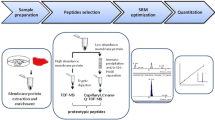
Abstract
Drug transporter expression in tissues (in vivo) usually differs from that in cell lines used to measure transporter activity (in vitro). Therefore, quantification of transporter expression in tissues and cell lines is important to develop scaling factor for in vitro to in vivo extrapolation (IVIVE) of transporter-mediated drug disposition. Since traditional immunoquantification methods are semiquantitative, targeted proteomics is now emerging as a superior method to quantify proteins, including membrane transporters. This superiority is derived from the selectivity, precision, accuracy, and speed of analysis by liquid chromatography tandem mass spectrometry (LC-MS/MS) in multiple reaction monitoring (MRM) mode. Moreover, LC-MS/MS proteomics has broader applicability because it does not require selective antibodies for individual proteins. There are a number of recent research and review papers that discuss the use of LC-MS/MS for transporter quantification. Here, we have compiled from the literature various elements of MRM proteomics to provide a comprehensive systematic strategy to quantify drug transporters. This review emphasizes practical aspects and challenges in surrogate peptide selection, peptide qualification, peptide synthesis and characterization, membrane protein isolation, protein digestion, sample preparation, LC-MS/MS parameter optimization, method validation, and sample analysis. In particular, bioinformatic tools used in method development and sample analysis are discussed in detail. Various pre-analytical and analytical sources of variability that should be considered during transporter quantification are highlighted. All these steps are illustrated using P-glycoprotein (P-gp) as a case example. Greater use of quantitative transporter proteomics will lead to a better understanding of the role of drug transporters in drug disposition.






Similar content being viewed by others
References
Giacomini KM, Huang SM, Tweedie DJ, Benet LZ, Brouwer KL, Chu X, et al. Membrane transporters in drug development. Nat Rev Drug Discov. 2010;9(3):215–36.
Keogh JP. Membrane transporters in drug development. Adv Pharmacol. 2012;63:1–42.
Yan Q. Membrane transporters and drug development: relevance to pharmacogenomics, nutrigenomics, epigenetics, and systems biology. Methods Mol Biol. 2010;637:1–21.
Eyal S, Chung FS, Muzi M, Link JM, Mankoff DA, Kaddoumi A, et al. Simultaneous PET imaging of P-glycoprotein inhibition in multiple tissues in the pregnant nonhuman primate. J Nucl Med. 2009;50(5):798–806.
Eyal S, Ke B, Muzi M, Link JM, Mankoff DA, Collier AC, et al. Regional P-glycoprotein activity and inhibition at the human blood–brain barrier as imaged by positron emission tomography. Clin Pharmacol Ther. 2010;87(5):579–85.
Muzi M, Mankoff DA, Link JM, Shoner S, Collier AC, Sasongko L, et al. Imaging of cyclosporine inhibition of P-glycoprotein activity using 11C-verapamil in the brain: studies of healthy humans. J Nucl Med. 2009;50(8):1267–75.
Harwood MD, Neuhoff S, Carlson GL, Warhurst G, Rostami-Hodjegan A. Absolute abundance and function of intestinal drug transporters: a prerequisite for fully mechanistic in vitro-in vivo extrapolation of oral drug absorption. Biopharm Drug Dispos. 2012;34(1):2–28.
Proctor NJ, Tucker GT, Rostami-Hodjegan A. Predicting drug clearance from recombinantly expressed CYPs: intersystem extrapolation factors. Xenobiotica. 2004;34(2):151–78.
Deo AK, Prasad B, Balogh L, Lai Y, Unadkat JD. Interindividual variability in hepatic expression of the multidrug resistance-associated protein 2 (MRP2/ABCC2): quantification by liquid chromatography/tandem mass spectrometry. Drug Metab Dispos. 2012;40(5):852–5.
Lee N, Hebert MF, Prasad B, Easterling TR, Kelly EJ, Unadkat JD, et al. Effect of gestational age on mRNA and protein expression of polyspecific organic cation transporters during pregnancy. Drug Metab Dispos. 2013.
Prasad B, Evers R, Gupta A, Hop CE, Salphati L, Shukla S, et al. Interindividual variability in hepatic organic anion-transporting polypeptides and P-glycoprotein (ABCB1) protein expression: quantification by liquid chromatography tandem mass spectroscopy and influence of genotype, age, and sex. Drug Metab Dispos. 2013.
