Abstract
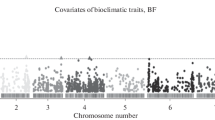
Genetic diversity of native populations of North Eurasia is investigated using a panel of genetic markers of candidate genes for cold climate adaptation. A high level of within- and between-population variability is detected. Comparative analysis of data on North Eurasian populations combined with data on worldwide populations from the 1000 Genomes and HDGP projects reveals correlations of genetic diversity in candidate genes for cold climate adaptation with key climate parameters, as well as the increase of genetic diversity in markers of this group of genes with the increase of latitude, that is, as modern humans migrated out of Africa. Using the method of searching for extreme empirical values of the coefficient of genetic diversity, signals of directional selection for markers of six genes adaptive to cold (MYOF, LONP2, IFNL4, MKL1, SLC2A12, and CPT1A) are found. The data are discussed in framework of the hypothesis of decanalization of genome–phenome relationships under the pressure of natural selection during human settlement throughout the world.
Similar content being viewed by others
References
Malyarchuk, B.A., Derenko, M.V., Denisova, G.A., and Litvinov, A.N., Distribution of the arctic variant of the CPT1A gene in indigenous populations of Siberia, Vavilovskii Zh. Genet. Sel., 2016, vol. 20, no. 5, pp. 571–575. doi 10.18699/VJ16.130
Babenko, V.N., Isakova, Zh.T., Talaibekova, E.T., et al., Polymorphism in the TRP8 gene in Kyrgyz population: putative association with highland adaptation, Russ. J. Genet.: Appl. Res., 2016, vol. 6, no. 5, pp. 605–612. doi 10.1134/S2079059716050038
Cardona, A., Pagani, L., Antao, T., et al., Genomewide analysis of cold adaptation in indigenous Siberian populations, PLoS One, 2014, vol. 9, no. 5, e98076. doi 10.1371/journal.pone.0098076
Lappalainen, T., Salmela, E., Andersen, P.M., et al., Genomic landscape of positive natural selection in Northern European populations, Eur. J. Hum. Genet., 2010, vol. 18, pp. 471–478. doi 10.1038/ejhg.2009.184
Rasmussen, M., Li, Y., Lindgreen, S., et al., Ancient human genome sequence of an extinct Palaeo-Eskimo, Nature, 2010, vol. 463, pp. 757–762. doi 10.1038/nature08835
Hancock, A.M., Witonsky, D.B., Alkorta-Aranburu, G., et al., Adaptations to climate-mediated selective pressures in humans, PLoS Genet., 2011, vol. 7, no. 4, e1001375. doi 10.1371/journal.pgen.1001375
Stepanov, V.A., Etnogenomika naseleniya Severnoi Evrazii (Population Ethnogenomics of Northern Eurasia), Tomsk: Pechatnaya Manufaktura, 2002.
Khitrinskaya, I.Yu., Khar’kov, V.N., Voevoda, M.I, and Stepanov, V.A., Genetic diversity and relationships of populations of northern Eurasia by polymorphic Alu insertions, Mol. Biol. (Moscow), 2014, vol. 48, no. 1, pp. 58–68. doi 10.1134/S0026893314010051
Cherednichenko, A.A., Trifonova, E.A., Vagaitseva, K.V., et al., Association of the genetic polymorphism of cytokines and their receptors with climate and geographic factors in human populations, Russ. J. Genet., 2014, vol. 50, no. 10, pp. 1112–1116. doi 10.1134/S1022795-414100020
The 1000 Genomes Project Consortium, A global reference for human genetic variation, Nature, 2015, vol. 526, pp. 68–74. doi 10.1038/nature15393
Sudmant, P.H., Rausch, T., Gardner, E.J., et al., An integrated map of structural variation in 2504 human genomes, Nature, 2015, vol. 526, pp. 75–81. doi 10.1038/nature15394
Cann, H.M., De Toma, C., Cazes, L., et al., A human genome diversity cell line panel, Science, 2002, vol. 296, no. 5566, pp. 261–262.
