Synthesis, Characterization and Anti Mycobacterial Activity of Novel Hydrazones
Kulandai Therese S and G Geethamalika
Department of Chemistry, Nirmala College for Women, Coimbatore, Tamil Nadu, India.
Corresponding Author E-mail: kulandaifspm@gmail.com
DOI : http://dx.doi.org/10.13005/ojc/330140
The hydrazone Schiff base analogues namely benzoic acid (4-allyloxy-benzylidene)-hydrazide and its series were designed and synthesized. The structures of newly synthesized compounds were characterized by analytical methods and spectral analysis and subjected to antibacterial and docking studies. The synthesized molecules were subjected to molecular docking studies using enoyl-acyl–carrier protein reductase (NADH) from Mycobacterium tuberculosis as the receptor. The docking results confirm the binding affinity of the synthesized compounds with the selected receptor. Preliminary in-vitro anti bacterial studies were carried out with Gram-positive (Staphylococcus aureus) and Gram-negative (Escherichia coli) bacteria. Benzoic acid (4-allyloxy-benzylidene)-hydrazide was found to be most potent anti tuberculosis agent at 200 and 100µg per ml, the percentage of inhibition was 98.7% and 97.9%. The designed synthesized molecules were considered for evaluation for the molecular docking studies of their binding pattern with enoyl-acyl-carrier protein reductase from Mycobacterium tuberculosis. The antimycobacterial screening was performed against MTB H37Rv an isoniazid-resistant clinical isolate of MTB was used for the study.
KEYWORDS:hydrazones; spectral studies; docking and anti mycobacterial activity
Download this article as:| Copy the following to cite this article: Therese S. K, Geethamalika G. Synthesis, Characterization and Anti Mycobacterial Activity of Novel Hydrazones. Orient J Chem 2017;33(1). |
| Copy the following to cite this URL: Therese S. K, Geethamalika G. Synthesis, Characterization and Anti Mycobacterial Activity of Novel Hydrazones. Orient J Chem 2017;33(1). Available from: http://www.orientjchem.org/?p=29782 |
Introduction
Tuberculosis (TB) is a worldwide pandemic caused by different species of mycobacteria. Mycobacterium tuberculosis is a successful pathogen, which causes tuberculosis and is the greatest single infectious cause of death worldwide, killing approximately two million people annually (1). Hydrazides are highly reactive bases with reducing properties. They have been used as a synthetic intermediate to produce different type of drugs in the treatment of different chronic illnesses. Hydrazides used in medicine include the anti-tuberculosis drug isoniazid and the antihypertensive and peripheral vasodilator drug hydralsane, whose hydrochloride is an effective drug for emergency reduction of blood pressure in hypertensive crisis. In the same way the hydrazine derivatives have been widely used in the treatment of tuberculosis and mental disorder. They can also be used as antimicrobial, antihypertensive, antimalarial and antitumoral agents (2-11).
Schiff bases are considered to be among the most important group of compounds in medicinal chemistry due to their preparative accessibility, structural variety and wide biological profile (12). The synthesized benzohydrazides were potent antimicrobial and anticancer agents (13). Numbers of hydrazide-hydrazone derivatives possess antituberculosis activity (14-21). The present study is aimed to carry out design, synthesis, in silico and in-vitro antimicrobial studies for the series of benzoic acid (4-allyloxy-benzylidene)-hydrazides.
Experimental
All the chemicals and solvents used are of analytical grade and were purchased from Royal scientific company and used without further purification. TLC was run on the silica coated aluminium sheets (TLC silica gel 60 F254, Analytical chromatography) and visualized in low UV light. IR spectra in KBr pellets were recorded on the FT-IR Perkin Elmer Spectrum spectrophotometer. 1H NMR and C13NMR spectra were recorded on a 300 M Hz spectrometer with DMSO-d6 as solvent and TMS as internal reference. Chemical shifts are quoted as δ ppm and the coupling constants J in Hz in signals are described as s (singlet), d (doublet), t (triplet), m (multiplet) and b (broad). Melting points were determined on a Sigma melting point apparatus without corrections. GC MS mass spectrum was taken in JEOL GCMATE II GC-MS with Data system in a high resolution, double focusing instrument.
General Procedure for the Synthesis of Benzohydrazides (3a-E)
The substituted benzoic acid (2.46 m mol) was refluxed with methanol in sulphuric acid for 4h. The ester formed reacts with hydrazine and gets converted to benzohydrazides by hydrozinolysis (22).
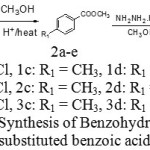 |
Scheme 1: Synthesis of Benzohydrazide from substituted benzoic acid |
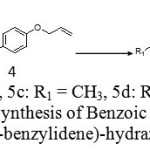 |
Scheme 2: Synthesis of Benzoic acid (4-allyloxy benzylidene)-hydrazides |
Synthesis of Benzoic Acid (4-Allyloxy-Benzylidene)-Hydrazide (5a-E)
Benzaldehyde undergoes nucleophilic addition reaction with aromatic hydrazides resulting in the formation of hydrazones. The completion of reaction and the homogeneity of the synthesized compounds were checked by TLC. Further the product formed was washed with petroleum ether, dried, recrystallized and characterized by spectral analysis.