Prasad B, Lai Y, Lin Y, Unadkat JD. Interindividual variability in the hepatic expression of the human breast cancer resistance protein (BCRP/ABCG2): effect of age, sex, and genotype. J Pharm Sci. 2013;102(3):787–93.
Prasad B, Unadkat J. Comparison of heavy labeled (SIL) peptide vs. SILAC protein internal standards for LC-MS/MS quantification of hepatic drug transporters. Int J Proteomics. 2014;2014:1–11.
Ohtsuki S, Schaefer O, Kawakami H, Inoue T, Liehner S, Saito A, et al. Simultaneous absolute protein quantification of transporters, cytochromes P450, and UDP-glucuronosyltransferases as a novel approach for the characterization of individual human liver: comparison with mRNA levels and activities. Drug Metab Dispos. 2011;40(1):83–92.
Agarwal S, Uchida Y, Mittapalli RK, Sane R, Terasaki T, Elmquist WF. Quantitative proteomics of transporter expression in brain capillary endothelial cells isolated from P-glycoprotein (P-gp), breast cancer resistance protein (Bcrp), and P-gp/Bcrp knockout mice. Drug Metab Dispos. 2012;40(6):1164–9.
Ohtsuki S, Uchida Y, Kubo Y, Terasaki T. Quantitative targeted absolute proteomics-based ADME research as a new path to drug discovery and development: methodology, advantages, strategy, and prospects. J Pharm Sci. 2011;100(9):3547–59.
Uchida Y, Ohtsuki S, Katsukura Y, Ikeda C, Suzuki T, Kamiie J, et al. Quantitative targeted absolute proteomics of human blood–brain barrier transporters and receptors. J Neurochem. 2011;117(2):333–45.
Uchida Y, Tachikawa M, Obuchi W, Hoshi Y, Tomioka Y, Ohtsuki S, et al. A study protocol for quantitative targeted absolute proteomics (QTAP) by LC-MS/MS: application for inter-strain differences in protein expression levels of transporters, receptors, claudin-5, and marker proteins at the blood–brain barrier in ddY, FVB, and C57BL/6 J mice. Fluids Barriers CNS. 2013;10(1):21.
Groer C, Bruck S, Lai Y, Paulick A, Busemann A, Heidecke CD, et al. LC-MS/MS-based quantification of clinically relevant intestinal uptake and efflux transporter proteins. J Pharm Biomed Anal. 2013;85:253–61.
Qiu X, Bi YA, Balogh LM, Lai Y. Absolute measurement of species differences in sodium taurocholate cotransporting polypeptide (NTCP/Ntcp) and its modulation in cultured hepatocytes. J Pharm Sci. 2013;102(9):3252–63.
Zhang Y, Li N, Brown PW, Ozer JS, Lai Y. Liquid chromatography/tandem mass spectrometry based targeted proteomics quantification of P-glycoprotein in various biological samples. Rapid Commun Mass Spectrom. 2011;25(12):1715–24.
Li N, Singh P, Mandrell KM, Lai Y. Improved extrapolation of hepatobiliary clearance from in vitro sandwich cultured rat hepatocytes through absolute quantification of hepatobiliary transporters. Mol Pharm. 2010;7(3):630–41.
Li N, Palandra J, Nemirovskiy OV, Lai Y. LC-MS/MS mediated absolute quantification and comparison of bile salt export pump and breast cancer resistance protein in livers and hepatocytes across species. Anal Chem. 2009;81(6):2251–9.
Li N, Nemirovskiy OV, Zhang Y, Yuan H, Mo J, Ji C, et al. Absolute quantification of multidrug resistance-associated protein 2 (MRP2/ABCC2) using liquid chromatography tandem mass spectrometry. Anal Biochem. 2008;380(2):211–22.
Maiolica A, Junger MA, Ezkurdia I, Aebersold R. Targeted proteome investigation via selected reaction monitoring mass spectrometry. J Proteome. 2012;75(12):3495–513.
Picotti P, Aebersold R. Selected reaction monitoring-based proteomics: workflows, potential, pitfalls and future directions. Nat Methods. 2013;9(6):555–66.
Kamiie J, Ohtsuki S, Iwase R, Ohmine K, Katsukura Y, Yanai K, et al. Quantitative atlas of membrane transporter proteins: development and application of a highly sensitive simultaneous LC/MS/MS method combined with novel in-silico peptide selection criteria. Pharm Res. 2008;25(6):1469–83.
