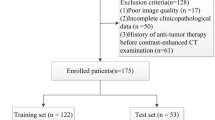
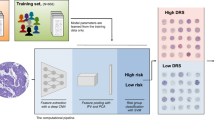
Abstract
Objective
The aim of this study was to develop quantitative feature-based models from histopathological images to distinguish hepatocellular carcinoma (HCC) from adjacent normal tissue and predict the prognosis of HCC patients after surgical resection.
Methods
A fully automated pipeline was constructed using computational approaches to analyze the quantitative features of histopathological slides of HCC patients, in which the features were extracted from the hematoxylin and eosin (H&E)-stained whole-slide images of HCC patients from The Cancer Genome Atlas and tissue microarray images from West China Hospital. The extracted features were used to train the statistical models that classify tissue slides and predict patients’ survival outcomes by machine-learning methods.
Results
A total of 1733 quantitative image features were extracted from each histopathological slide. The diagnostic classifier based on 31 features was able to successfully distinguish HCC from adjacent normal tissues in both the test [area under the receiver operating characteristic curve (AUC) 0.988] and external validation sets (AUC 0.886). The random-forest prognostic model using 46 features was able to significantly stratify patients in each set into longer- or shorter-term survival groups according to their assigned risk scores. Moreover, the prognostic model we constructed showed comparable predicting accuracy as TNM staging systems in predicting patients’ survival at different time points after surgery.
Conclusions
Our findings suggest that machine-learning models derived from image features can assist clinicians in HCC diagnosis and its prognosis prediction after hepatectomy.




Similar content being viewed by others
References
Mak LY, Cruz-Ramon V, Chinchilla-Lopez P, Torres HA, LoConte NK, Rice JP, et al. Global epidemiology, prevention, and management of hepatocellular carcinoma. Am Soc Clin Oncol Educ Book. 2018;38:262–79. https://doi.org/10.1200/edbk_200939.
Rastogi A. Changing role of histopathology in the diagnosis and management of hepatocellular carcinoma. World J Gastroenterol. 2018;24(35):4000–13. https://doi.org/10.3748/wjg.v24.i35.4000.
Qin LX, Tang ZY. The prognostic molecular markers in hepatocellular carcinoma. World J Gastroenterol. 2002;8(3):385–92. https://doi.org/10.3748/wjg.v8.i3.385.
Lauwers GY, Terris B, Balis UJ, Batts KP, Regimbeau JM, Chang Y, et al. Prognostic histologic indicators of curatively resected hepatocellular carcinomas: a multi-institutional analysis of 425 patients with definition of a histologic prognostic index. Am J Surg Pathol. 2002;26(1):25–34. https://doi.org/10.1097/00000478-200201000-00003.
Calderaro J, Couchy G, Imbeaud S, Amaddeo G, Letouze E, Blanc JF, et al. Histological subtypes of hepatocellular carcinoma are related to gene mutations and molecular tumour classification. J Hepatol. 2017;67(4):727–38. https://doi.org/10.1016/j.jhep.2017.05.014.
Ziol M, Pote N, Amaddeo G, Laurent A, Nault JC, Oberti F, et al. Macrotrabecular-massive hepatocellular carcinoma: a distinctive histological subtype with clinical relevance. Hepatology. 2018;68(1):103–12. https://doi.org/10.1002/hep.29762.
Calderaro J, Meunier L, Nguyen CT, Boubaya M, Caruso S, Luciani A, et al. ESM1 as a Marker of macrotrabecular-massive hepatocellular carcinoma. Clin Cancer Res. 2019;25(19):5859–5865. https://doi.org/10.1158/1078-0432.ccr-19-0859.
Calderaro J, Ziol M, Paradis V, Zucman-Rossi J. Molecular and histological correlations in liver cancer. J Hepatol. 2019;71(3):616–30. https://doi.org/10.1016/j.jhep.2019.06.001.
Cooper LA, Kong J, Gutman DA, Dunn WD, Nalisnik M, Brat DJ. Novel genotype-phenotype associations in human cancers enabled by advanced molecular platforms and computational analysis of whole slide images. Lab Invest. 2015;95(4):366–76. https://doi.org/10.1038/labinvest.2014.153.
van den Bent MJ. Interobserver variation of the histopathological diagnosis in clinical trials on glioma: a clinician’s perspective. Acta Neuropathol. 2010;120(3):297–304. https://doi.org/10.1007/s00401-010-0725-7.
