Abstract
Assessment of anti-drug antibodies (ADAs) for neutralizing activity is important for the clinical development of biopharmaceuticals. Two types of neutralizing antibody (NAb) assays (competitive ligand-binding assay [CLBA] and cell-based assay [CBA]) are commonly used to characterize neutralizing activities. To support the clinical development of benralizumab, a humanized, anti–interleukin-5 receptor α, anti-eosinophil monoclonal antibody, we developed and validated a CLBA and a CBA. The CLBA and CBA were compared for sensitivity, drug tolerance, and precision to detect NAbs in serum samples from clinical trials. The CLBA was more sensitive (27.1 and 37.5 ng/mL) than the CBA (1.02 and 1.10 μg/mL) in detecting NAbs to benralizumab for the polyclonal and monoclonal ADA controls, respectively. With the same polyclonal ADA control, the CLBA detected 250 ng/mL of ADA in the presence of 100 ng/mL of benralizumab, whereas the CBA detected 1.25 μg/mL of ADA in the presence of 780 ng/mL of benralizumab. In 195 ADA-positive samples from 5 studies, 63.59% (124/195) and 16.9% (33/195) were positive for NAb as measured by the CLBA and the CBA, respectively. ADA titers were strongly correlated (Pearson’s correlation coefficient r = 0.91; n = 195) with CLBA titers. Moreover, the CLBA titer correlated with CBA percentage inhibition in the CBA-positive samples (Spearman’s coefficient r = 0.50; n = 33). Our data demonstrated advantages of the CLBA in various aspects and supported the choice of the CLBA as a NAb assay for the phase III trials.
Similar content being viewed by others
INTRODUCTION
All biopharmaceuticals have the potential risk of eliciting an immune response. Immunogenicity assessment has become an essential element of safety and efficacy evaluations in the clinical development of biopharmaceuticals. Characterization of anti-drug antibodies (ADAs) for neutralizing activity is a crucial component of these evaluations. Neutralizing antibodies (NAbs) may have the ability to abolish drug-associated biologic activities (1) and cross-react with the endogenous counterpart of the drug product (2), potentially resulting in severe clinical consequences.
The aim of the NAb assay is to characterize the nature of the ADA response in patients participating in clinical trials. The corresponding NAb data, along with other clinical information, are used to evaluate drug efficacy and patients’ safety profiles. The principle of NAb assay development is to reflect the mechanism of action (MOA) of the respective biopharmaceutical. Because of the specific MOAs of individual biopharmaceuticals, the formats of NAb assays are more diverse than screening/confirmatory ADA assays, which usually employ a bridging format. NAb assay formats are commonly classified into two types: cell-based assays (CBAs) and non–cell-based assays including competitive ligand-binding assays (CLBAs). CLBAs are typically used to assess the binding of the drug to its target (e.g., soluble ligand) (3). For the evaluation of NAb activity, regulatory agencies recommend a CBA as the preferred platform because the cellular responses it exhibits more closely reflect the NAb’s effect on the drug in vivo (4). However, this approach may have its limitation when there are technical difficulties associated with a CBA (5,6). It is generally recognized that the CLBA displays advantages in sensitivity. Robustness and ease of operation of a CLBA when compared with a CBA make it an appealing substitute for CBA if strong and solid justifications are provided (7).
There is a growing effort by pharmaceutical companies and government agencies to compare the two NAb assay formats. For assay characteristics, CLBAs are comparable or superior to bioassays for detecting NAb to four types of therapeutic proteins (5). In a separate study, the two NAb assays demonstrated a strong correlation in NAb titer from clinical samples despite a weaker sensitivity observed with CLBAs (8). Hu et al. found that CLBAs and CBAs behaved comparably in detecting NAbs in clinical samples for two case studies, although CLBAs were inferior to CBAs for a third case (9). Thus, non–CBAs would be a viable option for detecting NAbs for various biopharmaceuticals. The choice of appropriate NAb format should be based primarily on the drug’s MOA, immunogenicity risk assessment, and technical feasibility.
Benralizumab is a humanized, afucosylated, anti–interleukin-5 receptor α (IL-5Rα) monoclonal antibody (mAb) with enhanced effector function. It potently induces antibody-dependent cell-mediated cytotoxicity (ADCC) of eosinophils and basophils rapidly, selectively depleting circulating and tissue-resident eosinophils and basophils (10). To support the clinical development of benralizumab for the treatment of asthma and chronic obstructive pulmonary disease, we developed NAb assays in both CLBA and CBA formats. We report on the validation of the two assays for benralizumab NAb assessment and compare their performance for sensitivity, drug tolerance, robustness, and data obtained from testing of a subset clinical samples.
