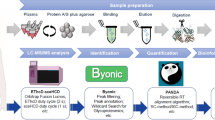
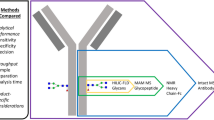
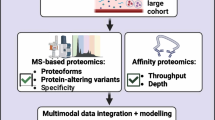
ABSTRACT
This paper represents the consensus views of a cross-section of companies and organizations from the USA and Canada regarding the validation and application of liquid chromatography tandem mass spectrometry (LC-MS/MS) methods for bioanalysis of protein biotherapeutics in regulated studies. It was prepared under the auspices of the AAPS Bioanalytical Focus Group’s Protein LC-MS Bioanalysis Subteam and is intended to serve as a guide to drive harmonization of best practices within the bioanalytical community and provide regulators with an overview of current industry thinking on applying LC-MS/MS technology for protein bioanalysis. For simplicity, the scope was limited to the most common current approach in which the protein is indirectly quantified using LC-MS/MS measurement of one or more of its surrogate peptide(s) produced by proteolytic digestion. Within this context, we considered a range of sample preparation approaches from simple in-matrix protein denaturation and digestion to complex procedures involving affinity capture enrichment. Consideration was given to the method validation experiments normally associated with traditional LC-MS/MS and ligand-binding assays. Our collective experience, thus far, is that LC-MS/MS methods for protein bioanalysis require different development and validation considerations than those used for small molecules. The method development and validation plans need to be tailored to the particular assay format being established, taking into account a number of important factors: the intended use of the assay, the test species or study population, the characteristics of the protein biotherapeutic and its similarity to endogenous proteins, potential interferences, as well as the nature, quality, and availability of reference and internal standard materials.



Similar content being viewed by others
REFERENCES
Dawes ML, Gu H, Wang J, Schuster AE, Haulenbeek J. Development of a validated liquid chromatography tandem mass spectrometry assay for a PEGylated adnectin in cynomolgus monkey plasma using protein precipitation and trypsin digestion. J Chromatogr B Anal Technol Biomed Life Sci. 2013;934:1–7.
Jiang H, Zeng J, Titsch C, Voronin K, Akinsanya B, Luo L, et al. Fully validated LC-MS/MS assay for the simultaneous quantitation of coadministered therapeutic antibodies in cynomolgus monkey serum. Anal Chem. 2013;85(20):9859–67.
Wang Y, Heilig JS. Differentiation and quantification of endogenous and recombinant-methionyl human leptin in clinical plasma samples by immunocapture/mass spectrometry. J Pharm Biomed Anal. 2012;70:440–6.
Xu K, Liu L, Maia M, Li J, Lowe J, Song A, et al. A multiplexed hybrid LC-MS/MS pharmacokinetic assay to measure two co-administered monoclonal antibodies in a clinical study. Bioanalysis. 2014;6(13):1781–94.
Huang L, Lu J, Wroblewski VJ, Beals JM, Riggin RM. In vivo deamidation characterization of monoclonal antibody by LC/MS/MS. Anal Chem. 2005;77(5):1432–9.
Hall MP, Gegg C, Walker K, Spahr C, Ortiz R, Patel V, et al. Ligand-binding mass spectrometry to study biotransformation of fusion protein drugs and guide immunoassay development: strategic approach and application to peptibodies targeting the thrombopoietin receptor. AAPS J. 2010;12(4):576–85.
U.S. Food and Drug Administration Guidance for Industry: Bioanalytical Method Validation, (2001).
European Medicines Agency Guideline on Bioanalytical Method Validation, (2011).
Brazilian ANVISA RESOLUÇÃO–RDC n 27, DE 17/05/2012, (2012).
Japanese MHLW Guideline on bioanalytical method validation in pharmaceutical development (2013).
U.S. Food and Drug Administration Guidance for Industry: Bioanalytical Method Validation-Draft Guidance, (2013).