Friemel J, Rechsteiner M, Frick L, Bohm F, Struckmann K, Egger M, et al. Intratumor heterogeneity in hepatocellular carcinoma. Clin Cancer Res. 2015;21(8):1951–61. https://doi.org/10.1158/1078-0432.ccr-14-0122.
Bishop JW, Marshall CJ, Bentz JS. New technologies in gynecologic cytology. J Reprod Med. 2000;45(9):701–19.
Hipp J, Flotte T, Monaco J, Cheng J, Madabhushi A, Yagi Y, et al. Computer aided diagnostic tools aim to empower rather than replace pathologists: Lessons learned from computational chess. J Pathol Inform. 2011;2:25. https://doi.org/10.4103/2153-3539.82050.
Cooper LA, Demicco EG, Saltz JH, Powell RT, Rao A, Lazar AJ. PanCancer insights from The Cancer Genome Atlas: the pathologist’s perspective. J Pathol. 2018;244(5):512–24. https://doi.org/10.1002/path.5028.
Beck AH, Sangoi AR, Leung S, Marinelli RJ, Nielsen TO, van de Vijver MJ, et al. Systematic analysis of breast cancer morphology uncovers stromal features associated with survival. Sci Transl Med. 2011;3(108):108ra13. https://doi.org/10.1126/scitranslmed.3002564.
Ji MY, Yuan L, Jiang XD, Zeng Z, Zhan N, Huang PX, et al. Nuclear shape, architecture and orientation features from H&E images are able to predict recurrence in node-negative gastric adenocarcinoma. J Transl Med. 2019;17(1):92. https://doi.org/10.1186/s12967-019-1839-x.
Luo X, Zang X, Yang L, Huang J, Liang F, Rodriguez-Canales J, et al. Comprehensive Computational Pathological Image Analysis Predicts Lung Cancer Prognosis. J Thorac Oncol. 2017;12(3):501–9. https://doi.org/10.1016/j.jtho.2016.10.017.
Sertel O, Kong J, Catalyurek UV, Lozanski G, Saltz JH, Gurcan MN. Histopathological Image Analysis Using Model-Based Intermediate Representations and Color Texture: Follicular Lymphoma Grading. J. Signal Process. Syst. 2008;55(1):169. https://doi.org/10.1007/s11265-008-0201-y.
Sertel O, Kong J, Shimada H, Catalyurek UV, Saltz JH, Gurcan MN. Computer-aided prognosis of neuroblastoma on whole-slide images: classification of stromal development. Pattern Recognit. 2009;42(6):1093–103. https://doi.org/10.1016/j.patcog.2008.08.027.
Yu KH, Zhang C, Berry GJ, Altman RB, Re C, Rubin DL, et al. Predicting non-small cell lung cancer prognosis by fully automated microscopic pathology image features. Nat Commun. 2016;7:12474. https://doi.org/10.1038/ncomms12474.
Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–4. https://doi.org/10.1158/2159-8290.cd-12-0095.
Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. https://doi.org/10.1126/scisignal.2004088.
Linkert M, Rueden CT, Allan C, Burel JM, Moore W, Patterson A, et al. Metadata matters: access to image data in the real world. J Cell Biol. 2010;189(5):777–82. https://doi.org/10.1083/jcb.201004104.
Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7(10):R100. https://doi.org/10.1186/gb-2006-7-10-r100.
Kamentsky L, Jones TR, Fraser A, Bray MA, Logan DJ, Madden KL, et al. Improved structure, function and compatibility for CellProfiler: modular high-throughput image analysis software. Bioinformatics 2011;27(8):1179–80. https://doi.org/10.1093/bioinformatics/btr095.
Breiman LJML. Random forests. Mach. Learn. 2001;45(1):5–32. https://doi.org/10.1023/a:1010933404324.
Lê S, Josse J, Husson F. FactoMineR: An R Package for Multivariate Analysis. J. Stat. Softw. 2008;25(1):1–18. https://doi.org/10.18637/jss.v025.i01.