MATERIALS AND METHODS
Materials
MedImmune LLC generated benralizumab and human IL-5R and produced and purified goat anti-benralizumab polyclonal antibody. Pooled normal human sera were purchased from Bioreclamation (Hicksville, NY). Meso Scale Discovery (MSD) plates and Sulfo-TAG™ Plus NHS Ester were purchased from MSD (Gaithersburg, MD). Amicon® Ultra Centrifugal Filter Units, UItra-50 membrane was purchased from Millipore (Billerica, MA). Protein Assay Kit was purchased from Pierce (Rockford, IL). MedImmune LLC prepared biotinylated benralizumab and ruthenium-labeled IL-5R.
MedImmune LLC generated the CTLL-2/IL-5Rα cell line and NK92/NFAT-luciferase cell line. RPMI 1640 with GlutaMax™, horse serum (heat-inactivated), human IL-5, geneticin, and mercaptoethanol were purchased from Invitrogen. Fetal bovine serum (heat-inactivated) was purchased from HyClone. Steady-Glo® Luciferase Assay System was purchased from Promega. Human IL-2 was purchased from Henry Schein.
Selection of ADA-Positive Samples from Clinical Studies for the Assessment of NAb Activity
We used both CLBA and CBA to test for NAbs in 195 ADA-positive samples from 5 studies: MI-CP166 (study 1, NCT00659659) (11), MI-CP186 (study 2, NCT00768079) (12), MI-CP197 (study 3, NCT00783289) (13), and MI-CP220 (studies 4 and 5, NCT01238861) (14). To assess NAb activity in both NAb assays, we chose all ADA-positive samples from studies 1 to 3, regardless of drug concentrations. For comparative assessment of the two NAb methods, we chose samples from studies 4 and 5 primarily based on the validation data of drug tolerance and availability of clinical samples.
Methods for Assessment of NAb to Benralizumab
CLBA Method
We evaluated serum samples for NAbs to benralizumab using a semi-solution phase competitive CLBA that employed biotinylated benralizumab for capture and sulfo-TAG–labeled human IL-5R for detection. The assay assessed the ability of the ADA to inhibit (neutralize) the interaction of benralizumab with IL-5R. Test samples were incubated overnight at room temperature (18–23 °C) in the presence of biotinylated benralizumab at a final sample dilution of 1:10 for binding of any ADAs present in the samples to the drug. Following incubation, we transferred the mixture to a streptavidin-coated MSD microtiter plate, to which biotinylated drug and any biotinylated drug-ADA complex would bind. Following a wash step, we added sulfo-TAG–labeled human recombinant IL-5R (ligand) for binding to benralizumab. If NAbs were present in the samples, binding of labeled IL-5R to benralizumab was blocked. Only NAbs that bound to benralizumab in regions with recognition sites for its antigen (IL-5R) would inhibit the interaction of the drug with its target (IL-5R). After a final wash step and addition of MSD read buffer, plates were read on an MSD Sector Imager. The electrochemiluminescence (ECL) signal generated was inversely proportional to the amount of NAbs present in the samples. We classified samples as positive or negative for the presence of NAbs to benralizumab relative to a cutoff ECL value that we calculated for each plate by multiplying the mean value of the negative control by a cutoff factor of 0.85 (15% inhibition of signal). Samples that measured positive for NAbs were subsequently titered to determine relative amounts of NAb present. We determined titers by serially diluting samples at a ratio of 1:2 with negative control matrix and reported the titers as the reciprocal of the greatest dilution (over the 1:10 minimum required sample dilution [MRD]) that produced a positive response.
CBA Method
Methods for quality control preparation, screening cutoff determination, sensitivity, drug tolerance, selectivity, cell culture, and assay procedures for the ADCC cell-based NAb assay have been described in detail (15). In brief, the validated CBA is an ADCC assay that consists of target cells (a murine cytotoxic cell line CTLL-2 stably transfected with human IL-5R) and an effector cell line (human NK92 cells). This effector cell line is engineered to express CD16 (FcRIIIγ) and to contain a luciferase reporter gene driven by the Nuclear Factor of Activated T (NFAT) promoter. Benralizumab binds to human IL-5R on the target cell, and the Fc region of the drug binds to the effector cell via CD16, thereby inducing ADCC of the target cell. ADCC activity is strongly correlated with activation of the luciferase reporter gene. We measured the amount of luciferase generated by using a Steady-Glo® Luciferase Assay System according to manufacturer’s instructions. In the presence of NAbs in serum samples, the ADCC is inhibited, causing the luciferase signal (relative luminescence unit [RLU] value) to decrease. The luciferase signal generated was inversely proportional to the amount of NAbs present in the samples. We classified samples as positive or negative for the presence of NAbs to benralizumab relative to a cutoff RLU value. We calculated the cutoff RLU value for each plate by multiplying the mean value of the negative control by a cutoff factor of 0.73 (27% inhibition of signal). We did not perform titer determination for the samples that were positive for NAbs in the CBA.