Ethanolic solution of 4-allyloxy benzaldehyde (10 m mol) was added to a 10 m mol of ethanolic solution of benzohydrazides and the reaction mixture was stirred for 30 minutes at room temperature in the presence of glacial acetic acid as catalyst. After refluxing for 3-5 h the completion of the reaction was monitored by TLC. The reaction mixture was cooled and placed in refrigerator overnight. The resulting solid was filtered and washed with petroleum ether and recrystallised with ethanol.
In Silico Charaterisation
The synthesized compounds were assessed in silico for their biological activity using PASS server (23). The three dimensional structure of the protein / enzymes was retrieved from Protein Data Bank (24). The compound under study was docked with the identified receptor using Autodock (25) software and further analyzed for interactions using Pymol.
Antimicrobial Activity
Antibacterial screening of the synthesized compounds was evaluated against Staphylococcus aureus and Escherichia coli, and the zone of inhibition was measured in mm. The broth media was inoculated and grown at 37ºC for 18 h to revive the bacteria from the stock cultures. The agar plates with wells were inoculated by spreading evenly with 18h old bacterial cultures (100 μl, 10-4 cfu). After 20 min, various concentrations of the compound and the antibiotic were filled in the wells. Finally, the plates were incubated at 37ºC for 24 h and the inhibition zone was observed (26).
Luciferase Reporter Phage (LRP) Assay (27)
The MTB H37Rv and clinical isolate of MTB were grown in Middlebrook 7H9 complete medium 12 with and without test sample for 3 days at 37˚C. LRP assay was done using concentrations of 200, 100 and 50 µg/mL of test samples. 50µL bacterial suspension equivalent to MacFarlands.No.2 standard was added to 400 µL of 7H9 with and without the test compound. For each sample, two drug-free controls and two drug concentrations were prepared and this set up was incubated for 72 h at 37˚C. After incubation, 50µL of the high titer luciferase reporter phage (phAETRC202) and 40 µL of 0.1 M CaCl2 were added to all the vials and this set up was incubated at 37˚C for 4 h. After incubation, 100 µL of the mixture was taken from each tube into a Luminometer cuvette and an equal amount of working D-luciferin (0.3 mM in 0.05 M sodium citrate buffer, pH 4.5) solution was added. The Relative Light Units (RLU) was measured after 10s of integration in the Luminometer. Readings were recorded in duplicate for each sample and the mean was calculated. The percentage reduction in the RLU was calculated for each test sample and compared with that of control.
Broth Microdilution Method (28)
The MIC for MTB H37Rv and isoniazid (INH) resistant clinical isolate of MTB was determined using a broth microdilution method (28) in Middlebrook 7H9 medium supplemented with OADC, with a final inoculum of 1×107cfu/mL. The compounds were dissolved in DMF (1.25 mg/ mL) and used as stock solution. Concentrations ranging from 1250 to 1 µg/mL were used to assess the effectiveness of the compounds. After inoculation the microtiter tubes were incubated at 37˚C for 72 h, and the growth inhibition was recorded for 14 and 21 days, respectively. The MIC value represents the lowest dilution of the compound at which no bacterial growth was detected.
Results and Discussion
Benzoic Acid (4-Allyloxy-Benzylidene)-Hydrazide (5a)
was prepared by the above mentioned method. benzohydrazide (0.136g 0.001mol 1 equivalent) 4-allyloxybenzaldehyde (0.15ml 0.001mol 1 equivalent), pale yellow amorphous solid, yield: 90%. m.p. 118-120˚C. FT-IR (υmax KBr pellets, cm-1) 935(N-N) str, 1256 (C-O-C), 1542 (C=C), 1650 (C=N) str, 3032 (C-H) str. 1H NMR (CDCl3, 300 MHz) δH: 5.29-5.45 (dd, 1.2Hz, 2 H), 5.9-6.1(m, 1H), 4.57(d, 2H, 5.1Hz), 6.9(d, 8.1Hz, 2H), 7.7 (d, 6.6Hz, 2H), 8.23 (1H, s), 9.2 (1H, s), 7.8 (d, 6.6 Hz, 2H), 7.4-7.5 (m, 3H). 13C NMR (DMSO 300 MHz): 160, 132, 131, 129, 128, 127, 126, 117, 114 and 68. m/z = 315 (M+1). Molecular Formula C17H16N2O2.
4-Chloro-Benzoic Acid (4-Allyloxy-Benzylidene)-Hydrazide (5b)
was prepared by the above mentioned method. 4-chlorobenzohydrazide (0.171g 0.001mol 1 equivalent) 4-allyloxybenzaldehyde (0.15ml 0.001mol 1 equivalent), pale yellow amorphous solid, yield: 92%. m.p. 158-160˚C. FT-IR (υmax KBr pellets, cm-1) 3434 or 3283 (N-H), 2930Ar (C-H) str, 2849Ali (CH) str, 1652 (C=O) str, 1506 (C=N) str, DMSO, 300 MHz) δH: 5.2-5.3 (d, d, 2H1.5Hz), 5.9-6.1 (CH 1H m), 4.63 (d 2H, 5.1 Hz), 7.0(d, 2H, 8.7 Hz), 7.61 (d, 2H, 8.7Hz), 5.3-5.4 (d, d 1.5-1.8Hz, 1H), 8.38 (1H, s), 7.9 (d, 2H, 8.7Hz), 7.6 (d, 2H, 9 Hz). 13C NMR (DMSO 300 MHz) 161, 159, 147 136, 133, 132, 129, 128, 128, 117, 115 and 68. m/z = 315 (M+1). Molecular Formula C17H15ClN2O2.