Ishwaran H, Kogalur UB, Blackstone EH, Lauer MS. Random survival forests. Ann Appl Stat. 2008;2(3):841–60. https://doi.org/10.1214/08-aoas169.
Ishwaran H, Kogalur UB, Gorodeski EZ, Minn AJ, Lauer MS. High-dimensional variable selection for survival data. J. Am. Stat. Assoc. 2010;105(489):205–17. https://doi.org/10.1198/jasa.2009.tm08622.
Martins-Filho SN, Paiva C, Azevedo RS, Alves VAF. Histological grading of hepatocellular carcinoma-a systematic review of literature. Front Med (Lausanne). 2017;4:193. https://doi.org/10.3389/fmed.2017.00193.
Sobin LH, Compton CC. TNM seventh edition: what’s new, what’s changed: communication from the International Union Against Cancer and the American Joint Committee on Cancer. Cancer. 2010;116(22):5336–9. https://doi.org/10.1002/cncr.25537.
Blanche P, Dartigues J-F, Jacqmin-Gadda H. Estimating and comparing time-dependent areas under receiver operating characteristic curves for censored event times with competing risks. 2013;32(30):5381–97. https://doi.org/10.1002/sim.5958.
Chiang C-T, Hung H. Non‐parametric estimation for time-dependent AUC. J. Stat. Plan. Inference. 2010;140(5):1162–74. https://doi.org/10.1016/j.jspi.2009.10.012.
Friemel J, Rechsteiner M, Frick L, Böhm F, Struckmann K, Egger M, et al. Intratumor heterogeneity in hepatocellular carcinoma. Clin. Cancer Res. 2015;21(8):1951. https://doi.org/10.1158/1078-0432.ccr-14-0122.
Kim HD, Song GW, Park S, Jung MK, Kim MH, Kang HJ, et al. Association between expression level of PD1 by tumor-infiltrating CD8(+) T cells and features of hepatocellular carcinoma. Gastroenterology. 2018;155(6):1936-50.e17. https://doi.org/10.1053/j.gastro.2018.08.030.
Calderaro J, Rousseau B, Amaddeo G, Mercey M, Charpy C, Costentin C, et al. Programmed death ligand 1 expression in hepatocellular carcinoma: relationship With clinical and pathological features. Hepatology. 2016;64(6):2038–46. https://doi.org/10.1002/hep.28710.
Patel KR, Liu TC, Vaccharajani N, Chapman WC, Brunt EM. Characterization of inflammatory (lymphoepithelioma-like) hepatocellular carcinoma: a study of 8 cases. Arch Pathol Lab Med. 2014;138(9):1193–202. https://doi.org/10.5858/arpa.2013-0371-oa.
Costentin CE, Ferrone CR, Arellano RS, Ganguli S, Hong TS, Zhu AX. Hepatocellular carcinoma with macrovascular invasion: defining the optimal treatment strategy. Liver Cancer 2017;6(4):360–74. https://doi.org/10.1159/000481315.
Stahl J, Voyvodic F. Biopsy diagnosis of malignant versus benign liver “nodules”: new helpful markers. An update. Adv Anat Pathol 2000;7(4):230–9. https://doi.org/10.1097/00125480-200007040-00005.
Wee A. Fine needle aspiration biopsy of the liver: Algorithmic approach and current issues in the diagnosis of hepatocellular carcinoma. Cytojournal. 2005;2:7. https://doi.org/10.1186/1742-6413-2-7.
Bergman S, Graeme-Cook F, Pitman MB. The usefulness of the reticulin stain in the differential diagnosis of liver nodules on fine-needle aspiration biopsy cell block preparations. Mod Pathol. 1997;10(12):1258–64.
Filmus J, Selleck SB. Glypicans: proteoglycans with a surprise. J Clin Invest. 2001;108(4):497–501. https://doi.org/10.1172/jci13712.
Capurro M, Wanless IR, Sherman M, Deboer G, Shi W, Miyoshi E, et al. Glypican-3: a novel serum and histochemical marker for hepatocellular carcinoma. Gastroenterology. 2003;125(1):89–97. https://doi.org/10.1016/s0016-5085(03)00689-9.
Coston WM, Loera S, Lau SK, Ishizawa S, Jiang Z, Wu CL, et al. Distinction of hepatocellular carcinoma from benign hepatic mimickers using Glypican-3 and CD34 immunohistochemistry. Am J Surg Pathol. 2008;32(3):433–44. https://doi.org/10.1097/pas.0b013e318158142f.