Assay Cutoffs
For cutoff determination in both NAb assays during validation, we used the same set of serum samples obtained from 80 men and women with asthma. Two operators analyzed each sample in duplicate wells in four assays over 2 days. The data set consisting of 320 normalized responses (signal to background values) was evaluated by statistical methods for the determination of the cutoff factor with a 1% false-positive rate (15).
ADA Assays
ADAs in clinical samples were analyzed using an ECL solution phase bridging method that MedImmune developed and validated. Samples at a final dilution of 1:50 overnight were incubated at room temperature in a solution with biotinylated benralizumab and sulfo-TAG–labeled benralizumab. Bridged biotin drug:ADA:sulfo-TAG drug complexes were subsequently captured onto streptavidin-coated MSD plates and measured on the MSD Sector Imager. Signal intensity was proportional to the amount of ADA present in the samples.
Samples were classified as potentially positive for ADA in the screening assay if the signal measured at or above the cutoff for the plate. The cutoff value was calculated using a cutoff factor (1.29) determined during validation from 200 measurements of serum samples from 50 patients with asthma. In a confirmatory (specificity) assay, positive samples were retested in both the absence and presence of excess (100 μg/mL) benralizumab. Samples were confirmed as positive for ADA if the percentage inhibition of response in the presence of excess drug was greater than or equal to the confirmatory cutoff (33.1%) established during validation to provide a 1% false-positive rate. Endpoint titers were measured in samples confirmed as positive and were reported as the reciprocal of the greatest dilution (over the 1:50 MRD) that measured positive in the assay.
Drug Tolerance Level Determination
A similar assay procedure for drug tolerance assessment was employed for all three assays except the concentrations of the polyclonal anti-benralizumab idiotype antibody and benralizumab in neat serum. For the CLBA, the concentrations of the polyclonal anti-benralizumab idiotype antibody were 2.0, 1.5, 1.0, 0.75, 0.5, 0.25, 0.1, 0.05, 0.025, 0.0125, and 0 μg/mL; and the benralizumab concentrations were 1.0, 0.5, 0.25, 0.1, 0.05, 0.025, and 0 μg/mL. For the CBA, the concentrations of the polyclonal anti-benralizumab idiotype antibodies were 10, 5, 2.5, 1.25, 0.63, 0.31, and 0 μg/mL, and the benralizumab concentrations were 12.5, 6.25, 3.13, 1.56, 0.78, 0.39, and 0 μg/mL (15). For the binding ADA assay, the concentrations of the polyclonal anti-benralizumab idiotype antibody were 4.0, 2.0, 1.0, 0.5, 0.25, and 0 μg/mL, and the benralizumab concentrations were 500, 250, 100, 50, 25, and 0 μg/mL. Pre-incubations of the polyclonal anti-benralizumab idiotype antibody with benralizumab were performed at room temperature for 60 ± 5 min prior to analysis.
Statistical Methods
Correlations between ADA/CLBA titer and several types of NAb results (titer and percentage inhibition) were calculated. If both results were titers, a Pearson correlation coefficient was used. If the correlation was between the ADA titer and the NAb percentage inhibition, a Spearman’s rank correlation coefficient was calculated. The scatterplots of correlation between ADA titer/CLBA titer, ADA titer/CLBA percentage inhibition, ADA titer/CBA percentage inhibition, CLBA titer/CBA percentage inhibition, and CLBA percentage inhibition/CBA percentage inhibition were presented. For the plots, the ADA titer and CLBA titer were transformed by using log (1 + titer). For the correlative analysis between ADA titer and CLBA percentage inhibition, a three-parameter logistic regression with an upper limit of 100% was used because there was a nonlinear logistic trend when CLBA inhibition was close to 100%. For the rest of the trend analyses, a linear regression was used to depict the relationship between the two correlated variables. The probability of being CLBA positive or CBA positive given ADA titer values was evaluated using logistical regression. All statistical analyses were performed using Statistical Analysis Systems Version 9.3 (SAS Institute, Cary NC) and R 3.0.1 (www.r-project.org).