4-Methyl-Benzoic Acid (4-Allyloxy-Benylidene)-Hydrazide (5c)
was prepared by the above mentioned method. 4-methyl benzohydrazide (0.15g 0.001mol 1 equivalent) 4-allyloxybenzaldehyde (0.15ml 0.001mol 1 equivalent), yellow amorphous solid, yield: 88%. m.p. 168˚C. FT-IR (υmax KBr pellets, cm-1) 937 (N=N), 1654 (C=O), 1526 (C=N). 1H NMR (DMSO, 300 MHz) δH: 5.23- 5.24 (d, 2H), 5.89(s, 1H), 4.61 (2H), 6.8(d, 2H), 7.5(d, 2H), 8.1(s, 1H), 8(s, 1H), 7.83(d, 2H), 7.24(d, 2H), 2.35(S, 3H). 13C NMR (DMSO 300 MHz) 115, 137, 75, 164, 114, 130, 123, 154, 127, 130.5, 129, 141 and 20. m/z = 292 (M+1). Molecular Formula C18H18N2O2.
4-Methoxy-Benzoic Acid (4-Allyloxy-Benylidenehydrazide (5d)
was prepared by the above mentioned method. 4-methoxy benzohydrazide (0.166g 0.001mol 1 equivalent) 4-allyloxybenzaldehyde (0.15ml 0.001mol 1 equivalent), light yellow amorphous solid, yield: 88%. m.p.162˚C. FT-IR (υmax KBr pellets, cm-1) 1640 (C=N), 937 (N-N), 1410 (C-O), 2918 Ar (C-H). 1H NMR (DMSO, 300 MHz) δH: 5.23- 5.24 (d, 2H), 5.89(s, 1H), 4.61 (2H), 6.8(d, 2H), 7.5(d, 2H), 8.1(s, 1H), 8(s, 1H), 7.84(d, 2H), 6.95(d, 2H), 3.73(s, 3H). 13C NMR (DMSO 300 MHz) 115, 137, 75, 164, 114, 130, 123, 154, 125, 128, 114, 165 and 56. m/z = 292 (M+1). Molecular Formula C18H18N2O2.
4- Hydroxy -Benzoic Acid (4-Allyloxy-Benylidene)- Hydrazide (5e)
was prepared by the above mentioned method. 4-hydroxy benzohydrazide (0.152g 0.001mol 1 equivalent) 4-allyloxybenzaldehyde (0.15ml 0.001mol 1 equivalent), yellow amorphous solid, yield: 84%. m.p. 178˚C. FT-IR (υmax KBr pellets, cm-1) 1535 (C=C), 1625 (C=N), 3250 (N-H), 1658 (C=O). 1H NMR (DMSO, 300 MHz) δH: 5.23- 5.24 (d, 2H), 5.89(s, 1H), 4.61 (2H), 6.8(d, 2H), 7.5(d, 2H), 8.1(s, 1H), 8(s, 1H), 7.78 (d, 2H), 6.91 (d, 2H), 5 (S, 1H). 13C NMR (DMSO 300 MHz) 115, 137, 75, 164, 114, 130, 123, 154, 126, 128, 115 and 160. m/z = 310 (M+1). Molecular Formula C18H18N2O3.
Synthesized compounds 5a, 5b, were subjected to a preliminary anti bacterial screening against the strain gram positive (Staphylococcus aureus) and Gram-negative (Escherichia coli) bacteria to assess the antibacterial susceptibility of the test compounds. The compounds showed good inhibition at lower concentration.
Table 1: Activity prediction for the synthesized compounds
|
Compound code |
Activity |
Pa |
Pi |
|
5a |
Antituberculosic |
0.831 |
0.003 |
|
5b |
Antituberculosic |
0.827 |
0.003 |
|
5c |
Antituberculosic | 0.794 | 0.003 |
|
5d |
Antituberculosic | 0.781 | 0.003 |
|
5e |
Antituberculosic | 0.805 | 0.003 |
It is evident from table-1 that the compound 5a exhibits maximum anti-tuberculosis activity among all the study compounds as its probability of being active (Pa) value is 0.831 and Probability of being inactive (Pi) is only 0.003. Also all the other compounds demonstrate anti-tuberculosic activity with Pa between 0.827 and 0.781. Hence, all the study compounds were docked with the target Enoyl-[acyl-carrier-protein] reductase [NADH] from Mycobacterium tuberculosis. The interaction of the synthesized compound with the target was evaluated using Docking studies. The docking scores along with the other parameters for the different poses of the docked molecules are presented in Tables 2(a) – 2(e).