Heudi O, Barteau S, Zimmer D, Schmidt J, Bill K, Lehmann N, et al. Towards absolute quantification of therapeutic monoclonal antibody in serum by LC-MS/MS using isotope-labeled antibody standard and protein cleavage isotope dilution mass spectrometry. Anal Chem. 2008;80(11):4200–7.
Li H, Ortiz R, Tran L, Hall M, Spahr C, Walker K, et al. General LC-MS/MS method approach to quantify therapeutic monoclonal antibodies using a common whole antibody internal standard with application to preclinical studies. Anal Chem. 2012;84(3):1267–73.
Li H, Ortiz R, Tran LT, Salimi-Moosavi H, Malella J, James CA, et al. Simultaneous analysis of multiple monoclonal antibody biotherapeutics by LC-MS/MS method in rat plasma following cassette-dosing. AAPS J. 2013;15(2):337–46.
Dufield DR, Radabaugh MR. Online immunoaffinity LC/MS/MS. A general method to increase sensitivity and specificity: how do you do it and what do you need? Methods. 2012;56(2):236–45.
Manuilov AV, Radziejewski CH, Lee DH. Comparability analysis of protein therapeutics by bottom-up LC-MS with stable isotope-tagged reference standards. MAbs. 2011;3(4):387–95.
Yang Z, Hayes M, Fang X, Daley MP, Ettenberg S, Tse FL. LC-MS/MS approach for quantification of therapeutic proteins in plasma using a protein internal standard and 2D-solid-phase extraction cleanup. Anal Chem. 2007;79(24):9294–301.
van den Broek I, Niessen WM, van Dongen WD. Bioanalytical LC-MS/MS of protein-based biopharmaceuticals. J Chromatogr B Anal Technol Biomed Life Sci. 2013;929:161–79.
European Medicines Agency Guideline on the Clinical Investigation of the Pharmacokinetics of Therapeutic Proteins, (2007).
Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics: MCP. 2002;1(5):376–86.
Ji C, Sadagopan N, Zhang Y, Lepsy C. A universal strategy for development of a method for absolute quantification of therapeutic monoclonal antibodies in biological matrices using differential dimethyl labeling coupled with ultra performance liquid chromatography-tandem mass spectrometry. Anal Chem. 2009;81(22):9321–8.
Wang S, Bobst CE, Kaltashov IA. Pitfalls in protein quantitation using acid-catalyzed O18 labeling: hydrolysis-driven deamidation. Anal Chem. 2011;83(18):7227–32.
Bronsema KJ, Bischoff R, van de Merbel NC. Internal standards in the quantitative determination of protein biopharmaceuticals using liquid chromatography coupled to mass spectrometry. J Chromatogr B Anal Technol Biomed Life Sci. 2012;893–894:1–14.
Staack RF, Stracke JO, Stubenrauch K, Vogel R, Schleypen J, Papadimitriou A. Quality requirements for critical assay reagents used in bioanalysis of therapeutic proteins: what bioanalysts should know about their reagents. Bioanalysis. 2011;3(5):523–34.
DeSilva B, Smith W, Weiner R, Kelley M, Smolec J, Lee B, et al. Recommendations for the bioanalytical method validation of ligand-binding assays to support pharmacokinetic assessments of macromolecules. Pharm Res. 2003;20(11):1885–900.
King LE, Farley E, Imazato M, Keefe J, Khan M, Ma M, et al. Ligand binding assay critical reagents and their stability: recommendations and best practices from the Global Bioanalysis Consortium Harmonization Team. AAPS J. 2014;16(3):504–15.
Bronsema KJ, Bischoff R, van de Merbel NC. High-sensitivity LC-MS/MS quantification of peptides and proteins in complex biological samples: the impact of enzymatic digestion and internal standard selection on method performance. Anal Chem. 2013;85(20):9528–35.