Di Tommaso L, Franchi G, Park YN, Fiamengo B, Destro A, Morenghi E, et al. Diagnostic value of HSP70, glypican 3, and glutamine synthetase in hepatocellular nodules in cirrhosis. Hepatology 2007;45(3):725–34. https://doi.org/10.1002/hep.21531.
Libbrecht L, Severi T, Cassiman D, Vander Borght S, Pirenne J, Nevens F, et al. Glypican-3 expression distinguishes small hepatocellular carcinomas from cirrhosis, dysplastic nodules, and focal nodular hyperplasia-like nodules. Am J Surg Pathol. 2006;30(11):1405–11. https://doi.org/10.1097/01.pas.0000213323.97294.9a.
Zhu ZW, Friess H, Wang L, Abou-Shady M, Zimmermann A, Lander AD, et al. Enhanced glypican-3 expression differentiates the majority of hepatocellular carcinomas from benign hepatic disorders. Gut. 2001;48(4):558–64. https://doi.org/10.1136/gut.48.4.558.
Garrido C, Gurbuxani S, Ravagnan L, Kroemer G. Heat shock proteins: endogenous modulators of apoptotic cell death. Biochem Biophys Res Commun. 2001;286(3):433–42. https://doi.org/10.1006/bbrc.2001.5427.
Helmbrecht K, Zeise E, Rensing L. Chaperones in cell cycle regulation and mitogenic signal transduction: a review. Cell Prolif 2000;33(6):341–65. https://doi.org/10.1046/j.1365-2184.2000.00189.x.
Chuma M, Sakamoto M, Yamazaki K, Ohta T, Ohki M, Asaka M, et al. Expression profiling in multistage hepatocarcinogenesis: identification of HSP70 as a molecular marker of early hepatocellular carcinoma. Hepatology 2003;37(1):198–207. https://doi.org/10.1053/jhep.2003.50022.
Di Tommaso L, Destro A, Seok JY, Balladore E, Terracciano L, Sangiovanni A, et al. The application of markers (HSP70 GPC3 and GS) in liver biopsies is useful for detection of hepatocellular carcinoma. J Hepatol. 2009;50(4):746–54. https://doi.org/10.1016/j.jhep.2008.11.014.
Haratake J, Horie A. An immunohistochemical study of sarcomatoid liver carcinomas. Cancer. 1991;68(1):93–7. https://doi.org/10.1002/1097-0142(19910701)68:1%3c93::aid-cncr2820680119%3e3.0.co;2-g.
Kakizoe S, Kojiro M, Nakashima T. Hepatocellular carcinoma with sarcomatous change. Clinicopathologic and immunohistochemical studies of 14 autopsy cases. Cancer. 1987;59(2):310–6. https://doi.org/10.1002/1097-0142(19870115)59:2%3c310::aid-cncr2820590224%3e3.0.co;2-s.
Hu L, Lau SH, Tzang CH, Wen JM, Wang W, Xie D, et al. Association of Vimentin overexpression and hepatocellular carcinoma metastasis. Oncogene. 2004;23(1):298–302. https://doi.org/10.1038/sj.onc.1206483.
Wang Z, Wu Q, Feng S, Zhao Y, Tao C. Identification of four prognostic LncRNAs for survival prediction of patients with hepatocellular carcinoma. PeerJ. 2017;5:e3575. https://doi.org/10.7717/peerj.3575.
Yuan S, Wang J, Yang Y, Zhang J, Liu H, Xiao J, et al. The prediction of clinical outcome in hepatocellular carcinoma based on a six-gene metastasis signature. Clin Cancer Res. 2017;23(1):289–97. https://doi.org/10.1158/1078-0432.ccr-16-0395.
Villanueva A, Portela A, Sayols S, Battiston C, Hoshida Y, Mendez-Gonzalez J, et al. DNA methylation-based prognosis and epidrivers in hepatocellular carcinoma. Hepatology. 2015;61(6):1945–56. https://doi.org/10.1002/hep.27732.