Table 2a: Docking results of Benzoic acid (4-allyloxy-benzylidene)-hydrazide with the receptor
|
Pose |
B.E |
L.E |
IC (nM) |
Int.E |
Vdw |
Elec.E |
Total.IE |
Tor.E |
Unb.E |
No. of H bonds |
Hydrogen bond |
|
1 |
-11.81 |
-0.56 |
2.22 |
-13.6 |
-13.59 |
-0.01 |
-0.92 |
1.79 |
-0.92 |
1 |
Met98:HN::Lig1:O |
|
2 |
-10.79 |
-0.51 |
12.37 |
-12.58 |
-12.58 |
0.0 |
-0.81 |
1.79 |
-0.81 |
0 |
– |
|
3 |
-10.73 |
-0.51 |
13.61 |
-12.52 |
-12.49 |
-0.03 |
-0.97 |
1.79 |
-0.97 |
0 |
– |
|
4 |
-10.72 |
-0.51 |
13.94 |
-12.51 |
-12.52 |
0.01 |
-1.03 |
1.79 |
-1.03 |
0 |
– |
|
5 |
-10.68 |
-0.51 |
14.91 |
-12.47 |
-12.42 |
-0.05 |
-1.14 |
1.79 |
-1.14 |
1 |
Ile194:HN::Lig1:N |
|
6 |
-10.44 |
-0.5 |
22.27 |
-12.23 |
-12.2 |
-0.03 |
-1.07 |
1.79 |
-1.07 |
0 |
– |
|
7 |
-10.33 |
-0.49 |
26.91 |
-12.12 |
-12.04 |
-0.07 |
-0.95 |
1.79 |
-0.95 |
1 |
Tyr158:HH::Lig1:O |
|
8 |
-9.84 |
-0.47 |
61.29 |
-11.63 |
-11.62 |
-0.01 |
-0.85 |
1.79 |
-0.85 |
2 |
Lig1:N::Ala201:O Lig1:N::Ala201:O |
|
9 |
-9.82 |
-0.47 |
63.87 |
-11.61 |
-11.58 |
-0.03 |
-1.23 |
1.79 |
-1.23 |
1 |
Ile194:HN::Lig1:N |
|
10 |
-9.56 |
-0.46 |
98.8 |
-11.35 |
-11.26 |
-0.09 |
-1.18 |
1.79 |
-1.18 |
1 |
Tyr158:HH::Lig1:N |
The docking results of Enoyl-[acyl-carrier-protein] reductase [NADH] from Mycobacterium tuberculosis with the ligand Benzoic acid (4-allyloxy-benzylidene)-hydrazide is presented in table 2a. The binding energies of the different binding conformations range between -11.81 and -9.56 kcal/mol. Among the ten different docked conformations conformation 8 had a binding energy of -9.84 kcal/mol with 2 hydrogen bonds between the ligand Benzoic acid (4-allyloxy-benzylidene)-hydrazide and Ala 201. The conformation 1, Conformation 5, Conformation 7, Conformation 9 and Conformation 10 had only one hydrogen bond formed with binding energies -11.81, -10.68, -10.33, -9.82 and -9.56 kcal/mol respectively. Hence, conformation 8 with 2 hydrogen bonds and binding energy of -9.84 kcal/mol is considered the best docked conformation and represented in figure-1
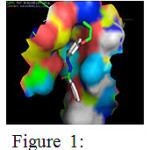 |
Figure 1: Binding interactions between Benzoic acid (4-allyloxy-benzylidene)-hydrazide with the receptor. Click here to View Figure |
Table 2b: Docking results of 4-chloro-benzoic acid (4-allyloxy-benzylidene)-hydrazide with the receptor
|
Pose |
B.E | L.E | IC (nM) | Int.E | Vdw | Elec.E | Total.IE | Tor.E | Unb.E | No. of H bonds | Hydrogen bond |
| 1 | -11.87 | -0.54 | 1.99 | -13.66 | -13.66 | -0.01 | -0.85 | 1.79 | -0.85 |
3 |
Lig1:O::4oim:Ala201:OLig1:N::4oim:Ala201:OLig1:N::4oim:Ala201:O |
| 2 | -11.31 | -0.51 | 5.09 | -13.1 | -13.11 | 0.01 | -0.81 | 1.79 | -0.81 |
3 |
Lig1:O::Gly205:OLig1:N::Ala201:OLig1:N::Ala201:O |
| 3 | -11.07 | -0.5 | 7.67 | -12.86 | -12.89 | 0.03 | -0.83 | 1.79 | -0.83 |
1 |
Lig1:O::Gly96:O |
| 4 | -11.04 | -0.5 | 8.12 | -12.83 | -12.86 | 0.04 | -0.79 | 1.79 | -0.79 |
3 |
Lig1:O::Ala201:OLig1:O::Gly96:OLig1:N::Met98:O |
| 5 | -11.01 | -0.5 | 8.44 | -12.8 | -12.8 | 0.0 | -0.81 | 1.79 | -0.81 |
0 |
– |
| 6 | -10.54 | -0.48 | 18.78 | -12.33 | -12.34 | 0.01 | -0.63 | 1.79 | -0.63 |
1 |
Met98:HN::Lig1:O |
| 7 | -10.51 | -0.48 | 19.66 | -12.3 | -12.36 | 0.05 | -0.54 | 1.79 | -0.54 |
0 |
– |
| 8 | -10.42 | -0.47 | 22.98 | -12.21 | -12.18 | -0.03 | -0.73 | 1.79 | -0.73 |
1 |
Lig1:O::Ile194:O |
| 9 | -9.86 | -0.45 | 58.91 | -11.65 | -11.63 | -0.02 | -0.8 | 1.79 | -0.8 |
1 |
Lig1:N::Ala201:O |