Knutsson M, Schmidt R, Timmerman P. LC-MS/MS of large molecules in a regulated bioanalytical environment—which acceptance criteria to apply? Bioanalysis. 2013;5(18):2211–4.
Kaur S, Xu K, Saad OM, Dere RC, Carrasco-Triguero M. Bioanalytical assay strategies for the development of antibody-drug conjugate biotherapeutics. Bioanalysis. 2013;5(2):201–26.
Gorovits B, Alley SC, Bilic S, Booth B, Kaur S, Oldfield P, et al. Bioanalysis of antibody-drug conjugates: American Association of Pharmaceutical Scientists Antibody-Drug Conjugate Working Group position paper. Bioanalysis. 2013;5(9):997–1006.
AAPS, editor AAPS Workshop Crystal City V: Quantitative Bioanalytical Methods Validation and Implementation: The 2013 Revised FDA Guidance. AAPS Workshop Crystal City V: Quantitative Bioanalytical Methods Validation and Implementation: The 2013 Revised FDA Guidance; 2013; Baltimore, MD.
ACKNOWLEDGMENTS
The authors wish to acknowledge the contributions of the many colleagues, within and outside their respective companies, who have provided valuable input with their reviews of the draft manuscript. We would also like to thank the AAPS organization and the Bioanalytical Focus Group for their support throughout this initiative. Additionally, we would like to recognize the Ligand-Binding Assay and Bioanalytical Focus Groups for their preliminary reviews of the draft manuscript. Their comments are greatly appreciated. Among all those that have provided valuable feedback, we would specifically like to acknowledge the following reviewers: Bradley L. Ackermann, Eli Lilly and Company; Lakshmi Amaravadi, Biogen Idec; Mark E. Arnold, Bristol-Myers Squibb Company; Michael J. Berna, Eli Lilly and Company; Suzanne Brignoli, Genentech; Margarete Brudny-Kloeppel, Bayer Pharma AG; Binodh DeSilva, Bristol-Myers Squibb Company; Jimmy Flarakos, Novartis Institutes for Biomedical Research; Eric Fluhler, Pfizer Inc.; Lindsay E. King, Pfizer Inc.; Stephen Lowes, Quintiles Bioanalytical and ADME Labs; An Song, Genentech; Lauren F. Stevenson, Biogen Idec; Matt Szapacs, GlaxoSmithKline; and Eric Yang, GlaxoSmithKline.
Author information
Authors and Affiliations
Corresponding author
GLOSSARY
- ADA
-
Anti-drug antibodies (also called ATAs, anti-therapeutic antibodies); ADAs are antibodies that are produced by the immune response (animal or human) against a dosed biotherapeutic.
- ADC
-
Antibody-drug conjugate; ADCs consist of a monoclonal antibody covalently conjugated with one or more drug molecules via a chemical linker.
- Affinity Capture
-
The purification and/or enrichment of an analyte (protein or peptide) from the sample (matrix or digest) onto a solid support (bead, column, membrane, or plate) via an immobilized reagent (e.g., antibody or target) having a high binding affinity.
- Immunoaffinity Capture
-
Specific form of affinity capture that uses an immunological reagent, such as an antibody. Immunopurification or immunoprecipitation (IP) and immunoenrichment (IE) are synonymous terms.
- LC-MS/MS
-
Technology or instrument combining liquid chromatographic separation with tandem mass spectrometric detection
- LBA
-
Ligand-binding assay refers to any analytical method, in which quantification is based on macromolecular interactions. The complex formed between a ligand and a binding molecule (either of which may be the analyte) is typically detected using enzyme-linked colorimetric, electrochemiluminescence or fluorescence methods. Immunoassay is one form of ligand-binding assay in which antibodies are used as binding reagents.
- Matrix Effect
-
The effect (i.e., suppression or enhancement) of chromatographically co-eluting residual matrix components of a biological sample on the ionization signal (typically ESI) of the detected analyte in the mass spectrometer.
