Abstract
Background
Earlier studies have identified the minimal overlapping region of amplification at 3q26 in esophageal squamous cell carcinoma (ESCC) by comparative genomic hybridization (CGH) analysis. These include PIK3CA which encodes the p110α catalytic subunit of phosphatidylinositol (PI) 3-kinase, a telomerase RNA component (TERC), a squamous cell carcinoma-related oncogene (SCCRO), ecotropic viral integration site-1 (EVI-1), and a Ski-related novel oncogene (SnoN). In the present study, we investigated the mRNA levels of four candidate genes (TERC, SCCRO, EVI-1, and SnoN) to determine whether genes other than PIK3CA are targets for amplification at 3q26 in ESCC. And also, we examined SnoN expression in ESCC samples.
Methods
Fifty-nine representative cases with ESCC were selected from our archives. We performed quantitative RT-PCR of four candidate genes (TERC, SCCRO, EVI-1, and SnoN) and immunohistochemistry for SnoN. Finally, we correlated these findings with the clinicopathological characteristics to determine their interrelationship.
Results
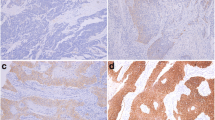
Among the four genes we tested, only SnoN mRNA was consistently overexpressed in primary ESCC, compared with those in corresponding nontumorous esophageal epithelia (P < 0.001). Immunoreactive SnoN was detectable in 31 of 59 (52.5%) esophageal squamous cell carcinoma specimens. The levels of SnoN expression were found to correlate with the depth of invasion and recurrence (P < 0.05). Furthermore, patients with positive staining for SnoN displayed more unfavorable outcomes than patients with negative staining (P < 0.05).
Conclusion
SnoN is likely to be the target of the amplification at 3q26 in ESCC and plays an important role in the development of ESCC, influencing disease-specific survival.





Similar content being viewed by others
References
Shinomiya T, Mori T, Ariyama Y, et al. Comparative genomic hybridization of squamous cell carcinoma of the esophagus: the possible involvement of the DPI gene in the 13q34 amplicon. Genes Chromosomes Cancer 1999; 24:337–44.
Pack SD, Karkera JD, Zhuang Z, et al. Molecular cytogenetic fingerprinting of esophageal squamous cell carcinoma by comparative genomic hybridization reveals a consistent pattern of chromosomal alterations. Genes Chromosomes Cancer 1999; 25:160–8.
Pimkhaokham A, Shimada Y, Fukuda Y, et al. Nonrandom chromosomal imbalances in esophageal squamous cell carcinoma cell lines: possible involvement of the ATF3 and CENPF genes in the 1q32 amplicon. Jpn J Cancer Res 2000; 91:1126–33.
Heselmeyer K, Schrock E, du Manoir S, et al. Gain of chromosome 3q defines the transition from severe dysplasia to invasive carcinoma of the uterine cervix. Proc Natl Acad Sci USA 1996; 93:479–84.
Sugita M, Tanaka N, Davidson S, et al. Molecular definition of a small amplification domain within 3q26 in tumors of cervix, ovary, and lung. Cancer Genet Cytogenet 2000; 117:9–18.
Sonoda G, Palazzo J, du Manoir S, et al. Comparative genomic hybridization detects frequent overrepresentation of chromosomal material from 3q26, 8q24, and 20q13 in human ovarian carcinomas. Genes Chromosomes Cancer 1997; 20:320–8.
Brass N, Racz A, Heckel D, et al. Amplification of the genes BCHE and SLC2A2 in 40% of squamous cell carcinoma of the lung. Cancer Res 1997; 57:2290–4.
Racz A, Brass N, Heckel D, et al. Expression analysis of genes at 3q26-q27 involved in frequent amplification in squamous cell lung carcinoma. Eur J Cancer 1999; 35:641–6.
Bergamo NA, Rogatto SR, Poli-Frederico RC, et al. Comparative genomic hybridization analysis detects frequent over-representation of DNA sequences at 3q, 7p, and 8q in head and neck carcinomas. Cancer Genet Cytogenet 2000; 119:48–55.
Sattler HP, Lensch R, Rohde V, et al. Novel amplification unit at chromosome 3q25-q27 in human prostate cancer. Prostate 2000; 45:207–15.
Shayesteh L, Lu Y, Kuo WL, et al. PIK3CA is implicated as an oncogene in ovarian cancer. Nat Genet 1999; 21:99–102.
Ma YY, Wei SJ, Lin YC, et al. PIK3CA as an oncogene in cervical cancer. Oncogene 2000; 19:2739–44.
Zhang L, Yang N, Katsaros D, et al. The oncogene phosphatidylinositol 3′-kinase catalytic subunit alpha promotes angiogenesis via vascular endothelial growth factor in ovarian carcinoma. Cancer Res 2003; 63:4225–31.
Woenckhaus J, Steger K, Werner E, et al. Genomic gain of PIK3CA and increased expression of p110alpha are associated with progression of dysplasia into invasive squamous cell carcinoma. J Pathol 2002; 198:335–42.
Yokoi S, Yasui K, Iizasa T, et al. TERC identified as a probable target within the 3q26 amplicon that is detected frequently in non-small cell lung cancers. Clin Cancer Res 2003; 9 :4705–13.
Sarkaria I, O-Charoenrat P, Talbot SG, et al. Squamous cell carcinoma related oncogene/DCUN1D1 is highly conserved and activated by amplification in squamous cell carcinomas. Cancer Res 2006; 66:9437–44.