| 10 | -8.71 | -0.4 | 414.7 | -10.5 | -10.47 | -0.02 | -0.78 | 1.79 | -0.78 |
3 |
Ala157:HN::Lig1:N,NAsn 159:HD21::Lig1:OLig1:N::Asn159:OD1 |
The above table presents the ten different binding conformations of Enoyl-[acyl-carrier-protein] reductase [NADH] from Mycobacterium tuberculosis with the ligand 4-chloro-benzoic acid (4-allyloxy-benzylidene)-hydrazide. The binding energy ranges from -11.87 kcal/mol to -8.71 kcal/mol. The conformations 1,2,4 and 10 had three hydrogen bonds between the receptor, Enoyl-[acyl-carrier-protein] reductase [NADH] and ligand 4-chloro-benzoic acid (4-allyloxy-benzylidene)-hydrazide. In the first binding conformation, all the three hydrogen bonds are formed between the ligand and Alanine 201 in Enoyl-[acyl-carrier-protein] reductase [NADH] protein. But in the second binding conformation two hydrogen bonds are formed between the ligand and Alanine 201 and the third hydrogen bond is formed between the ligand and Glycine 205 in the receptor. Alternatively in the binding conformation 4, Alanine 201, Glycine 96 and Methionine 98 in the receptor form one hydrogen bond each with the ligand. Finally, the conformation 10 had 3 hydrogen bonds formed each between Alanine 157, Asparagine 159, Asparagine 159 and the ligand with a binding energy of -8.71 kcal/mol. Among the ten different binding conformations, the conformation 1 with the least binding energy and three hydrogen bonds is chosen is as the best possible orientation which is displayed in Figure-2
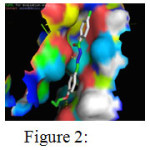 |
Figure 2: Binding interactions between 4-chloro-benzoic acid (4-allyloxy-benzylidene)-hydrazide with the receptor Click here to View Figure |
Table 2c: Docking results of 4-methyl-benzoic acid (4-allyloxy-benylidene)- hydrazide with the receptor
|
Pose |
B.E | L.E | IC (nM) | Int.E | Vdw | Elec.E | Total.IE | Tor.E | Unb.E | No. of H bonds | Hydrogen bond |
|
1 |
-11.37 | -0.52 | 4.64 | -13.46 | -13.48 | 0.03 | -0.8 | 2.09 | -0.8 | 2 | LIG1:N::MET98:OLIG1:N::MET98:O |
|
2 |
-11.17 | -0.51 | 6.48 | -13.26 | -13.25 | -0.01 | -0.85 | 2.09 | -0.85 | 0 | – |
|
3 |
-10.69 | -0.49 | 14.59 | -12.78 | -12.78 | 0.0 | -0.85 | 2.09 | -0.85 | 0 | – |
|
4 |
-10.61 | -0.48 | 16.73 | -12.7 | -12.68 | -0.02 | -0.91 | 2.09 | -0.91 | 2 | LIG1:O::MET98:OMET98:HN::LIG1:O |
|
5 |
-10.4 | -0.47 | 23.61 | -12.49 | -12.47 | -0.03 | -0.92 | 2.09 | -0.92 | 0 | – |
|
6 |
-10.3 | -0.47 | 28.01 | -12.39 | -12.34 | -0.05 | -0.83 | 2.09 | -0.83 | 1 | ILE194:HN::LIG1:O |
|
7 |
-10.02 | -0.46 | 45.14 | -12.11 | -12.08 | -0.02 | -0.94 | 2.09 | -0.94 | 0 | – |
|
8 |
-9.98 | -0.45 | 48.37 | -12.07 | -12.01 | -0.05 | -1.09 | 2.09 | -1.09 | 1 | TYR158:HH::LIG1:O |
|
9 |
-9.74 | -0.44 | 72.73 | -11.83 | -11.72 | -0.11 | -1.05 | 2.09 | -1.05 | 0 | – |
|
10 |
-9.7 | -0.44 | 77.03 | -11.79 | -11.76 | -0.03 | -0.94 | 2.09 | -0.94 | 0 | – |
The above table depicts the different binding orientations of the ligand K1b with the receptor Enoyl-[acyl-carrier-protein] reductase [NADH] from Mycobacterium tuberculosis. Poses 1 and 4 have two hydrogen bonds formed by ‘O’ of Met98 and ‘N’ and ‘O’ atoms in the ligand. Poses 6 and 8 had one hydrogen bond each between ‘H’ atom of Ile 194 and Tyr 158 respectively with the ‘O’ atom of the Ligand. Among the ten different binding conformations, the conformation 1 with binding energy of -11.37 kcal/mol and two hydrogen bonds is chosen as the best possible orientation which is displayed in Figure 3.
