Abstract—
A serious problem in the treatment of HIV infection is the emergence of drug-resistant forms of the virus. One promising approach to solving this problem is the development of inhibitors of the interaction between viral proteins with cellular co-factors. However, the development of this approach is hampered due to the lack of knowledge about the involvement of cellular proteins in the pathogenesis of HIV infection. In particular, it is known that the integration of viral DNA into the host genome generates numerous lesions in the cellular DNA, the repair of which is absolutely necessary for successful replication of the virus. However, it is still unknown which cellular proteins are involved in repairing this damage. In this review, we summarize what is known to date about the role of cellular repair systems in the replication of HIV-1 in general, and in the repair of damage that occurs during the integration of viral DNA into a cell’s genome, in particular.

Similar content being viewed by others
REFERENCES
Laskey S.B., Siliciano R.F. 2014. A mechanistic theory to explain the efficacy of antiretroviral therapy. Nat. Rev. Microbiol. 12, 772–780.
Domingo P., Vidal F. 2011. Combination antiretroviral therapy. Expert. Opin. Pharmacother. 12, 995–998.
Cihlar T., Fordyce M. 2016. Current status and prospects of HIV treatment. Curr. Opin. Virol. 18, 50–56.
Solomon D.A., Sax P.E. 2015. Current state and limitations of daily oral therapy for treatment. Curr. Opin. HIV AIDS. 10, 219–225.
Iyidogan P., Anderson K.S. 2014. Current perspectives on HIV-1 antiretroviral drug resistance. Viruses. 6, 4095–4139.
Adamson C.S., Freed E.O. 2010. Novel approaches to inhibiting HIV-1 replication. Antiviral Res. 85, 119–141.
Tintori C., Brai A., Fallacara A.L., Fazi R., Schenone S., Botta M. 2014. Protein–protein interactions and human cellular cofactors as new targets for HIV therapy. Curr. Opin. Pharmacol. 18, 1–8.
Skalka A.M., Katz R.A. 2005. Retroviral DNA integration and the DNA damage response. Cell Death Differ. 12, 971–978.
Sükösd Z., Andersen E.S., Seemann S.E., Jensen M.K., Hansen M., Gorodkin J., Kjems J. 2015. Full-length RNA structure prediction of the HIV-1 genome reveals a conserved core domain. Nucleic Acids Res. 43, 10168–10179.
Muriaux D., Darlix J.L. 2010. Properties and functions of the nucleocapsid protein in virus assembly. RNA Biol. 7, 744–753.
Engelman A. 2009. Isolation and analysis of HIV-1 preintegration complexes. Methods Mol. Biol. 485, 135–149.
Agapkina Yu.Yu., Prikazchikova T.A., Smolov M.A., Gottikh M.B. 2005. Structure and functions of HIV-1 integrase. Usp. Biol. Khim. 45, 87–122.
Knyazhanskaya e.s., Shadrina O.A., Anisenko A.N., Gottikh M.B. 2016. Role of DNA-dependent protein kinase in the HIV-1 replication cycle. Mol. Biol. (Moscow). 50 (4), 567–579.
Jackson S.P., Bartek J. 2009. The DNA-damage response in human biology and disease. Nature. 461, 1071–1078.
Ryan E.L., Hollingworth R., Grand R.J. 2016. Activation of the DNA damage response by RNA viruses. Biomolecules. 6, 2.
Norbury C.J., Zhivotovsky B. 2004. DNA damage-induced apoptosis. Oncogene. 23, 2797–2808.
Yang J., Yu Y., Hamrick H.E., Duerksen-Hughes P.J. 2003. ATM, ATR and DNA-PK: Initiators of the cellular genotoxic stress responses. Carcinogenesis. 24, 1571–1580.
Ciccia A., Elledge S.J. 2010. The DNA damage response: Making it safe to play with knives. Mol. Cell. 40, 179–204.
Gagné J.P., Rouleau M., Poirier G.G. 2012. Structural biology. PARP-1 activation – bringing the pieces together. Science. 336, 678–679.
Khodyreva S.N., Lavrik O.I. 2016. Poly(ADP-ribose) polymerase 1 as a key regulator of DNA repair. Mol. Biol. (Moscow). 50 (4), 580–595.
Ko H.L., Ren E.C. 2012. Functional aspects of PARP1 in DNA repair and transcription. Biomolecules. 2, 524–548
Ceccaldi R., Rondinelli B., D’Andrea A.D. 2016. Repair pathway choices and consequences at the double-strand break. Trends Cell Biol. 26, 52–64.