Sarkaria IS, Stojadinovic A, Talbot SG, et al. Squamous cell carcinoma-related oncogene is highly expressed in developing, normal, and adenomatous adrenal tissue but not in aggressive adrenocortical carcinomas. Surgery 2004; 136:1122–8.
Sarkaria IS, Pham D, Ghossein RA, et al. SCCRO expression correlates with invasive progression in bronchioloalveolar carcinoma. Ann Thorac Surg 2004; 78:1734–41.
Talbot SG, O-Charoenrat P, Sarkaria IS, et al. Squamous cell carcinoma related oncogene regulates angiogenesis through vascular endothelial growth factor-A. Ann Surg Oncol 2004; 11:530–4.
Estilo CL, O-Charoenrat P, Ngai I, et al. The role of novel oncogenes squamous cell carcinoma-related oncogene and phosphatidylinositol 3-kinase p110alpha in squamous cell carcinoma of the oral tongue. Clin Cancer Res 2003; 9:2300–6.
Hirai H, Izutsu K, Kurokawa M, et al. Oncogenic mechanisms of Evi-1 protein. Cancer Chemother Pharmacol 2001; 48:S35–40.
Nomura T, Khan MM, Kaul SC, et al. Ski is a component of the histone deacetylase complex required for transcriptional repression by Mad and thyroid hormone receptor. Genes Dev 1999; 13:412–23.
Imoto I, Pimkhaokham A, Fukuda Y, et al. SNO is a probable target for gene amplification at 3q26 in squamous-cell carcinomas of the esophagus. Biochem Biophys Res Commun 2001; 286:559–65.
Nomura N, Sasamoto S, Ishii S, et al. Isolation of human cDNA clones of ski and the ski-related gene, sno. Nucleic Acids Res 1989; 17:5489–500.
Pearson-White S. SnoI, a novel alternatively spliced isoform of the ski protooncogene homolog, sno. Nucleic Acids Res 1993; 21:4632–8.
Zhang F, Lundin M, Ristimaki A, et al. Ski-related novel protein N (SnoN), a negative controller of transforming growth factor-beta signaling, is a prognostic marker in estrogen receptor-positive breast carcinomas. Cancer Res 2003; 63:5005–10.
Liu X, Sun Y, Weinberg RA, et al. Ski/Sno and TGF-beta signaling. Cytokine Growth Factor Rev 2001; 12:1–8.
Osawa H, Shitara Y, Shoji H, et al. Mutation analysis of transforming growth factor beta type II receptor, Smad2, Smad3 and Smad4 in esophageal squamous cell carcinoma. Int J Oncol 2000; 17:723–8.
Fukai Y, Fukuchi M, Masuda N, et al. Reduced expression of transforming growth factor-beta receptors is an unfavorable prognostic factor in human esophageal squamous cell carcinoma. Int J Cancer 2003; 104 :161–6.
Fukuchi M, Nakajima M, Fukai Y, et al. Increased expression of c-Ski as a co-repressor in transforming growth factor-beta signaling correlates with progression of esophageal squamous cell carcinoma. Int J Cancer 2004; 108 :818–24.
Edmiston JS, Yeudall WA, Chung TD, et al. Inability of transforming growth factor-beta to cause SnoN degradation leads to resistance to transforming growth factor-beta-induced growth arrest in esophageal cancer cells. Cancer Res 2005; 65:4782–8.
Fukuchi M, Miyazaki T, Fukai Y, et al. Plasma level of transforming growth factor beta1 measured from the azygos vein predicts prognosis in patients with esophageal cancer. Clin Cancer Res 2004; 10 :2738–41.
Gorospe M, Wang X, Holbrook NJ. Functional role of p21 during the cellular response to stress. Gene Expr 1999; 7:377–85.
Krakowski AR, Laboureau J, Mauviel A, et al. Cytoplasmic SnoN in normal tissues and nonmalignant cells antagonizes TGF-beta signaling by sequestration of the Smad proteins. Proc Natl Acad Sci USA 2005; 102:12437–42.
Bonni S, Wang HR, Causing CG, et al. TGF-beta induces assembly of a Smad2-Smurf2 ubiquitin ligase complex that targets SnoN for degradation. Nat Cell Biol 2001; 3:587–95.
Stroschein SL, Bonni S, Wrana JL, et al. Smad3 recruits the anaphase-promoting complex for ubiquitination and degradation of SnoN. Genes Dev 2001; 15:2822–36.
Sun Y, Liu X, Ng-Eaton E, et al. SnoN and Ski protooncoproteins are rapidly degraded in response to transforming growth factor beta signaling. Proc Natl Acad Sci USA 1999; 96:12442–7.
Sarker KP, Wilson SM, Bonni S. SnoN is a cell type-specific mediator of transforming growth factor-beta responses. J Biol Chem 2005; 280:13037–46.
Acknowledgements
We thank Takuji Kosuge for his expert technical assistance. We also thank Dr. T. Nishihira and Tohoku University for providing the TE-series cell lines.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Akagi, I., Miyashita, M., Makino, H. et al. SnoN Overexpression is Predictive of Poor Survival in Patients with Esophageal Squamous Cell Carcinoma. Ann Surg Oncol 15, 2965–2975 (2008). https://doi.org/10.1245/s10434-008-9986-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1245/s10434-008-9986-y