RESULTS
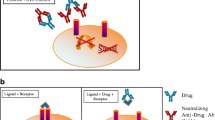
We developed and validated two NAb assays in our laboratory for the detection of anti-benralizumab NAbs in human serum. Because benralizumab binds to the IL-5 receptor on cells, which prevents IL-5 binding to the receptor and the subsequent signaling through this receptor and induces ADCC in IL-5 receptor–expressing eosinophils and basophils, the CLBA and CBA were designed to reflect the MOA of benralizumab. Thus, the assays were designed to assess the ability of the ADA to inhibit (neutralize) the interaction of benralizumab with the IL-5 receptor. The CLBA measures the reduction of benralizumab–IL-5R interaction caused by the presence of NAb. The CBA determines the reduction of benralizumab-mediated ADCC activity caused by the presence of NAb for the inhibition of the binding of benralizumab to the IL-5R–bearing target cell. The assay principles for the CLBA and CBA (15) are described in detail in the “Methods” section, and the assay formats are provided in Fig. 1a, b, respectively.
Design of NAb assays. a Design of the ligand-binding–based NAb assay. Biotinylated benralizumab and Sulfo-TAG–conjugated IL-5R form a complex that is captured on streptavidin-coated MSD standard plates. The presence of ADA blocks complex formation. The ECL signal generated by the ligand-IL-5R complex is detected by an MSD SECTOR™ Imager and is inversely proportional to the amount of ADA present in the samples. b Design of the ADCC-based reporter gene NAb assay. We incubated CTLL-2/hIL-5R cells (a murine cytotoxic cell line CTLL-2 stably transfected with human IL-5R) and NK92/NFAT cells expressing CD16 (FcRIIIγ) and containing a luciferase gene reporter driven by NFAT-containing promoter, with diluted serum sample and benralizumab. Benralizumab binds to human IL-5R on the target cell, and the Fc region of the drug binds to the effector cell via CD16, thereby inducing ADCC of the target cell. The ADCC activity is strongly correlated with activation of the luciferase reporter gene. The presence of ADA blocks binding of benralizumab to IL-5R. ADA anti-drug antibody, ADCC antibody-dependent cell-mediated cytotoxicity, ECL electrochemiluminescence, IL-5R interleukin-5 receptor, MSD Meso Scale Discovery, NAb neutralizing antibody, NFAT nuclear factor of activated T
Summary of Assay Parameters for the CLBA and CBA
Critical assay parameters are summarized in Table I. In this study, we evaluated correlations between the two NAb assays, as well as the relationship between the NAb assays and the binding ADA assay (Table I). Intra-assay precisions were comparable for the CLBA and CBA (8.6 vs. 11.0%). The CBA achieved an inter-assay variability of 24.6%, which the authors believe is common for a CBA given its complexity. Inter-assay variability of the CLBA (12.5%) was much less than that of the CBA, suggesting that the CLBA is more robust and less variable than the CBA.
To better compare assay sensitivity, we used the same surrogate anti-benralizumab idiotype polyclonal and monoclonal ADA controls (Kd approximately 1 nM) across three assays (Table I). The CLBA displayed comparable sensitivity to the binding ADA assay. The estimated sensitivity of the CLBA at the cutoff using the anti-benralizumab polyclonal and monoclonal ADA controls was 27.1 and 37.5 ng/mL, respectively (Table I). We determined the sensitivity of the assay also by using a second lot of the polyclonal antibody surrogate control (the same used in the binding ADA assay), and this control yielded a similar sensitivity result of 41.7 ng/mL. Because of matrix interference, the MRD for the CBA was 40, which was four times greater than that for the CLBA (MRD of 10) (Table I). As a result, estimated sensitivity of the CBA at the cutoffs using the polyclonal and monoclonal ADA controls was 1.02 and 1.10 μg/mL, respectively. Thus, the CLBA was far more sensitive (approximately 38-fold for the polyclonal ADA control and 29-fold for the monoclonal ADA control) than the CBA.
