Abstract
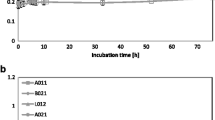
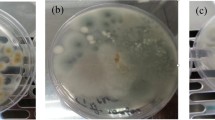
Removal of petroleum benzene, toluene, and xylene compounds from the environment is necessary to ensure quality life. In this research, 41 yeasts were isolated from oily soils. Among them, nine yeasts named KKUs (A5, A6, A12, A20, A23, A24, A26, A29, and A38) were selected based on their use of benzene, toluene, and xylene as a sole carbon and energy source. Based on their growth rates, all selected yeasts displayed a high efficiency for toluene degradation, but had no ability to degrade benzene and a low ability to degrade xylene, except A29 and A38, which could not degrade xylene. HPLC analysis for toluene removal indicated that A6, A12, A20, A23, A24, and A26 almost completely removed the toluene compound after 3 days of incubation (92.74, 94.61, 95.05, 91.74, 91.85, and 97.29%, respectively). In addition, strains A29 and A38 showed moderate degradation (88.29 and 85.30%, respectively), while the ability of A5 was low (39.00%). The isolates were identified based on amplifying and sequencing the D1/D2 domain of the 26S rRNA gene. Alignments and comparisons of the 26S rRNA gene sequences of the isolates with those available in GenBank, plus phylogenetic analysis, proved isolates as Rhodotorula lactose KKU-A5, Rhodotorula nymphaeae KKU-A6, Rhodotorula graminis KKU-A12, Rhodotorula minuta KKU-A20, Exophiala dermatitidis KKU-A23, Candida davisiana KKU-A24, Rhodotorula slooffiae KKU-A26, Rhodotorula mucilaginosa KKU-A29, and Rhodosporidium diobovatum KKU-A38. Random amplified polymorphic DNA-PCR fingerprinting was accomplished within seven toluene-degrading red yeasts (A5, A6, A12, A20, A26, A29, and A38). The results indicated no correlation between the random amplified polymorphic DNA profile and the geographic origin of the isolates.
Similar content being viewed by others
References
Mitra, S. and Roy, P., BTEX: a serious ground-water contaminant, Res. J. Environ. Sci., 2011, vol. 5, pp. 394–398.
Singh, S., Kang, S.H., Mulchandani, A., and Chen, W., Bioremediation: environmental clean-up through pathway engineering. Curr. Opin. Biotechnol., 2008, vol.19, pp. 437–444.
Díaz, E., 2008. Microbial Biodegradation: Genomics and Molecular Biology, Horizon Scientific Press, 2008.
Dua, M., Singh A., Sethunathan N., and Johri A., Biotechnology and bioremediation: successes and limitations, Appl. Microbiol. Biotechnol., 2002, vol. 59, pp. 143–152.
Hesham, A., Khan, S., Liu, X., et al., Application of PCR–DGGE to analyze the yeast population dynamics in slurry reactors during degradation of polycyclic aromatic hydrocarbons in weathered oil, Yeast, 2006, vol. 23, pp. 879–887.
Cappello, S., Caruso, G., Zampino, D., et al., Microbial community dynamics during assays of harbour oil spill bioremediation: a microscale simulation study, J. Appl. Microbiol., 2007, vol. 102, pp. 184–194.
Hesham, A., Alrumman, A.S., and Jawaher, A., 16S rDNA phylogenetic and RAPD–PCR analysis of petroleum polycyclic aromatic hydrocarbons degrading bacteria enriched from oil-polluted soils, Arab. J. Sci. Eng., 2015, vol. 41, pp. 2095–2106.
Hesham, A., Wang, Z., Zhang, Y., et al., Isolation and identification of a yeast strain capable of degrading four and five ring aromatic hydrocarbons, Ann. Microbiol., 2006, vol. 56, pp. 109–112.
Abari, A.H., Emtiazi, G., Ghasemi S.M., et al., Isolation and characterization of a novel toluene-degrading bacterium exhibiting potential application in bioremediation, J. Microbiol., 2013, vol. 6, pp. 256–261.
Farag, S. and Soliman, N.A., Biodegradation of crude petroleum oil and environmental pollutants by Candida tropicalis strain, Braz. Arch. Biol. Technol., 2011, vol. 54, pp. 821–830.
Hesham, A., New safety and rapid method for extraction of genomic DNA from bacteria and yeast strains suitable for PCR amplifications, J. Pure Appl. Microbiol., 2014, vol. 8, pp. 383–388.
Kurtzman, C. and Robnett, C., Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie Van Leeuwenhoek, 1998, vol. 73, pp. 331–371.
Hesham, A., Khan, S., Tao, Y., et al., Biodegradation of high molecular weight PAHs using isolated yeast mixtures: application of meta-genomic methods for community structure analyses. Environ. Sci. Pollut. Res., 2012, vol. 19, pp. 3568–3578.
Coates, J.D., Chakraborty, R., and Lack, J.G., Anaerobic benzene oxidation coupled to nitrate reduction in pure culture by two strains of Dechloromonas, Nature, 2001, vol. 411, pp. 1039–1043.