- Monitoring Peptide
-
A peptide that is unique to the target protein analyte and selected to monitor to confirm or assess its structural integrity and evaluate assay robustness.
- Parallelism
-
Assessment of dilutional linearity using incurred samples, intended to demonstrate that the analyte concentration vs. response relationship observed for incurred samples is sufficiently similar to that measured for non-dosed standard and QC samples prepared in pooled matrix.
- Protein Therapeutic
-
Proteins used for treating diseases. A similar, but broader term, Biotherapeutics, applies to all types of biomolecules that can be used for treating diseases.
- Proteolytic Digestion
-
Procedure by which proteins are digested, primarily using enzymes such as trypsin, at cleavage points specifically related to their amino acid sequence, thereby reproducibly producing predictable peptide fragments. Chemical proteolysis with acid or cyanogen bromide is also sometimes used.
- Selectivity/Specificity
-
The ability of the bioanalytical method to measure and differentiate the analytes in the presence of components that may be expected to be present. These could include metabolites, impurities, degradants, or matrix components (11).
- Selected Reaction Monitoring
-
Selected reaction monitoring (SRM) refers to a tandem mass spectrometric method in which a precursor ion of a particular mass-to-charge ratio is selected in the first stage of a tandem mass spectrometer and a product ion produced by fragmentation of the precursor ion is selected in the second mass spectrometer stage for detection
- SISCAPA®
-
Stable isotope standards and capture by anti-peptide antibodies, a technique for simultaneous and specific affinity capture of a signature peptide and its SIL-peptide IS from a protein digest sample.
- Signature Peptide
-
General term for a specially selected peptide that is unique to the target protein analyte and can be used for quantification (i.e., as a surrogate) of the target protein or for other qualitative analytical purposes.
- SIL-IS, SIL-Protein IS, and SIL-Peptide IS
-
SIL-IS: stable isotope-labeled internal standard; a compound that has the same chemical structure as the analyte, except that some of atoms of the molecule are replaced with the corresponding stable isotopes, such as, 13C, 15N, 2H, and 18O. They can be used in quantitative LC-MS/MS assays to normalize the response of an analyte for variations in sample preparation, injection volume, matrix effects, and other instrumental conditions.
SIL-Protein IS: stable isotope-labeled protein internal standard, typically made by incorporating stable isotope containing amino acids into the protein during recombinant synthesis.
SIL-Peptide IS: stable-isotope-labeled peptide internal standard, typically made by incorporating stable isotope containing amino acids into the surrogate peptide during chemical synthesis.
- Soluble Target
-
An endogenous receptor or its soluble fragment that can be found in the circulation or excretion, to which a biotherapeutic drug is specifically designed to bind.
- Surrogate Peptide
-
A peptide that is specific to the target protein analyte, in the context of the assay and the intended sample matrix, and quantified in lieu of the target protein analyte in a LC-MS/MS assay
- Overall Recovery, Process Recovery, and Digestion Efficiency
-
Overall recovery: the combination of recoveries through all processes of an LC-MS/MS protein bioanalysis method, including pre-digestion, proteolytic digestion, and post-digestion treatments. It is typically calculated from the ratio of the detector responses obtained from matrix samples spiked with intact protein analyte prior to any processing, against blank matrix sample extracts spiked after final processing with a stoichiometric amount of surrogate peptide.
Process recovery: the recovery determined for each individual process step.
Digestion efficiency: the completeness or recovery of the digestion process. This is usually expressed as the molar equivalent ratio of surrogate peptide recovered from the final bioanalytical process to that of the nominal molar concentration of the biotherapeutic spiked into the QC sample pre-digestion.
Rights and permissions
About this article
Cite this article
Jenkins, R., Duggan, J.X., Aubry, AF. et al. Recommendations for Validation of LC-MS/MS Bioanalytical Methods for Protein Biotherapeutics. AAPS J 17, 1–16 (2015). https://doi.org/10.1208/s12248-014-9685-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1208/s12248-014-9685-5