Xu RH, Wei W, Krawczyk M, Wang W, Luo H, Flagg K, et al. Circulating tumour DNA methylation markers for diagnosis and prognosis of hepatocellular carcinoma. Nat. Mater. 2017;16(11):1155–61. https://doi.org/10.1038/nmat4997.
Miao R, Luo H, Zhou H, Li G, Bu D, Yang X, et al. Identification of prognostic biomarkers in hepatitis B virus-related hepatocellular carcinoma and stratification by integrative multi-omics analysis. J Hepatol. 2014;61(4):840–9. https://doi.org/10.1016/j.jhep.2014.05.025.
Gillies RJ, Kinahan PE, Hricak H. Radiomics: Images are more than pictures, they are data. Radiology. 2016;278(2):563–77. https://doi.org/10.1148/radiol.2015151169.
Lambin P, Rios-Velazquez E, Leijenaar R, Carvalho S, van Stiphout RG, Granton P, et al. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer. 2012;48(4):441–6. https://doi.org/10.1016/j.ejca.2011.11.036.
Limkin EJ, Sun R, Dercle L, Zacharaki EI, Robert C, Reuze S, et al. Promises and challenges for the implementation of computational medical imaging (radiomics) in oncology. Ann Oncol. 2017;28(6):1191–206. https://doi.org/10.1093/annonc/mdx034.
Liao H, Zhang Z, Chen J, Liao M, Xu L, Wu Z, et al. Preoperative radiomic approach to evaluate tumor-infiltrating CD8(+) T cells in hepatocellular carcinoma patients using contrast-enhanced computed tomography. Ann Surg Oncol. 2019;26(13):4537–4547. https://doi.org/10.1245/s10434-019-07815-9.
Xu X, Zhang HL, Liu QP, Sun SW, Zhang J, Zhu FP, et al. Radiomic analysis of contrast-enhanced CT predicts microvascular invasion and outcome in hepatocellular carcinoma. J Hepatol. 2019;70(6):1133–44. https://doi.org/10.1016/j.jhep.2019.02.023.
Kim S, Shin J, Kim DY, Choi GH, Kim MJ, Choi JY. Radiomics on gadoxetic acid-enhanced magnetic resonance imaging for prediction of postoperative early and late recurrence of single hepatocellular carcinoma. Clin Cancer Res. 2019;25(13):3847–55. https://doi.org/10.1158/1078-0432.ccr-18-2861.
Chen S, Li J, Wang D, Fung H, Wong LY, Zhao L. The hepatitis B epidemic in China should receive more attention. Lancet. 2018;391(10130):1572. https://doi.org/10.1016/s0140-6736(18)30499-9.
Ma WJ, Wang HY, Teng LS. Correlation analysis of preoperative serum alpha-fetoprotein (AFP) level and prognosis of hepatocellular carcinoma (HCC) after hepatectomy. World J Surg Oncol. 2013;11:212. https://doi.org/10.1186/1477-7819-11-212.
Bruix J, Castells A, Bosch J, Feu F, Fuster J, Garcia-Pagan JC, et al. Surgical resection of hepatocellular carcinoma in cirrhotic patients: prognostic value of preoperative portal pressure. Gastroenterology. 1996;111(4):1018–22. https://doi.org/10.1016/s0016-5085(96)70070-7.
Poon RT, Fan ST, Lo CM, Liu CL, Ng IO, Wong J. Long-term prognosis after resection of hepatocellular carcinoma associated with hepatitis B-related cirrhosis. J Clin Oncol. 2000;18(5):1094–101. https://doi.org/10.1200/jco.2000.18.5.1094.
Sasaki Y, Imaoka S, Masutani S, Ohashi I, Ishikawa O, Koyama H, et al. Influence of coexisting cirrhosis on long-term prognosis after surgery in patients with hepatocellular carcinoma. Surgery. 1992;112(3):515–21.
Shimozawa N, Hanazaki K. Longterm prognosis after hepatic resection for small hepatocellular carcinoma. J Am Coll Surg. 2004;198(3):356–65. https://doi.org/10.1016/j.jamcollsurg.2003.10.017.
Fuster J, Garcia-Valdecasas JC, Grande L, Tabet J, Bruix J, Anglada T, et al. Hepatocellular carcinoma and cirrhosis. Results of surgical treatment in a European series. Ann Surg. 1996;223(3):297–302. https://doi.org/10.1097/00000658-199603000-00011.