Oswald S, Groer C, Drozdzik M, Siegmund W. Mass spectrometry-based targeted proteomics as a tool to elucidate the expression and function of intestinal drug transporters. AAPS J. 2013;15(4):1128–40.
Vuckovic D, Dagley LF, Purcell AW, Emili A. Membrane proteomics by high performance liquid chromatography-tandem mass spectrometry: analytical approaches and challenges. Proteomics. 2013;13(3–4):404–23.
Shukla HD, Vaitiekunas P, Cotter RJ. Advances in membrane proteomics and cancer biomarker discovery: current status and future perspective. Proteomics. 2012;12(19–20):3085–104.
Aebersold R, Burlingame AL, Bradshaw RA. Western blots versus selected reaction monitoring assays: time to turn the tables? Mol Cell Proteomics. 2013;12(9):2381–2.
Hoofnagle AN, Wener MH. The fundamental flaws of immunoassays and potential solutions using tandem mass spectrometry. J Immunol Methods. 2009;347(1–2):3–11.
Razavi M, Frick LE, LaMarr WA, Pope ME, Miller CA, Anderson NL, et al. High-throughput SISCAPA quantitation of peptides from human plasma digests by ultrafast, liquid chromatography-free mass spectrometry. J Proteome Res. 2012;11(12):5642–9.
Anderson L, Hunter CL. Quantitative mass spectrometric multiple reaction monitoring assays for major plasma proteins. Mol Cell Proteomics. 2006;5(4):573–88.
Schmidt C, Lenz C, Grote M, Luhrmann R, Urlaub H. Determination of protein stoichiometry within protein complexes using absolute quantification and multiple reaction monitoring. Anal Chem. 2010;82(7):2784–96.
Brant SR, Panhuysen CI, Nicolae D, Reddy DM, Bonen DK, Karaliukas R, et al. MDR1 Ala893 polymorphism is associated with inflammatory bowel disease. Am J Hum Genet. 2003;73(6):1282–92.
Kuzyk MA, Parker CE, Domanski D, Borchers CH. Development of MRM-based assays for the absolute quantitation of plasma proteins. Methods Mol Biol. 2013;1023:53–82.
Kioka N, Tsubota J, Kakehi Y, Komano T, Gottesman MM, Pastan I, et al. P-glycoprotein gene (MDR1) cDNA from human adrenal: normal P-glycoprotein carries Gly185 with an altered pattern of multidrug resistance. Biochem Biophys Res Commun. 1989;162(1):224–31.
Chen G, Duran GE, Steger KA, Lacayo NJ, Jaffrezou JP, Dumontet C, et al. Multidrug-resistant human sarcoma cells with a mutant P-glycoprotein, altered phenotype, and resistance to cyclosporins. J Biol Chem. 1997;272(9):5974–82.
Rodriguez J, Gupta N, Smith RD, Pevzner PA. Does trypsin cut before proline? J Proteome Res. 2008;7(1):300–5.
Lai M, Skanchy D, Stobaugh J, Topp E. Capillary electrophoresis separation of an asparagine containing hexapeptide and its deamidation products. J Pharm Biomed Anal. 1998;18(3):421–7.
Neta P, Pu QL, Kilpatrick L, Yang X, Stein SE. Dehydration versus deamination of N-terminal glutamine in collision-induced dissociation of protonated peptides. J Am Soc Mass Spectrom. 2007;18(1):27–36.
Suzuki Y, Motoi H, Sato K. Quantitative analysis of pyroglutamic acid in peptides. J Agric Food Chem. 1999;47(8):3248–51.
Crimmins DL, Mische SM, Denslow ND. Chemical cleavage of proteins in solution. Curr Protoc Protein Sci. 2005;Chapter 11:Unit 11 4.
Lange V, Picotti P, Domon B, Aebersold R. Selected reaction monitoring for quantitative proteomics: a tutorial. Mol Syst Biol. 2008;4:222.
Sprung RW, Martinez MA, Carpenter KL, Ham AJ, Washington MK, Arteaga CL, et al. Precision of multiple reaction monitoring mass spectrometry analysis of formalin-fixed, paraffin-embedded tissue. J Proteome Res. 2012.
Li S, Schoneich C, Borchardt RT. Chemical instability of protein pharmaceuticals: mechanisms of oxidation and strategies for stabilization. Biotechnol Bioeng. 1995;48(5):490–500.