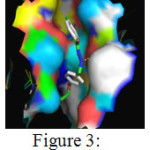 |
Figure 3: Binding interactions between 4-methyl-benzoic acid (4-allyloxy-benylidene)- hydrazide with the receptor Click here to View Figure |
Table 2d: Docking results of 4-methoxy-benzoic acid (4-allyloxy-benylidene)- hydrazide with the receptor
|
Pose |
B.E | L.E | IC (nM) | Int.E | Vdw | Elec.E | Total.IE | Tor.E | Unb.E | No. of H bonds | Hydrogen bond |
|
1 |
-11.89 | -0.52 | 1.93 | -13.98 | -13.97 | -0.01 | -0.85 | 2.09 | -0.85 | 1 | LIG1:N::ALA201:O |
|
2 |
-10.96 | -0.48 | 9.29 | -13.05 | -13.09 | 0.04 | -0.82 | 2.09 | -0.82 | 1 | LIG1:O::PRO156:O |
|
3 |
-10.89 | -0.47 | 10.41 | -12.98 | -12.98 | 0.0 | -0.85 | 2.09 | -0.85 | 2 | LIG:N::ALA201:OLIG1:N::ALA201:O |
|
4 |
-10.8 | -0.47 | 12.02 | -12.89 | -12.89 | 0.0 | -0.91 | 2.09 | -0.91 | 4 | LIG1:N::ALA201:OLIG1:O::ASN106:OD1LIG1:O::GLY96:OLIG1:O::ALA201:O |
|
5 |
-10.62 | -0.46 | 16.4 | -12.71 | -12.67 | -0.03 | -0.87 | 2.09 | -0.87 | 0 | – |
|
6 |
-10.41 | -0.45 | 23.36 | -12.5 | -12.46 | -0.04 | -0.93 | 2.09 | -0.93 | 1 | LIG1:O::MET98:O |
|
7 |
-10.16 | -0.44 | 35.45 | -12.25 | -12.27 | 0.02 | -0.73 | 2.09 | -0.73 | 1 | LIG1:O::PRO156:O |
|
8 |
-9.7 | -0.42 | 77.07 | -11.79 | -11.67 | -0.12 | -0.84 | 2.09 | -0.84 | 1 | MET98:HN::LIG1:O |
|
9 |
-9.41 | -0.41 | 125.97 | -11.5 | -11.46 | -0.04 | -0.9 | 2.09 | -0.9 | 1 | LIG1:O::ALA201:O |
|
10 |
-8.98 | -0.39 | 259.88 | -11.07 | -10.94 | -0.13 | -0.85 | 2.09 | -0.85 | 0 | – |
The above table presents the ten different binding conformations of Enoyl-[acyl-carrier-protein] reductase [NADH] from Mycobacterium tuberculosis with the ligand 5d. The binding energy ranges from -11.89 kcal/mol to -8.98 kcal/mol. The poses 1, 2, 6, 7, 8 and 9 had one hydrogen bond between the receptor, Enoyl-[acyl-carrier-protein]reductase [NADH] and ligand 5d. But pose 3 and 4 had 2 and four hydrogen bonds between the receptor and the ligand. Hence, among the ten different binding conformations, the conformation 4 with the binding energy of -10.8 and 4 hydrogen bonds is chosen as the best possible orientation which is displayed in Figure 4.
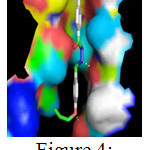 |
Figure 4: Binding interactions between 4-methoxy-benzoic acid (4-allyloxy-benylidene)- hydrazide with the receptor Click here to View Figure |
Table 2e: Docking results of 4- hydroxy -benzoic acid (4-allyloxy-benylidene)- hydrazide with the receptor
|
Pose |
B.E | L.E | IC (nM) | Int.E | Vdw | Elec.E | Total.IE | Tor.E | Unb.E | No. of H bonds | Hydrogen bond |
|
1 |
-11.35 | -0.52 | 4.83 | -13.14 | -13.12 | -0.02 | -0.61 | 1.79 | -0.61 | 1 | LIG:N::ALA201:O |
|
2 |
-10.26 | -0.47 | 30.34- | -12.05 | -12.03 | -0.02 | -0.81 | 1.79 | -0.81 | 0 | – |
|
3 |
-10.25 | -0.47 | 30.41 | -12.04 | -12.08 | 0.03 | -0.81 | 1.79 | -0.81 | 0 | – |
|
4 |
9.97 | -0.45 | 49.19 | -11.76 | -11.74 | -0.02 | -0.58 | 1.79 | -0.58 | 0 | – |
|
5 |
-9.95 | -0.45 | 50.58 | -11.74 | -11.74 | -0.01 | -0.64 | 1.79 | -0.64 | 2 | LIG1:N::ALA201:OLIG1:O::ASN106:OD1 |
|
6 |
-9.78 | -0.44 | 67.28 | -11.57 | -11.59 | 0.02 | -0.73 | 1.79 | -0.73 | 0 | – |
|
7 |
-9.76 | -0.44 | 70.34 | -11.55 | -11.62 | 0.07 | -0.79 | 1.79 | -0.79 | 1 | LIG:O::MET98:O |
|
8 |
-9.74 | -0.44 | 72.95 | -11.53 | -11.49 | -0.04 | -0.77 | 1.79 | -0.77 | 2 | LIG1:O::MET98:OMET98:HN::LIG1:O |
|
9 |
-9.73 | -0.44 | 73.19 | -11.52 | -11.53 | 0.01 | -0.84 | 1.79 | -0.84 | 1 | LIG1:N::ALA201:O |
|
10 |
-7.1 | -0.32 | 6.29 | -8.89 | -8.84 | -0.05 | -0.77 | 1.79 | -0.77 | 0 | – |
The above table presents the ten different binding conformations of Enoyl-[acyl-carrier-protein] reductase [NADH] from Mycobacterium tuberculosis with the ligand 5e. The binding energy ranges from -11.35 kcal/mol to -7.1 kcal/mol. The poses 1, 7 and 9 had one hydrogen bond between the receptor, Enoyl-[acyl-carrier-protein]reductase [NADH] and ligand 5e, whereas conformations 5 and 8 had two hydrogen bonds. Among the ten different binding conformations, the conformation 5 with binding energy of -9.95 kcal/mol and two hydrogen bonds is chosen as the best possible orientation which is displayed in Figure 5