Chang H.H., Pannunzio N.R., Adachi N., Lieber M.R. 2017. Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat. Rev. Mol. Cell Biol. 18, 495–506.
Cannan W.J., Pederson D.S. 2016. Mechanisms and consequences of double-strand DNA break formation in chromatin. J. Cell Physiol. 231, 3–14.
Shinagawa H., Iwasaki H. 1996. Processing the Holliday junction in homologous recombination. Trends Biochem. Sci. 21, 107–111.
Abbotts R., Wilson D.M. 2017. Coordination of DNA single strand break repair. Free Radic. Biol. Med. 107, 228–244.
Krokan H.E., Bjørås M. 2013. Base excision repair. Cold Spring Harb. Perspect. Biol. 5, a012583
Lans H., Marteijn J.A., Vermeulen W. 2012. ATP-dependent chromatin remodeling in the DNA-damage response. Epigenetics Chromatin. 5, 4.
Kunkel T.A., Erie D.A. 2015. Eukaryotic mismatch repair in relation to DNA replication. Annu. Rev. Genet. 49, 291–313.
Brin E., Yi J., Skalka A.M., Leis J. 2000. Modeling the late steps in HIV-1 retroviral integrase-catalyzed DNA integration. J. Biol. Chem. 275, 39287–39295.
Miller M.D., Wang B., Bushman F.D. 1995. Human immunodeficiency virus type 1 preintegration complexes containing discontinuous plus strands are competent to integrate in vitro. J. Virol. 69, 3938–3944.
Daniel R., Katz R.A., Skalka A.M. 1999. A role for DNA-PK in retroviral DNA integration. Science. 284, 644–647.
Sakurai Y., Komatsu K., Agematsu K., Matsuoka M. 2009. DNA double strand break repair enzymes function at multiple steps in retroviral infection. Retrovirology. 6, 114.
Daniel R., Greger J.G., Katz R.A., Taganov K.D., Wu X., Kappes J.C., Skalka A.M. 2004. Evidence that stable retroviral transduction and cell survival following DNA integration depend on components of the nonhomologous end joining repair pathway. J. Virol. 78, 8573–8581.
Coffin J.M., Rosenberg N. 1999. Retroviruses. Closing the joint. Nature. 399, 413–416.
Baekelandt V., Claeys A., Cherepanov P., De Clercq E., De Strooper B., Nuttin B., Debyser Z. 2000. DNA-dependent protein kinase is not required for efficient lentivirus integration. J. Virol. 74, 11278–11285.
Daniel R., Katz R.A., Merkel G., Hittle J.C., Yen T.J., Skalka A.M. 2001. Wortmannin potentiates integrase-mediated killing of lymphocytes and reduces the efficiency of stable transduction by retroviruses. Mol. Cell Biol. 21, 1164–1172.
Li L., Olvera J.M., Yoder K.E., Mitchell R.S., Butler S.L., Lieber M., Martin S.L., Bushman F.D. 2001. Role of the non-homologous DNA end joining pathway in the early steps of retroviral infection. EMBO J. 20, 3272–3281.
Studamire B., Goff S.P. 2008. Host proteins interacting with the Moloney murine leukemia virus integrase: multiple transcriptional regulators and chromatin binding factors. Retrovirology. 5, 48.
Zheng Y., Ao Z., Wang B., Jayappa K.D., Yao X. 2011. Host protein Ku70 binds and protects HIV-1 integrase from proteasomal degradation and is required for HIV replication. J. Biol. Chem. 286, 17722–17735.
Anisenko A.N., Knyazhanskaya E.S., Zalevsky A.O., Agapkina J.Y., Sizov A.I., Zatsepin T.S., Gottikh M.B. 2017. Characterization of HIV-1 integrase interaction with human Ku70 protein and initial implications for drug targeting. Sci. Rep. 7, 5649.
Jeanson L., Subra F., Vaganay S., Hervy M., Marangoni E., Bourhis J., Mouscadet J.F. 2002. Effect of Ku80 depletion on the preintegrative steps of HIV-1 replication in human cells. Virology. 300, 100–108.
Waninger S., Kuhen K., Hu X., Chatterton J.E., Wong-Staal F., Tang H. 2004. Identification of cellular cofactors for human immunodeficiency virus replication via a ribozyme-based genomics approach. J. Virol. 78, 12829–12837.
