Abstract
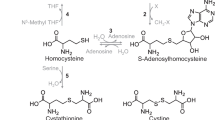
The purpose of this study was to identify the seleno-l-methionine (l-SeMet) α,γ-elimination enzyme that catalyzes l-SeMet to generate methylselenol (CH3SeH), a notable intermediate for the metabolism of selenium compounds, in mammalian tissues. The enzyme purified from ICR mouse liver was separated by one-dimensional gel electrophoresis, and the specific band was subjected to in-gel trypsin digestion followed by matrix-assisted laser desorption/ionization-time-of-flight mass spectrometric analysis. In the peptide mass fingerprinting search, the mass numbers of 14 peptides produced by tryptic digestion of the enzyme were consistent with the theoretical mass numbers calculated from the amino acid sequence of murine cystathionine γ-lyase (E.C. 4.4.1.1). The peptide sequence tags search was also performed to obtain the amino acid sequence data of five tryptic peptides. These peptides were significantly identical to the partial amino acid sequences of cystathionine γ-lyase. This enzyme was clearly shown to catalyze the α, γ-elimination reaction of l-cystathionine by the enzymological research. The K m value for the catalysis of l-cystathionine was 0.81 mM and V max was. 0.0013 unit/mg protein. These results suggested that cystathionine γ-lyase catalyzes l-SeMet to generate CH3SeH by its α,γ-elimination reaction.
Similar content being viewed by others
References
K. Yasumoto, T. Suzuki, and M. Yoshida, Identification of selenomethionine in soybean protein, J. Agric. Food Chem. 36, 463–467 (1988).
H. Chassaigne, C. C. Chery, G. Bordin, et al., Development of new analytical methods for selenium speciation in selenium-enriched yeast material, J. Chromatogr. A 976, 409–22 (2002).
G. N. Schrauzer, The nutritional signficance, metabolism and toxicology of selenomethionine, Adv. Tood Nutr. Res. 47, 73–112 (2003).
K. Takahashi, N. Avissar, J. Whitin, et al., Purification and characterization of human plasma glutathione peroxidase: a selenoglycoprotein distinct from the known cellular enzyme, Arch. Biochem. Biophys. 256, 677–686 (1987).
J. Yarimizu, H. Nakamjra, I. Yodoi, et al., Efficiency of selenocyteine incorporation in human thioredoxin redustase, Antixid Redox. Signal. 2, 643–651 (2000).
N. Esaki, T. Nakamura, H. Tanaka, et al., Enzymatic synthesis of selenocyteine in rat liver, Biochemistry 20, 4492–4496 (1981).
E. O. Kajander, R. J. Harvima, T. O. Eloranta, et al., Metabolism cellular actions and cytotoxicity of selenomethionine in cuitured cells. Biol. Trace Element Res. 28, 57–68 (1991).
N. Esaki, T. Nakamura, H. Tanaka, et al., Selenocyteine lyase, a novel enzyme that specifically acts on selenocystene, J. Biol. Chem. 257, 4386–4391 (1982).
T. Hasegawa, T. Okuno, K. Nakamuro, et al., Identification and metabolism of seleno-cysteine-glutathione selenenyl sulfide (CySeSG) in small intestine of mice orally exposed to selenocyteine, Arch. Technol. 71, 39–44 (1996).
H. E. Ganther, Pathways of selenium metabolism including respiratory excretory products, J. Am. Coll. Toxicol. 5, 1–5 (1986).
Y. Ogra, K. Ishiwata, H. Takayama, et al., Identification of novel selenium metabolite, Se-methyl-n-acetyl-selenohexosamine, in rat urine by the HPLC-ICP MS and-ESI MS/MS methods, J Chromatogr, B 767, 301–312 (2002).
Y. Kobayashi, Y. Ogra, K. Ishiwata, et al., Selenosugars are key and urinary metabolites for selenium excretion within the required to low-toxic range, Proc. Natl Acad. Sci. USA 99, 15,932–15,936 (2002).
C. Ip, H. J. Thompson, Z. Zhu, et al., In vitro and in vivo studies of methylseleninic acid: evidence that a monomethylated selenium metabolite is critical for cancer chemoprevention, Cancer Res. 60, 2882–2886 (2000).
T. Okuno, T. Kuroda, T. Kuroda, et al., Contribution of enzymic α,γ-elimination reaction in detoxification pathway of selenomethionine in mouse liver, Toxicol. Appl. Pharmacol. 176, 18–23 (2001).
T. Okuno, S. Motobayashi, H. Ueno, et al., Purification and characterization of mouse hepatic enzyme that converts selenomethionine to methylselenol by its α,γ-elimination, Biol. Trace Element Res., in press.
W. J. Henzel, T. M. Billeci, J. T. Stults, et al., Identifying proteins from two-dimensional gels by molecular mass searching of peptide fragments in protein sequence databases, Proc. Natl. Acad. Sci. USA 90, 5011–5015 (1993).
M. Mann, P. Hojrup, and P. Roepstorff, Use of mass spectrometric molecular weight information to dentify proteins in sequence databases, Biol. Mass Spectrom. 22, 338–345 (1993).