Both NAb assays displayed limited drug tolerance. The CLBA detected 250 ng/mL of polyclonal ADA control in the presence of 100 ng/mL of benralizumab, whereas the CBA detected 1.25 μg/mL of polyclonal ADA control in the presence of 780 ng/mL of benralizumab (Table I). By comparison, the ADA assay can detect 250 ng/mL of polyclonal ADA control in the presence of 100 μg/mL benralizumab (Table I). Additional drug tolerance for the ADA assay was assessed in another laboratory and data demonstrated that the polyclonal ADA levels of 62.5 and 250 ng/mL were reproducibly detectable in the presence of up to 50 and 125 μg/mL of benralizumab, respectively, indicating that the ADA assay was strongly drug-tolerant.
Comparison of the Abilities of the CLBA and CBA to Detect NAbs in Clinical Samples
From 5 studies, we selected 195 samples that were confirmed to be ADA positive to compare the abilities of the CLBA and CBA to detect NAb in clinical samples. CLBA was much more sensitive at detecting NAb than the CBA, with the NAb activity against benralizumab detected by CLBA consistently greater than by CBA across all five studies (Fig. 2 and Table II). Of 195 samples, 124 (63.59%) and 33 (16.92%) measured positive for NAb in the CLBA and CBA, respectively (Table II).
Comparison of NAb measured by the CLBA and the CBA in five studies. We analyzed a panel of a different number of individual serum samples from each study in the CLBA (a) and CBA (b) assays according to the assay procedures. Each value represents the S:B ratio. Samples below cutoff in each assay are defined NAb positive. CBA cell-based assay, CLBA competitive ligand-binding assay, NAb neutralizing antibody, S:B signal to noise
To evaluate whether drug presence in serum samples would interfere with NAb detection, we classified these 195 samples into 2 groups: no measurable benralizumab concentration (n = 130) and measurable (n = 65) benralizumab concentration (Table II). Of these 65 samples, detectable concentrations of benralizumab ranged from 5.20 ng/mL up to 5132.69 ng/mL, indicating that in some cases, NAb could be detected in the presence of measurable amounts of benralizumab, even when above the drug tolerance level determined in NAb assay validation. For both assays combined, the NAb incidence rate was greater in samples with no measurable drug (103/130, 79.2%) than those with measurable drug (28/65, 43.1%). Similarly, for CLBA alone, the NAb incidence rate was greater in samples with no measurable drug (99/130, 76.15%) compared with samples with measurable drug (25/65, 38.46%). The NAb incidence by the CBA was 28/130 (21.54%) for samples with no measurable drug and 5/65 (7.69%) for samples with measurable drug. Thus, drug presence in samples interfered with NAb detection in both NAb assays, with the CLBA displaying relatively better drug tolerance than the CBA.
We used a logistical model approach to project the probability of a sample measuring positive for NAb in the CLBA and CBA in a broader spectrum of ADA titers (Table III). Based on this model, ADA titers ≥ 400 had a ≥ 48% probability of being NAb positive in the CLBA compared with > 6% for the CBA. To achieve a probability of ≥ 47% that a sample will measure positive in the CBA, a titer of ≥ 3200 was required (Table III). CLBA was more sensitive than the CBA in detecting NAb (Tables I and III and Fig. 2).
Correlation of ADA Titer and NAb Responses Measured by CLBA
The potential relationship between ADA titer responses and NAb responses (CLBA titer and percentage inhibition) was examined in the samples that measured positive for both binding ADA and NAb. ADA titer and NAb titer responses were strongly correlated (Pearson’s correlation coefficient, r = 0.91, n = 195, Fig. 3a). ADA titer and the percentage inhibition measured in the CLBA were directly related (Fig. 3b, Spearman’s coefficient r = 0.77, n = 195). A logistic regression was also fitted to estimate the relationship between ADA titer and percentage inhibition because this model provides a better fit than the linear regression. The percentage inhibition increased with ADA titer but became flat at the greater ADA titers. The correlation coefficient of ADA titer with percentage inhibition was smaller than with the CLBA titer because several samples had near-maximum percentage inhibition.
NAb CLBA titer and CLBA percentage inhibition correlation with ADA titer in ADA-positive samples. a Correlation between ADA titer and NAb CLBA titer. b Correlation between ADA titer and CLBA percentage inhibition. Plot of log-transformed ADA titer and CLBA titer of ADA-positive samples from five clinical studies (n = 195, some titer values overlap). NAb percentage inhibition was calculated by (1 − the mean ECL serum sample/the mean ECL pooled normal human serum) × 100. ADA anti-drug antibody, CLBA competitive ligand-binding assay, ECL electrochemiluminescence, NAb neutralizing antibody
Correlation of ADA Titer and NAb Responses Measured by CBA
We examined the potential relationship between ADA titer and NAb responses measured by CBA by using the same set of 195 samples. Because samples positive for NAb in the CBA were not titered, only percentage inhibition measured in the CBA was compared with ADA titers. ADA titers were less correlated with CBA percentage inhibition (Fig. 4, Spearman’s coefficient r = 0.51, n = 195) than with CLBA percentage inhibition (Spearman’s coefficient r = 0.77) or CLBA titers (Pearson’s correlation coefficient r = 0.91). ADA titer was more predictive of the CLBA titers than the percentage inhibition of the CBA.