Izumi R, Shimizu K, Ii T, Yagi M, Matsui O, Nonomura A, et al. Prognostic factors of hepatocellular carcinoma in patients undergoing hepatic resection. Gastroenterology. 1994;106(3):720–7. https://doi.org/10.1016/0016-5085(94)90707-2.
Lim KC, Chow PK, Allen JC, Chia GS, Lim M, Cheow PC, et al. Microvascular invasion is a better predictor of tumor recurrence and overall survival following surgical resection for hepatocellular carcinoma compared to the Milan criteria. Ann Surg. 2011;254(1):108–13. https://doi.org/10.1097/sla.0b013e31821ad884.
Pandey D, Lee KH, Wai CT, Wagholikar G, Tan KC. Long term outcome and prognostic factors for large hepatocellular carcinoma (10 cm or more) after surgical resection. Ann Surg Oncol. 2007;14(10):2817–23. https://doi.org/10.1245/s10434-007-9518-1.
Roayaie S, Blume IN, Thung SN, Guido M, Fiel MI, Hiotis S, et al. A system of classifying microvascular invasion to predict outcome after resection in patients with hepatocellular carcinoma. Gastroenterology. 2009;137(3):850–5. https://doi.org/10.1053/j.gastro.2009.06.003.
Nanashima A, Morino S, Yamaguchi H, Tanaka K, Shibasaki S, Tsuji T, et al. Modified CLIP using PIVKA-II for evaluating prognosis after hepatectomy for hepatocellular carcinoma. Eur J Surg Oncol. 2003;29(9):735–42. https://doi.org/10.1016/j.ejso.2003.08.007.
Suehiro T, Sugimachi K, Matsumata T, Itasaka H, Taketomi A, Maeda T. Protein induced by vitamin K absence or antagonist II as a prognostic marker in hepatocellular carcinoma. Comparison with alpha-fetoprotein. Cancer 1994;73(10):2464–71. https://doi.org/10.1002/1097-0142(19940515)73:10%3c2464::aid-cncr2820731004%3e3.0.co;2-9.
Grieco A, Pompili M, Caminiti G, Miele L, Covino M, Alfei B, et al. Prognostic factors for survival in patients with early-intermediate hepatocellular carcinoma undergoing non-surgical therapy: comparison of Okuda, CLIP, and BCLC staging systems in a single Italian centre. Gut 2005;54(3):411–8. https://doi.org/10.1136/gut.2004.048124.
Acknowledgments
The authors would like to thank TCGA working group for offering the slide images and the corresponding cancer information, and are most grateful to the Core Facility of WCH for their technique support on the experiments.
Funding
This work was supported by grants from the National Key Technologies R&D Program (2018YFC1106800), the Natural Science Foundation of China (81972747, 81872004, 81800564, 81770615, 81700555, and 81672882), the Science and Technology Support Program of Sichuan Province (2019YFQ0001, 2018SZ0115, 2017SZ0003), the Science and Technology Program of Tibet Autonomous Region (XZ201801-GB-02), and the 1.3.5 Project for Disciplines of Excellence, West China Hospital, Sichuan University (ZYJC18008).
Author information
Authors and Affiliations
Contributions
KY and YZ designed the whole project and participated in result evaluation; ML, LX, and ZZ collected the clinical data of the WCH candidates; TX, HL and ZW participated in the IF extraction; HL and JP performed IF selection and construction of the machine-learning based models; HL and TX conducted the data analysis and wrote the manuscript; and KY and YZ modified the structure of the manuscript. All authors reviewed the manuscript and approved the final version.
Corresponding authors
Ethics declarations
Disclosures
Haotian Liao, Tianyuan Xiong, Jiajie Peng, Lin Xu, Mingheng Liao, Zhen Zhang, Zhenru Wu, Kefei Yuan and Yong Zeng declare no competing interests in this work.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Liao, H., Xiong, T., Peng, J. et al. Classification and Prognosis Prediction from Histopathological Images of Hepatocellular Carcinoma by a Fully Automated Pipeline Based on Machine Learning. Ann Surg Oncol 27, 2359–2369 (2020). https://doi.org/10.1245/s10434-019-08190-1
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1245/s10434-019-08190-1