Isidro-Llobet A, Alvarez M, Albericio F. Amino acid-protecting groups. Chem Rev. 2009;109(6):2455–504.
Krchnak V, Flegelova Z, Vagner J. Aggregation of resin-bound peptides during solid-phase peptide synthesis. Prediction of difficult sequences. Int J Pept Protein Res. 1993;42(5):450–4.
Orsburn BC, Stockwin LH, Newton DL. Challenges in plasma membrane phosphoproteomics. Exp Rev Proteome. 2011;8(4):483–94.
Wang Z, Fu M, Wang L, Liu J, Li Y, Brakebusch C, et al. p21-Activated kinase 1 (PAK1) can promote ERK activation in a kinase-independent manner. J Biol Chem. 2013;288(27):20093–9.
Bi YA, Qiu X, Rotter CJ, Kimoto E, Piotrowski M, Varma MV, et al. Quantitative assessment of the contribution of sodium-dependent taurocholate co-transporting polypeptide (NTCP) to the hepatic uptake of rosuvastatin, pitavastatin and fluvastatin. Biopharm Drug Dispos. 2013;34(8):452–61.
Li N, Bi YA, Duignan DB, Lai Y. Quantitative expression profile of hepatobiliary transporters in sandwich cultured rat and human hepatocytes. Mol Pharm. 2009;6(4):1180–9.
Li M, Yuan H, Li N, Song G, Zheng Y, Baratta M, et al. Identification of interspecies difference in efflux transporters of hepatocytes from dog, rat, monkey and human. Eur J Pharm Sci. 2008;35(1–2):114–26.
Noack A, Noack S, Hoffmann A, Maalouf K, Buettner M, Couraud PO, et al. Drug-induced trafficking of p-glycoprotein in human brain capillary endothelial cells as demonstrated by exposure to mitomycin C. PLoS ONE. 2014;9(2):e88154.
Howell GJ, Holloway ZG, Cobbold C, Monaco AP, Ponnambalam S. Cell biology of membrane trafficking in human disease. Int Rev Cytol. 2006;252:1–69.
Ameen N, Silvis M, Bradbury NA. Endocytic trafficking of CFTR in health and disease. J Cyst Fibros. 2007;6(1):1–14.
Proc JL, Kuzyk MA, Hardie DB, Yang J, Smith DS, Jackson AM, et al. A quantitative study of the effects of chaotropic agents, surfactants, and solvents on the digestion efficiency of human plasma proteins by trypsin. J Proteome Res. 2010;9(10):5422–37.
Burkhart JM, Schumbrutzki C, Wortelkamp S, Sickmann A, Zahedi RP. Systematic and quantitative comparison of digest efficiency and specificity reveals the impact of trypsin quality on MS-based proteomics. J Proteome. 2011;75(4):1454–62.
Ye X, Li L. Microwave-assisted protein solubilization for mass spectrometry-based shotgun proteome analysis. Anal Chem. 2012;84(14):6181–91.
Rial-Otero R, Carreira RJ, Cordeiro FM, Moro AJ, Santos HM, Vale G, et al. Ultrasonic assisted protein enzymatic digestion for fast protein identification by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Sonoreactor versus ultrasonic probe. J Chromatogr A. 2007;1166(1–2):101–7.
Lopez-Ferrer D, Petritis K, Hixson KK, Heibeck TH, Moore RJ, Belov ME, et al. Application of pressurized solvents for ultrafast trypsin hydrolysis in proteomics: proteomics on the fly. J Proteome Res. 2008;7(8):3276–81.
Krenkova J, Lacher NA, Svec F. Highly efficient enzyme reactors containing trypsin and endoproteinase LysC immobilized on porous polymer monolith coupled to MS suitable for analysis of antibodies. Anal Chem. 2009;81(5):2004–12.
Saveliev S, Bratz M, Zubarev R, Szapacs M, Budamgunta H, Urh M. Trypsin/Lys-C protease mix for enhanced protein mass spectrometry analysis. Nature Methods. 2013;10.
Leon IR, Schwammle V, Jensen ON, Sprenger RR. Quantitative assessment of in-solution digestion efficiency identifies optimal protocols for unbiased protein analysis. Mol Cell Proteomics. 2013;12(10):2992–3005.