Correlation between ADA titer and NAb CBA percentage inhibition of ADA-positive samples. Values represent the values of percentage inhibition in the NAb CBA at the corresponding measured ADA titer (log transformed; n = 195, some titer values overlap). We calculated NAb percentage inhibition as (1 − the mean RLU serum sample/the mean RLU pooled normal human serum spiked with benralizumab 35 ng/mL) × 100. ADA anti-drug antibody, CBA cell-based assay, NAb neutralizing antibody, RLU relative luminescence unit
Correlation Between the NAb CLBA Responses and CBA Percentage Inhibition in the CBA-Positive Clinical Samples
CLBA titer and percentage inhibition was moderately correlated with CBA percentage inhibition in the CBA-positive samples (Fig. 5a, b, Spearman’s coefficient r = 0.50 and r = 0.51, respectively; n = 33). The results indicate that NAb detected by the CLBA were representative of the ability of the CBA to detect neutralizing activity.
Correlation between NAb CLBA titer (a) and percentage inhibition (b) and CBA percentage inhibition of CBA-positive samples. Values represent percentage inhibition in the NAb CBA at the corresponding measured CLBA titer (log transformed; n = 33, some titer values overlap) and percentage inhibition. We calculated NAb percentage inhibition as (1 − the mean RLU serum sample/the mean RLU pooled normal human serum spiked with 35 ng/mL benralizumab) × 100. CBA cell-based assay, CLBA competitive ligand-binding assay, NAb neutralizing antibody, RLU relative luminescence unit
DISCUSSION
To support the clinical trial program for benralizumab, we developed and validated two NAb assays in CLBA and CBA formats. We subsequently applied them to assess neutralizing activities of benralizumab in patient samples from five clinical studies. Validation data demonstrated that CLBA was far more sensitive (approximately 38-fold for the polyclonal ADA control and 29-fold for the monoclonal control) than the CBA. Of 195 ADA-positive samples that we tested for NAb, 63.59% and 16.92% were positive for NAb as measured by the CLBA and the CBA, respectively. These results indicate that the CLBA is superior to the CBA in detecting NAb activities in patient sera. Our study clearly demonstrated that validation data were consistent with the clinical observation of neutralizing ADAs in clinical samples tested, supporting the concept that CLBA is more capable of detecting NAb in samples than the CBA. The CLBA is a more suitable assay format than the CBA for phase III trials.
Drug presence in samples can pose an additional challenge with NAb assays. Moreover, drug interference with an assay can potentially lead to a false negative for NAb detection. Therefore, we conducted validation assays by spiking samples with much lower drug concentrations for NAb assays as opposed to the binding ADA assay. This approach enabled us to determine drug tolerance of the two assays and to compare the capabilities of assays to detect NAbs. In general, validation data obtained from drug tolerance can serve as primary guidance in selecting samples from clinical trials for NAb testing. The validation data revealed that the NAb assays displayed poor or little drug tolerance relative to the binding ADA assay, in that NAbs were detectable when they were 1.6- to 2.5-fold in excess over drug for both NAb assays. Despite validation data indicating that both NAb assays have limited drug tolerance, data from clinical samples demonstrated that both assays were still capable of detecting NAb activities in samples with measurable drug concentrations.
