process. 34, 344–371.
Bota, M., Dong, H. W., and Swanson, L. W. (2005) Brain architecture management system. Neuroinformatics 3, 15–48.
Bowden D. M., and Dubach, M. F. (2003) Neuro Names 2002. Neuroinformatics 1, 43–59.
Brevik A., Leergaard T. B., Svanevik, M., and Bjaalie, J. G. (2001) Three-dimensional computerized atlas of the rat brain stem precerebellar system: approaches for mapping, visualization, and comparison of spatial distribution data. Anat. Embryol. (Berl.) 204, 319–332.
Funka-Lea, G. D. and Schwaber, J. S. (1994) A digital brain atlas and its application to the visceral neuraxis. J. Neurosci. Methods 54, 253–260.
Geiger, B., (1993) PhD thesis, Three-dimensional modeling of human organs and its application to diagnosis and surgical planning, Ecole Nationale Supérieure des Mines de Paris, Paris, France.
Leergaard T. B., Bjaalie, J. G., Devor A., Wald, L. L. and Dale, A. M. (2003) In vivo tracing of major rat brain pathways using manganese-enhanced magnetic resonance imaging and three-dimensional digital atlasing. Neuroimage 20, 1591–1600.
Paxinos, G. (2004) The Rat Nervous System, 3rd ed., Elsevier, New York.
Paxinos, G. and Watson, C. (2005) The rat brain in stereotaxic coordinates, 5th ed., Academic Press. New York.
Rosenfeld, A. and Platz, J. L. (1966) Sequential operations in digital picture processing. J. ACM 13, 471–494.
Schroeder, W., Martin, K., and Lorensen, B. (2004) The Visualization Toolkit, An Object-Oriented Approach To 3D Graphics, 3rd ed., Kitware, Inc., New York
Toga, A. W., Samaie, M., and Payne, B. A. (1989) Digitalrat brain: a computerized atlas. Brain Res. Bull. 22, 323–333.
Toga, A. W., Santori, E. M., Hazani, R., and Ambach, K. (1995) A3D digitalmap of rat brain. Brain Res. Bull. 38, 77–85.
Vercelli, A., Repici, M., Garbossa, D., and Grimaldi, A. (2000) Recent techniques for tracing pathways in the central nervous system of developing and adult mammals. Brain Res. Bull. 51, 11–28.
Vittone, J. (1999) PhD thesis, Caractérisation et reconnaissance de droites et de plans en géométrie discrète, Université Joseph Fourrier, Grenoble, France.
Author information
Authors and Affiliations
Corresponding author
Additional information
Note NAVIS software V1.0 (for Microsoft Windows) is available for download at the URL: http://www-evasion.imag.fr/Membres/Olivier.Palombi
Rights and permissions
About this article
Cite this article
Palombi, O., Shin, JW., Watson, C. et al. Neuroanatomical affiliation visualization-interface system. Neuroinform 4, 299–317 (2006). https://doi.org/10.1385/NI:4:4:299
Issue Date:
DOI: https://doi.org/10.1385/NI:4:4:299