Stepanov, V.A., Vagajceva, K.V., Bocharova, A.V., and Khar’kov, V.N., Development of multiplex genotyping method of polymorphic markers for genes involved in human adaptation to cold climate, Sci. Evol., 2016, vol. 2, no. 2, pp. 92–101. doi 10.21603/2500–1418–2016–1–2–92–101
Stepanov, V.A. and Trifonova, E.A., Multiplex SNP genotyping by MALDI-TOF mass spectrometry: frequencies of 56 immune response gene SNPs in human populations, Mol. Biol. (Moscow), 2013, vol. 47, no. 6, pp. 852–862. doi 10.1134/S0026893313060149
Excoffier, L. and Lischer, H.E.L., Arlequin Suite ver. 3.5: a new series of programs to perform population genetics analyses under Linux and Windows, Mol. Ecol. Resour., 2010, vol. 10, pp. 564–567. doi 10.1111/j.1755- 0998.2010.02847.x
Stepanov, V.A., Khar’kov, V.N., Trifonova, E.A., and Marusin, A.V., Metody statisticheskogo analiza v populyatsionnoi i evolyutsionnoi genetike cheloveka: uchebnometodicheskoe posobie (Methods of Statistical Analysis in Population and Evolutionary Human Genetics: A Study Guide), in Ser. Nasledstvennost’ i zdorov’e, (Heredity and Health), Tomsk: Pechatnaya Manufaktura, 2014, issue 12.
Schlotterer, C., A microsatellite-based multilocus screen for the identification of local selective sweeps, Genetics, 2002, vol. 160, pp. 753–763.
Beaumont, M.A. and Balding, D.J., Identifying adaptive genetic divergence among populations from genome scans, Mol. Ecol., 2004, vol. 13, pp. 969–980.
Foll, M. and Gaggiotti, O.E., A genome scan method to identify selected loci appropriate for both dominant and codominant markers: a Bayesian perspective, Genetics, 2008, vol. 180, pp. 977–993. doi 10.1534/genetics.108.092221
Excoffier, L., Hofer, T., and Foll, M., Detecting loci under selection in a hierarchically structured population, Heredity, 2009, vol. 103, pp. 285–298. doi 10.1038/hdy.2009.74
Stepanov, V.A., Kandelariya, P., Kkho, S., et al., Immune response decanalization in the resettlement of modern humans: the relationship of genetic diversity in the genes of the immune system with climatic and geographical factors, Med. Genet., 2013, no. 12, pp. 8–18.
Stepanov, V.A., Evolution of genetic diversity and human diseases, Russ. J. Genet., 2016, vol. 52, no. 7, pp. 746–756. doi 10.1134/S1022795416070103
Pilewski, J.M., Latoche, J.D., Arcasoy, S.M., and Albelda, S.M., Expression of integrin cell adhesion receptors during human airway epithelial repair in vivo, Am. J. Physiol., 1997, vol. 273, pp. L256–L263.
Demonbreun, A.R., Lapidos, K.A., Heretis, K., et al., Myoferlin regulation by NFAT in muscle injury, regeneration and repair, J. Cell Sci., 2010, vol. 123, pp. 2413–2422. doi 10.1242/jcs.065375
Doherty, K.R., Cave, A., Davis, D.B., et al., Normal myoblast fusion requires myoferlin, Development, 2005, vol. 132, pp. 5565–5575. doi 10.1242/dev.02155
Zhang, Y. and Storey, K.B., Expression of nuclear factor of activated T cells (NFAT) and downstream muscle-specific proteins in ground squirrel skeletal and heart muscle during hibernation, Mol. Cell. Biochem., 2016, vol. 412, no. 27, pp. 27–40. doi 10.1007/s11010-015-2605-x
Venkatesh, S., Lee, J., Singh, K., et al., Multitasking in the mitochondrion by the ATP-dependent Lon protease, Biochim. Biophys. Acta, 2012, vol. 1823, no. 1, pp. 56–66. doi 10.1016/j.bbamcr.2011.11.003
Nordgren, M., Wang, B., Apanasets, O., and Fransen, M., Peroxisome degradation in mammals: mechanisms of action, recent advances, and perspectives, Front. Physiol., 2013, vol. 4, no. 145, pp. 92–103.