Notably, measured NAb incidence rates for the samples with measurable benralizumab concentrations in both NAb assays were much lower than those for samples with no benralizumab concentrations. This finding indicates that NAb may not have been detected in some ADA-positive samples because of the presence of benralizumab. Moreover, the presence of benralizumab leading to interference with detecting NAb activities was more apparent in the CBA than that in the CLBA. This interference could potentially account for the lower incidence of NAb in the CBA, along with the lower sensitivity of the CBA. Seven samples measured positive for NAb only by CBA. Of these seven samples, four (two each in study 2 and study 5) were from the placebo or pre-dose group; one (with drug concentration of 5132.29 ng/mL in study 3) had a very low assay response (close to assay cutoff); and two had measurable drug and an inhibition response of 58.1% (395.38 ng/mL of benralizumab, ADA titer 200 in study 3) and 48.9% (90.82 ng/mL of benralizumab, ADA titer 50 in study 5), respectively. The NAb positive results from the CBA that were negative in the CLBA could be false positives, because four of the seven samples were from baseline or the placebo group. However, a mechanistic explanation for NAb-positive results from just the CBA cannot be fully ruled out (e.g., NAbs directed against the Fc region of benralizumab perhaps prevent binding of benralizumab to CD16 [FcRIIIγ] and subsequent activation of the effector cells). Nevertheless, most samples that were positive in the CBA were also positive in the CLBA.
During drug development, researchers have commonly used a tiered approach to measure ADA. Although assay platforms/formats and reagents used in the binding ADA and NAb assays are deemed different, the results from the assays are anticipated to be interrelated. To further compare the two NAb methods, we assessed the correlation between the ADA and NAb results as measured by the CLBA (titers and percentage inhibitions) and the CBA (percentage inhibitions). Our data demonstrated that ADA titer was strongly correlated with NAb CLBA titers, while ADA titers were less correlated with NAb CBA percentage inhibition versus CLBA percentage inhibition. A strong correlation between ADA titer and NAb CLBA titer indicates that ADA response is against variable regions of benralizumab and, more specifically, against the antigen-binding region (idiotype). The ADA titer mimics the CLBA titer as an assessment of the relative magnitude of the neutralizing activity of the ADA. This concept was further supported by the validation data, which the binding ADA and the CLBA NAb assays exhibit comparable sensitivities. In addition, a moderate correlation exists between the CLBA titer and CBA percentage inhibition in the CBA-positive samples. This finding indicates that the CLBA NAb assay can detect neutralizing activity detected by the CBA. The commonality of the results of CLBA titer and CBA percentage inhibition further supports the concept that the neutralizing activity is directed primarily against the antigen-binding region of benralizumab rather than the Fc portion of the molecule, given the format of the CLBA detects only NAbs against the antigen-binding region.
For study 5 (n = 128), we evaluated the effect of measured concentration of NAb to benralizumab on serum drug concentration and blood eosinophil concentrations (pharmacodynamics [PD] marker) in ADA-negative, ADA-positive, and NAb-positive patients. The presence of NAb reduced median benralizumab serum concentrations during the drug steady-state period, but the effects lessened as the dosage increased. Longitudinal data showed that the median eosinophil concentration of the NAb-positive population was greater than the corresponding median eosinophil concentration of the ADA-negative and ADA-positive (including both NAb-negative and NAb-positive) populations (unpublished data).
Unlike previous publications that either compared NAb assay formats for assay characteristics or performed a retrospective comparison of NAb methods using clinical samples (5,8,9), this study took an integrated approach in which we selected a subset of clinical samples from phase II and applied them to the two validated NAb assays. We also assessed the correlation between ADA/CLBA titers and PD readout. Although this approach requires upfront commitment in terms of resources on assay development/validation and sample testing, this is a data-driven and logistically efficient approach to selecting a NAb assay needed for pivotal phase III clinical trials.
Limitations in validating a NAb assay include (1) surrogate positive controls, because neither polyclonal antibodies produced in animals nor mouse anti-drug mAbs used in ADA assays are fully representative of a human ADA response, and (2) limited drug tolerance, a characteristic of most NAb assays. When one develops NAb assays that demonstrate poor drug tolerance, it might be worth further evaluating the assays’ feasibility. Conducting a comparative assessment by testing a subset of clinical samples if available, as in this study, may be a reliable approach for choosing an assay format.
CONCLUSION
Data from assay validation and clinical sample testing demonstrated that the CLBA is more sensitive in detecting NAb activities and more drug-tolerant than the CBA. Moreover, the data from clinical samples demonstrated that the CLBA yielded more positive samples than the CBA. The CLBA detects neutralizing activity in common with the CBA. These results, along with the more precise nature of the CLBA, support the choice of the CLBA over the CBA for future clinical trials for benralizumab. The strong correlation between ADA titer and NAb CLBA titer supports the conclusion that a qualitative readout from the CLBA NAb (positive/negative) is sufficient for the assessment of NAb in future clinical trials.
References
Koren E, Smith HW, Shores E, Shankar G, Finco-Kent D, Rup B, et al. Recommendations on risk-based strategies for detection and characterization of antibodies against biotechnology products. J Immunol Methods. 2008;333:1–9.