 |
Figure 5: Binding interactions between 4- hydroxy -benzoic acid (4-allyloxy-benylidene)- hydrazide with the receptor Click here to View Figure |
Table 2f: Docking results of isoniazid with the receptor
|
Pose |
B.E | L.E | IC (uM) | Int.E | Vdw | Elec.E | Total.IE | Tor.E | Unb.E | No. of H bonds | Hydrogen bond |
| 1 | -6.73 | -0.67 | 11.61 | -7.03 | -7.0 | -0.04 | -0.09 | 0.3 | -0.09 |
1 |
Iso:O::Ile 202:O |
| 2 | -6.73 | -0.67 | 11.69 | -7.03 | -6.99 | -0.04 | -0.09 | 0.3 | -0.09 |
1 |
Iso:O::Ile 202:O |
| 3 | -6.71 | -0.67 | 12.11 | -7.01 | -6.97 | -0.03 | -0.09 | 0.3 | -0.09 |
1 |
Iso:O::Ile 202:O |
| 4 | -6.67 | -0.67 | 12.94 | -6.97 | -6.98 | 0.01 | -0.09 | 0.3 | -0.09 |
1 |
Iso:O::Ala 201:O |
| 5 | -6.67 | -0.67 | 13.01 | -6.96 | -6.98 | 0.02 | -0.09 | 0.3 | -0.09 |
1 |
Iso:O::Ala 201:O |
| 6 | -6.54 | -0.65 | 16.15 | -6.84 | -6.73 | -0.11 | -0.09 | 0.3 | -0.09 |
0 |
– |
| 7 | -6.04 | -0.60 | 37.35 | -6.34 | -6.26 | -0.08 | -0.09 | 0.3 | -0.09 |
2 |
Tyr158:HH::Iso:O Ile194:HN::Iso:N |
| 8 | -6.04 | -0.60 | 37.62 | -6.33 | -6.25 | -0.08 | -0.09 | 0.3 | -0.09 |
2 |
Tyr158:HH::Iso:O Ile194:HN::Iso:N |
| 9 | -6.00 | -0.60 | 39.78 | -6.30 | -6.21 | -0.09 | -0.09 | 0.3 | -0.09 |
1 |
Ile194:HN::Iso:N |
| 10 | -5.83 | -0.58 | 53.14 | -6.13 | -6.06 | -0.07 | -0.09 | 0.3 | -0.09 |
1 |
Ile194:HN::Iso:O |
The docking results of Enoyl-[acyl-carrier-protein] reductase [NADH] from Mycobacterium tuberculosis with the standard drug Isoniazid is presented in table 7. The binding energies of the different binding conformations range between -6.73 and -5.83 kcal/mol. Among the ten different docked conformations conformation 7 and 8 show 2 hydrogen bonds between the drug and Tyr158 and Ile194. The conformation 1, Conformation 2, Conformation 3, Conformation 4 and Conformation 5 had only one hydrogen bond formed with binding energies -11.81, -10.68, -10.33, -9.82 and -9.56 kcal/mol respectively. Hence, conformation 8 with 2 hydrogen bonds and binding energy of -9.84 kcal/mol is considered the best docked conformation and represented in figure-6
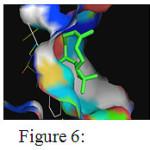 |
Figure 6: Binding interactions between isoniazid with the receptor Click here to View Figure |
Synthesized compounds were evaluated for antimycobacterial activity using Luciferase Reporter Phase (LRP) assay on MTB H37Rv and one isoniazid-resistant (INH-resistant) clinical isolate at National Institute for Research in Tuberculosis (NIRT), Chennai (28).
The percentage growth inhibition was recorded at the concentrations of 200, 100 and 50 µg/mL (Table 3). Seeing the good inhibition of the synthesized compound further biological assays were conducted to determine their MIC values on different strains of Mycobacterium.
Table 3: Antimicrobial activity data of 5a and5b by LPR assay
|
Compound code |
Concentration (mL) |
CFU(col/mL) |
% inhibition |
|
5a |
10 20 50 200 |
3.2×106 |
19.1 0 0 |
|
5b |
100 200 400 |
4.4×106 2.4×106 |
97.9 98.7 |
|
Control-1 |
– |
5.8×106 |
– |
|
Control-2 |
– |
5.0×106 |
– |
Control-1 -H37Rv
control-2 -H37Rv
Results of these investigations indicate that some of the test compounds exhibited significant antimycobacterial activity along with antibacterial activity against all the tested strains.
Summary and Conclusion
A series of hydrazone Schiff base analogues were synthesized in good yield adopting standard procedures. The compounds were characterized by spectral studies. Docking studies of the above mentioned compounds revealed scope for antibacterial activity especially anti-tuberculosis activity. Further, all the study compounds exhibit a binding better than the existing drug isoniazid for Tuberculosis. This was further confirmed using in vivo studies where compound 5b had maximum inhibition (97.9%) 100µmL. Hence, it can be concluded from the study that compound 5b can be further evaluated for its efficiency in treating Tuberculosis.