J. R. Yates III, S. Speicher, P. R. Griffin, et al., Peptide mass maps: a highly informative approach to protein indentification, Anal. Biochem. 214, 397–408 (1993).
P. R. Griffin, M. J. MacCoss, J. K. Eng, et al., Direct database searching with MALDI-PSD spectra of peptides, Rapid. Commun. Mass Spectrom 9, 1546–1551 (1995).
M. Mann and M. Wilm, Error-tolerant identification of peptides in sequence database by peptide sequence tag, Anal. Chem. 66, 4390–4399 (1994).
U. K. Leammli, Cleavage of structural proteins during the assembly of the head of bacteriophage T4, Nature 227, 680–685 (1970).
F. Gharahdaghi, C. R. Weinberg, D. A. Meagher, et al., Mass spectrometric identification of proteins from silver-stained polyacrylamide gel: a method for the removal of silver ions to enhance sensitivity, Electrophoresis 20, 601–605 (1999).
M. Yanagida, Y. Miura, K. Yagasaki, et al., Matrix assisted laser desorption/ionization time of flight-mass spectrometry analysis of proteins detected by anti-phosphotyrosine antibody on two-dimensional-gels of fibroblast cell lysates after tumor necrosis factor-α stimulation, Electrophoresis 21, 1890–1898 (2000).
M. Kussmann, E. Nordhoff, H. Rahbek-Nielsen, et al., Matrix-assisted laser desorption/ionization mass spectrometry sample preparation techniques designed for various peptide and protein analysis. J. Mass Spectrom. 32, 593–601 (1997).
H. Lineweaver and D. Burk, The detemination of enzyme dissociation constants, J. Am. Chem. Soc. 56, 658–666 (1934).
H. E. Ganther and C. Ip, Thioredoxin reducides activity in rat liver is not affected by supranutritional levels of monomethy lated selenum in vivo and is inhibited only by high levels of selenium in vitro, J. Nutr. 131, 301–304 (2001).
M. M. Bradford, A rapid and sensitive method for the quantiation of microgram quantities of protein utilizing the principle, Anal. Biochem. 72, 248–254 (1976).
D. J. C. Pappin, P. Hojrup, and A. J. Bleasby, Rapid identification of proteins by peptidemass fingerprinting, Curr. Biol. 3, 327–332 (1993).
R. S. Johnson, S. A. Martin, K. Birmann, et al., Novel fragmentation process of peptides by collision induced decomposition in a tandem mass spectrometer: differention of leucine and isoleucine, Anal. Chem. 59, 2621–2625 (1987).
P. Edman, Sequence determination, Mol. Biol. Biochem. Biophys. 8, 211–255 (1970).
A. Shevchenko, M. Wilm, O. Vorm, et al., Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels, Anal. Chem. 68, 850–858 (1996).
R. Deutzmann, Structural characterization of proteins and peptides. Methods Mol Med 94, 269–297 (2004).
I. Bikel, T. N. Pavlatos, and D. M. Livingston, Purification and subunit structure of mouse liver crystathionase, Arch. Biochem. Biophys. 186, 168–174 (1978).
W. R. Carroll, G. W. Stacy, and V. du Vigneaud, α-ketobutyric acid as production in the enzymatic cleavage of cystathionine, J. Biol. Chem. 180, 375–382 (1949).
Y. Matsuo and D. M. Greenberg, A crystalline enzyme that cleaves homoserine and cystathionine. I. Isolation procedure and some physicochemical properties, J. Biol. Chem. 230, 545–560 (1958).
B. J. Lee, S. G. Kang, and D. L. Hatfield, Transcription of Xenopus selenocysteine tRNA Ser (formerly designated opal suppressor phosphoserine tRNA) gene is directed by multiple 5′-extragenic regulatory elements. J. Biol. Chem. 264, 9696–9702 (1989).
B. J. Lee, M. Rajagopalan, Y. S. Kim, et al., Selenocyteine tRNA [Ser]Sec gene is ubiquitous within the animal king dom, Mol. Cell. Biol. 10, 1940–1949 (1990).
T. Mizutani, H. Kurata, and K. Yamada, Study of mammalian selenocysteyl-tRNA synthesis with [75Se]H Se−1, FEBS Lett. 289, 59–63 (1991).
T. Mizutani, H. Kurata, and K. Yamada, et al., Some properties of murine selenocysteine synthase, Biochem. J. 284, 827–834 (1992).
H. E. Ganther, Metabolism of hydrogen selenide and methylated selenides, Adv. Nutr. Res. 2, 107–128 (1979).
T. A. Pascal, G. E. Gaull, N. G. Bertis, et al., Vitamin B6-responsive and-unresponsive cystathioninuria: two variant molecular forms, Science 190, 1209–1211 (1975).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Okuno, T., Motobayashi, S., Ueno, H. et al. Identification of mouse selenomethionine α, γ-elimination enzyme. Biol Trace Elem Res 108, 245–257 (2005). https://doi.org/10.1385/BTER:108:1-3:245
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1385/BTER:108:1-3:245