Casadevall N, Nataf J, Viron B, Kolta A, Kiladjian JJ, Martin-Dupont P, et al. Pure red-cell aplasia and antierythropoietin antibodies in patients treated with recombinant erythropoietin. N Engl J Med. 2002;346:469–75.
U.S. Pharmacopeal Convention. Immunogenicity assays-design and validation of assays to detect anti-drug neutralizing antibody. 2014. http://www.uspnf.com/ Accessed 11 July 2017,
U.S. Department of Health and Human Services. Food and Drug Administration. Guidance for industry. Assay development for immunogenicity testing of therapeutic proteins. Draft guidance (2009). July 11, 2017. Available from: https://www.regulations.gov/contentStreamer?documentId=FDA-2009-D-0539-0002&attachmentNumber=1&contentType=pdf
Finco D, Baltrukonis D, Clements-Egan A, Delaria K, Gunn GR III, Lowe J, et al. Comparison of competitive ligand-binding assay and bioassay formats for the measurement of neutralizing antibodies to protein therapeutics. J Pharm Biomed Anal. 2011;54:351–8.
Deehan M, Garces S, Kramer D, Baker MP, Rat D, Roettger Y, et al. Managing unwanted immunogenicity of biologicals. Autoimmun Rev. 2015;14:569–74.
Wu B, Chung S, Jiang XR, McNally J, Pedras-Vasconcelos J, Pillutla R, et al. Strategies to determine assay format for the assessment of neutralizing antibody responses to biotherapeutics. AAPS J. 2016;18:1335–50.
Cludts I, Meager A, Thorpe R, Wadhwa M. Development and characterization of a non-cell-based assay to assess the presence of neutralizing antibodies to interferon-beta in clinical samples. J Immunol Methods. 2013;395:37–44.
Hu J, Wala I, Han H, Nagatani J, Barger T, Civoli F, et al. Comparison of cell-based and non-cell-based assay platforms for the detection of clinically relevant anti-drug neutralizing antibodies for immunogenicity assessment of therapeutic proteins. J Immunol Methods. 2015;419:1–8.
Kolbeck R, Kozhich A, Koike M, Peng L, Andersson CK, Damschroder MM, et al. MEDI-563, a humanized anti-IL-5 receptor alpha mAb with enhanced antibody-dependent cell-mediated cytotoxicity function. J Allergy Clin Immunol. 2010;125:1344–53.e2.
Laviolette M, Gossage DL, Gauvreau G, Leigh R, Olivenstein R, Katial R, et al. Effects of benralizumab on airway eosinophils in asthmatic patients with sputum eosinophilia. J Allergy Clin Immunol. 2013;132:1086–96.e5.
Nowak RM, Parker JM, Silverman RA, Rowe BH, Smithline H, Khan F, et al. A randomized trial of benralizumab, an antiinterleukin 5 receptor alpha monoclonal antibody, after acute asthma. Am J Emerg Med. 2015;33:14–20.
Pham TH, Damera G, Newbold P, Ranade K. Reductions in eosinophil biomarkers by benralizumab in patients with asthma. Respir Med. 2016;111:21–9.
Castro M, Wenzel SE, Bleecker ER, Pizzichini E, Kuna P, Busse WW, et al. Benralizumab, an anti-interleukin 5 receptor alpha monoclonal antibody, versus placebo for uncontrolled eosinophilic asthma: a phase 2b randomised dose-ranging study. Lancet Respir Med. 2014;2:879–90.
Wu Y, Li JJ, Kim HJ, Liu X, Liu W, Akhgar A, et al. A neutralizing antibody assay based on a reporter of antibody-dependent cell-mediated cytotoxicity. AAPS J. 2015;17:1417–26.
Acknowledgments
This study was funded by AstraZeneca/MedImmune. Editing assistance was provided by Alan Saltzman, PhD, of JK Associates, Inc. (Conshohocken, PA, USA), and Michael A. Nissen, ELS, of AstraZeneca (Gaithersburg, MD, USA). This support was funded by AstraZeneca.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Wu, Y., Akhgar, A., Li, J.J. et al. Selection of a Ligand-Binding Neutralizing Antibody Assay for Benralizumab: Comparison with an Antibody-Dependent Cell-Mediated Cytotoxicity (ADCC) Cell-Based Assay. AAPS J 20, 49 (2018). https://doi.org/10.1208/s12248-018-0207-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1208/s12248-018-0207-8