Elberg, G., Chen, L., Elberg, D., et al., MKL1 mediates TGF-beta1-induced alpha-smooth muscle actin expression in human renal epithelial cells, Am. J. Physiol. Renal Physiol., 2008, vol. 294, no. 5, pp. F1116–F1128. doi 10.1152/ajprenal.00142.2007
Amengual, J., Petrov, P., Bonet, M.L., et al., Induction of carnitine palmitoyl transferase 1 and fatty acid oxidation by retinoic acid in HepG2 cells, Int. J. Biochem. Cell Biol., 2012, vol. 44, no. 11, pp. 2019–2027. doi 10.1016/j.biocel.2012.07.026
Lemas, D.J., Wiener, H.W., O’Brien, D.M., et al., Genetic polymorphisms in carnitine palmitoyltransferase 1A gene are associated with variation in body composition and fasting lipid traits in Yup’ik Eskimos, J. Lipid Res., 2012, vol. 53, no. 1, pp. 175–184. doi 10.1194/jlr.P018952
Biswas, S. and Akey, J.M., Genomic insights into positive selection, Trends Genet., 2006, vol. 22, no. 8, pp. 437–446. doi 10.1016/j.tig.2006.06.005
Sabeti, P.C., Varilly, P., Fry, B., et al., The International HapMap Consortium: genome-wide detection and characterization of positive selection in human populations, Nature, 2007, vol. 449, pp. 913–918. doi 10.1038/nature06250
Izagirre, N. García, I., Junquera, C., et al., A scan for signatures of positive selection in candidate loci for skin pigmentation in humans, Mol. Biol. Evol., 2006, vol. 23, no. 9, pp. 1697–1706. doi 10.1093/molbev/msl030
Duforet-Frebourg, N., Luu, K., Laval, G., et al., Detecting genomic signatures of natural selection with principal component analysis: application to the 1000 genomes data, Mol. Biol. Evol., 2016, vol. 33, no. 4, pp. 1082–1093. doi 10.1093/molbev/msv33
Prokunina-Olsson, L., Muchmore, B., Tang, W., et al., A variant upstream of IFNL3 (IL28B) creating a new interferon gene IFNL4 is associated with impaired clearance of hepatitis C virus, Nat. Genet., 2013, vol. 45, pp. 164–171. doi 10.1038/ng.2521
Key, F.M., Peter, B., Dennis, M.Y., et al., Selection on a variant associated with improved viral clearance drives local, adaptive pseudogenization of interferon lambda 4 (IFNL4), PLoS Genet., 2014, vol. 10, e1004681. doi 10.1371/journal.pgen.1004681
Prokunina-Olsson, L., Tang, W., Gilyazova, I., et al., Population analysis of a frame-shift genetic variant that creates a novel interferon lambda 4 protein (ifnl4), in Genetics and Genomics of Global Health and Sustainability (Proc. Joint Conf. HGM 2013 and 21st International Congress of Genetics), 2013, p. 309.
Gibson, G., Decanalization and the origin of complex disease, Nat. Rev. Genet., 2009, vol. 10, no. 2, pp. 134–140. doi 10.1038/nrg2502
McGrath, J.J., Hannan, A.J., and Gibson, G., Decanalization, brain development and risk of schizophrenia, Transl. Psychiatry, 2011, vol. 1, e14. doi 10.1038/tp.2011.16
Author information
Authors and Affiliations
Corresponding author
Additional information
Original Russian Text © V.A. Stepanov, V.N. Kharkov, K.V. Vagaitseva, A.V. Bocharova, A.Yu. Kazantsev, A.A. Popovich, I.Yu. Khitrinskaya, 2017, published in Genetika, 2017, Vol. 53, No. 11, pp. 1254–1266.
Rights and permissions
About this article
Cite this article
Stepanov, V.A., Kharkov, V.N., Vagaitseva, K.V. et al. Search for genetic markers of climatic adaptation in populations of North Eurasia. Russ J Genet 53, 1172–1183 (2017). https://doi.org/10.1134/S1022795417110114
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S1022795417110114