Tanca A, Biosa G, Pagnozzi D, Addis MF, Uzzau S. Comparison of detergent-based sample preparation workflows for LTQ-Orbitrap analysis of the Escherichia coli proteome. Proteomics. 2013;13(17):2597–607.
Adrait A, Lebert D, Trauchessec M, Dupuis A, Louwagie M, Masselon C, et al. Development of a Protein Standard Absolute Quantification (PSAQ) assay for the quantification of Staphylococcus aureus enterotoxin A in serum. J Proteome. 2011;75(10):3041–9.
Brownridge PJ, Harman VM, Simpson DM, Beynon RJ. Absolute multiplexed protein quantification using QconCAT technology. Methods Mol Biol. 2012;893:267–93.
Castro-Borges W, Simpson DM, Dowle A, Curwen RS, Thomas-Oates J, Beynon RJ, et al. Abundance of tegument surface proteins in the human blood fluke Schistosoma mansoni determined by QconCAT proteomics. J Proteome. 2011;74(9):1519–33.
Russell MR, Achour B, McKenzie EA, Lopez R, Harwood MD, Rostami-Hodjegan A, et al. Alternative fusion protein strategies to express recalcitrant QconCAT proteins for quantitative proteomics of human drug metabolizing enzymes and transporters. J Proteome Res. 2013;12(12):5934–42.
Vertommen A, Panis B, Swennen R, Carpentier SC. Evaluation of chloroform/methanol extraction to facilitate the study of membrane proteins of non-model plants. Planta. 2010;231(5):1113–25.
Chernokalskaya E, Gutierrez S, Pitt AM, Leonard JT. Ultrafiltration for proteomic sample preparation. Electrophoresis. 2004;25(15):2461–8.
Wang H, Qian WJ, Mottaz HM, Clauss TR, Anderson DJ, Moore RJ, et al. Development and evaluation of a micro- and nanoscale proteomic sample preparation method. J Proteome Res. 2005;4(6):2397–403.
Amelina H, Sjodin MO, Bergquist J, Cristobal S. Quantitative subproteomic analysis of age-related changes in mouse liver peroxisomes by iTRAQ LC-MS/MS. J Chromatogr B Anal Technol Biomed Life Sci. 2011;879(30):3393–400.
Bergmann U, Ahrends R, Neumann B, Scheler C, Linscheid MW. Application of metal-coded affinity tags (MeCAT): absolute protein quantification with top-down and bottom-up workflows by metal-coded tagging. Anal Chem. 2012;84(12):5268–75.
Bettmer J. Application of isotope dilution ICP-MS techniques to quantitative proteomics. Anal Bioanal Chem. 2010;397(8):3495–502.
Butler GS, Dean RA, Morrison CJ, Overall CM. Identification of cellular MMP substrates using quantitative proteomics: isotope-coded affinity tags (ICAT) and isobaric tags for relative and absolute quantification (iTRAQ). Methods Mol Biol. 2010;622:451–70.
Chen Z, Wang Q, Lin L, Tang Q, Edwards JL, Li S, et al. Comparative evaluation of two isobaric labeling tags, DiART and iTRAQ. Anal Chem. 2012;84(6):2908–15.
Colaert N, Gevaert K, Martens L. RIBAR and xRIBAR: methods for reproducible relative MS/MS-based label-free protein quantification. J Proteome Res. 2011;10(7):3183–9.
Anderson NL, Jackson A, Smith D, Hardie D, Borchers C, Pearson TW. SISCAPA peptide enrichment on magnetic beads using an in-line bead trap device. Mol Cell Proteomics. 2009;8(5):995–1005.
Anderson NL, Anderson NG, Haines LR, Hardie DB, Olafson RW, Pearson TW. Mass spectrometric quantitation of peptides and proteins using Stable Isotope Standards and Capture by Anti-Peptide Antibodies (SISCAPA). J Proteome Res. 2004;3(2):235–44.
Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1(5):376–86.
Acknowledgments
This study was supported in part by The University of Washington Research Affiliate program on Transporters (UWRAPT).
Author information
Authors and Affiliations
Corresponding author
Additional information
Guest Editors: Marilyn E. Morris and Yurong Lai
Rights and permissions
About this article
Cite this article
Prasad, B., Unadkat, J.D. Optimized Approaches for Quantification of Drug Transporters in Tissues and Cells by MRM Proteomics. AAPS J 16, 634–648 (2014). https://doi.org/10.1208/s12248-014-9602-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1208/s12248-014-9602-y