Martorell, P., Fernández-Espinar, M.T., and Querol, A., Molecular monitoring of spoilage yeasts during the production of candied fruit nougats to determine food contamination sources, Int. J. Food Microbiol., 2005, vol. 101, pp. 293–302.
Yeh, F.C. and Boyle, T.B., Population genetic analysis of co-dominant and dominant markers and quantitative traits. Belg. J. Bot., 1997, pp. 129–157.
Hunter, P.R and Gaston, M. A., Numerical index of the discriminatory ability of typing systems: an application of Simpson’s index of diversity, J. Clin. Microbiol., 1988, vol. 26, pp. 2465–2466.
Larik, I., Qazi, M., Kanhar, A., et al., Biodegradation of petrochemical hydrocarbons using an efficient bacterial consortium: A2457, Arab. J. Sci. Eng., 2015, pp. 1–10.
Chaillan, F., Le Flèche, A., Bury, E., et al., Identification and biodegradation potential of tropical aerobic hydrocarbon-degrading microorganisms, Res. Microbiol., 2004, vol. 155, pp. 587–595.
Heider., J., Spormann, A.M., Beller, H.R., et al., Anaerobic bacterial metabolism of hydrocarbons, FEMS Microbiol. Rev., 1998, vol. 22, pp. 459–473.
Hassanshahian, M., Tebyanian, H., and Cappello, S., Isolation and characterization of two crude oil-degrading yeast strains, Yarrowia lipolytica PG-20 and PG-32, from the Persian Gulf, Mar. Pollut. Bull., 2012, vol. 64, pp.1386–1391.
Leahy, J.G., Tracy, K.D., and Eley, M.H., Degradation of mixtures of aromatic and chloroaliphatic hydrocarbons by aromatic hydrocarbon-degrading bacteria, FEMS Microbiol. Ecol., 2003, vol. 43, pp. 271–276.
Daniel, H.M. and Meyer, W., Evaluation of ribosomal RNA and actin gene sequences for the identification of ascomycetous yeasts, Int. J. Food Microbiol., 2003, vol. 86, pp. 61–78.
Frutos, R., Fernández-Espinar, M.T., and Querol, A., Identification of species of the genus Candida by analysis of the 5.8S rRNA gene and the two ribosomal internal transcribed spacers, Antonie Van Leeuwenhoek, 2004, vol. 85, pp. 175–185.
Kurtzman, C.P., Four new yeasts in the Pichia anomala clade, Int. J. Syst. Evol. Microbiol., 2000, vol. 50, pp. 395–404.
Abliz, P., Fukushima, K., Takizawa, K., and Nishimura, K., Identification of pathogenic dematiaceous fungi and related taxa based on large subunit ribosomal DNA D1/D2 domain sequence analysis, FEMS Immunol. Med. Microbiol., 2004, vol. 40, pp. 41–49.
Bento, F.M., Camargo, F.A.O., Okeke, B.C., et al., Comparative bioremediation of soils contaminated with diesel oil by natural attenuation, biostimulation and bioaugmentation, Bioresour. Technol., 2005, vol. 96, pp. 1049–1055.
Kumari, M. and Abraham, J., Biodegradation of diesel oil using yeast Rhodosporidium toruloides, Res. J. Environ. Toxicol., 2011, vol. 5, pp. 369–377.
Molnár, E., Baude, A., Richmond, S.A., et al., Biochemical and immunocytochemical characterization of antipeptide antibodies to a cloned GluR1 glutamate receptor subunit: cellular and subcellular distribution in the rat forebrain, Neuroscience, 1993, vol. 53, pp. 307–326.
Quesada, M. and Cenis, J., Use of random amplified polymorphic DNA (RAPD–PCR) in the characterization of wine yeasts, Am. J. Enol. Vitic., 1995, vol. 46, pp. 204–208.
Tofalo, R., Chaves-López, C., Di Fabio, F., et al., Molecular identification and osmotolerant profile of wine yeasts that ferment a high sugar grape must, Int. J. Food Microbiol., 2009, vol. 130, pp. 179–187.
Meroth, B., Hammes, W.P., and Hertel, C., Identification and population dynamics of yeasts in sourdough fermentation processes by PCR-denaturing gradient gel electrophoresis, Appl. Environ. Microbiol., 2003, vol. 69, pp. 7453–7461.
EL-Fiky, Z.A., Hassan, G.M., and Emam, A.M., Quality parameters and RAPD–PCR differentiation of commercial baker’s yeast and hybrid strains, J. Food Sci., 2012, vol. 77, pp. 312–317.
Author information
Authors and Affiliations
Corresponding author
Additional information
The article is published in the original.
Rights and permissions
About this article
Cite this article
Hesham, A.EL., Alrumman, S.A. & ALQahtani, A.D.S. Degradation of Toluene Hydrocarbon by Isolated Yeast Strains: Molecular Genetic Approaches for Identification and Characterization. Russ J Genet 54, 933–943 (2018). https://doi.org/10.1134/S1022795418080070
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S1022795418080070