References
- Ritesh P Bhole, Deepak. D. Borkar, Kishore. P. Bhusari and Prashant. A. Patil, Journal of Korean Chemical Society, 2012, 56(2) 236-245
- Liang Wang, Da-Gang Guo, Yan-Yan Wang and Chang-ZhengZheng, RSC Adv., 2014, 4, 58895-58901
- Vonburg R and Stout T, J. Appl. Toxicol., 1991, 11, 447-450
CrossRef - Chester E O, Norris G and Raymond D D, Anal. Profiles Drug Subt., 1979, 8, 283-314
CrossRef - Bedia K K, Elcin O, Seda U, Fatma K, Nathaly S, Sevim R and Dimogio A, Eur. J. Med. Chem., 2006, 41, 1253-1261
CrossRef - Immanuel N N, Ahmed H, Felicite M M, innocent N N,Offiong E O and Susan A B, Polyhedron, 2013, 63, 207-213
- Kajal A, Bala S, Kamboj S and Saini V, Med. Chem. Res., 2014, 23, 2676-2689
CrossRef - Cikla P, Tatar E, Kucukguzel I, Sahin F, Yurdakul D, Basu A, Krishnan R, Nichols D B, Kaushik-Basu N and Kucukguzel S G, Med. Chem. Res., 2013, 22, 5685-5699
CrossRef - Yildirim L T and Atakol O, Cryst. Res. Technol., 2002, 37, 1352-1359
CrossRef - Yildirim L T, Emregul K C, Kurtaran R, and Atakol O, Cryst. Res. Technol., 2002, 37, 1344-1351
CrossRef - Bedia K K, Oruc- Emre E E, Unsalan S and Rollaas S, Med. Chem. Res., 2009, 18, 277-286
- Rosu, T., Negoiu, M., Pasculescu, S., Pahontu, E., Porier, D., Gulea, A., Eur. J.Med. Chem,2010, 45, 774-789
CrossRef - MahakSaini, Pradeep Kumar, Mahesh kumar, KalavathyRamasamy, Vasudevan Mani, Rakesh Kumar Mishra, Abu Bakar Abdul Majeed, BalasubramanianNarasimhan, Arab. J. of chem., 2014, 7, 448-460
- Kucukguzel S G, Rollas S, Kucukguzel I, Kiraz M, Eur. J.Med. Chem., 1999, 34, 1093-1100
CrossRef - Patole J, Sandbhor U, Padhye S, Deobagkar D N, Anson C E, Powell A, Bioorg. Med.Chem. Lett. 2003, 13, 51-55
CrossRef - Maccari R, Ottana R and Vigorita M G, Bioorg. Med.Chem. Lett. 2005, 15, 2509-2513
CrossRef - Cocco M T, Congiu C, Onnis V, Pusceddu M C, Schivo M L, logu A, Eur. J. Med. Chem.34, 1999, 1071-1076
CrossRef - N. Karali, A. Kocabalkanli, A. Gursoy, O. Ates, Farmaco, , 2002 ,57, 589-593
CrossRef - Rando D G, Santo D N, siqueira L, Malvezzi A, Leite C Q F, Amaral A T, Ferreira E I, Tavares L C, Boiorg. Med. Chem. 2002, 10, 557-560
CrossRef - Kocyigit-KaymakciogluBedia, orucElcin, UnsalanSeda, KandermiriliFatma, ShvetsNathaly, RollasSevim, AnatholyDimoglo, Eur, j. Med. Chem.2006, 41, 1253-1261
- NarenderMalothu, Jaswanth S. Bhandaru, UmasankarKulandaivelub, MalathiJojula, Raghuram Reddy Adidala, Umadevi K. R., Dusthackeer A. V. N, VenkatRao Kaki, Raghuram R. Akkinepally, Bioorganic & Medicinal Chemistry Letters,2016, 26, 836–840
- MonazzaSerwar,TashfeenAkhtar, ShahidHameed and Khalid M. Khan, ARKIVOC 2009 (vii) 210-221
- Langumin A, StepanchikovaAlla, FilimonovDmitrii and Poroikov Vladimir, 2000, PASS: Prediction of activity spectra for biologically active substances, Bioinformatics, 16(8), 747-748
CrossRef - F.C.Bernstein, T.F Koetzle, G.T.Williams, E.E.MeyerJr, M.D.Brice, J.R.Rodgers, O.Kennard, T.Shimanouchi, M.Tasumi, “The Protein Data Bank: A computer based Archival File for Macromolecular Structure” J of Mol.Biol., 1997, 112, 535.
CrossRef - Morris, GM, Huey R, Lindstorm W, Sanner MF, Belew RK, Goodsell, DS and Olson,AJ (2009), Autodock 4 and Autodock Tool 54: automated docking with selective receptor flexibility, J. Computational Chemistry, 2009, 16, 2785-2791.
CrossRef - WALKER R.D.. Antimicrobial susceptibility testing and interpretation of
- results. In: Antimicrobial Therapy in Veterinary Medicine, Prescott J.F., Baggot J.D., Walker R.D., eds. Ames, IA, Iowa State University Press, 2000.,12–26.
- (a) Shawar, R. M.; Humble, D. J.; van Dalfsen, J. M.; Stover, C. K.; Hickey, M. J.; Steele, S.; Mitscher, L. A.; Baker, W. Antimicrob. Agents Chemother.1997, 41, 570;
CrossRef
(b) Prabu, A.; Seenivasan, P.; Kumar, V. Asian J. Med. Sci. 2014, 5, 54;
(c) Dusthackeer, A.; Kumar, V.; Subbian, S.; Sivaramakrishnan, G.; Zhu, G.; Subramanyam, B.; Hassan, S.; Nagamaiah, S.; Chan, J.; Rama, N. P. J. Microbiol. Methods2008, 73, 18.
CrossRef - Tran, T.; Saheba, E.; Arcerio, A. V.; Chavez, V.; Li, Q.-Y.; Martinez, L. E.; Primm, T.P. Bioorg. Med. Chem. 2004, 12, 4809.
CrossRef

This work is licensed under a Creative Commons Attribution 4.0 International License.