Manic G., Maurin-Marlin A., Laurent F., Vitale I., Thierry S., Delelis O., Dessen P., Vincendeau M., Leib-Mösch C., Hazan U., Mouscadet J.F., Bury-Moné S. 2013. Impact of the Ku complex on HIV-1 expression and latency. PLoS One. 8, e69691.
Espeseth A.S., Fishel R., Hazuda D., Huang Q., Xu M., Yoder K., Zhou H. 2011. siRNA screening of a targeted library of DNA repair factors in HIV infection reveals a role for base excision repair in HIV integration. PLoS One. 6, e17612.
Hultquist J.F., Schumann K., Woo J.M., Manganaro L., McGregor M.J., Doudna J., Simon V., Krogan N.J., Marson A. 2016. A Cas9 ribonucleoprotein platform for functional genetic studies of HIV–host interactions in primary human T cells. Cell Rep. 17, 1438–1452.
Appelqvist H., Johansson A.C., Linderoth E., Johansson U., Antonsson B., Steinfeld R., Kågedal K., Ollinger K. 2012. Lysosome-mediated apoptosis is associated with cathepsin D-specific processing of bid at Phe24, Trp48, and Phe183. Ann. Clin. Lab. Sci. 42, 231–242.
Cooper A., García M., Petrovas C., Yamamoto T., Koup R.A., Nabel G.J. 2013. HIV integration and T cell death: Additional commentary. Retrovirology. 10, 150.
Skalka A.M. 2013. HIV: Integration triggers death. Nature. 498, 305–306.
Estaquier J., Zaunders J., Laforge M. 2013. HIV integrase and the swan song of the CD4 T cells? Retrovirology. 10, 149.
Dehart J.L., Andersen J.L., Zimmerman E.S., Ardon O., An D.S., Blackett J., Kim B., Planelles V. 2005. The ataxia telangiectasia-mutated and Rad3-related protein is dispensable for retroviral integration. J. Virol. 79, 1389–1396.
Lau A., Swinbank K.M., Ahmed P.S., Taylor D.L., Jackson S.P., Smith G.C., O’Connor M.J. 2005. Suppression of HIV-1 infection by a small molecule inhibitor of the ATM kinase. Nat. Cell Biol. 7, 493–500.
Smith J.A., Wang F.X., Zhang H., Wu K.J., Williams K.J., Daniel R. 2008. Evidence that the Nijmegen breakage syndrome protein, an early sensor of double-strand DNA breaks (DSB), is involved in HIV-1 post-integration repair by recruiting the ataxia telangiectasia-mutated kinase in a process similar to, but distinct from, cellular DSB repair. Virol. J. 5, 11.
Awasthi P., Foiani M., Kumar A. 2016. ATM and ATR signaling at a glance. J. Cell Sci. 129, 1285.
Yan S., Sorrell M., Berman Z. 2014. Functional interplay between ATM/ATR-mediated DNA damage response and DNA repair pathways in oxidative stress. Cell Mol. Life Sci. 71, 3951–3967.
Durocher D., Jackson S.P. 2001. DNA-PK, ATM and ATR as sensors of DNA damage: Variations on a theme? Curr. Opin. Cell Biol. 13, 225–231.
Daniel R., Kao G., Taganov K., Greger J.G., Favorova O., Merkel G., Yen T.J., Katz R.A., Skalka A.M. 2003. Evidence that the retroviral DNA integration process triggers an ATR-dependent DNA damage response. Proc. Natl. Acad. Sci. U. S. A. 100, 4778–4783.
Ariumi Y., Turelli P., Masutani M., Trono D. 2005. DNA damage sensors ATM, ATR, DNA-PKcs, and PARP-1 are dispensable for human immunodeficiency virus type 1 integration. J. Virol. 79, 2973–2978.
Kameoka M., Nukuzuma S., Itaya A., Tanaka Y., Ota K., Ikuta K., Yoshihara K. 2004. RNA interference directed against poly(ADP-ribose) polymerase 1 efficiently suppresses human immunodeficiency virus type 1 replication in human cells. J. Virol. 78, 8931–8934.
Kameoka M., Nukuzuma S., Itaya A., Tanaka Y., Ota K., Inada Y., Ikuta K., Yoshihara K. 2005. Poly(ADP-ribose) polymerase-1 is required for integration of the human immunodeficiency virus type 1 genome near centromeric alphoid DNA in human and murine cells. Biochem. Biophys. Res. Commun. 334, 412–417.
Gäken J.A., Tavassoli M., Gan S.U., Vallian S., Giddings I., Darling D.C., Galea-Lauri J., Thomas M.G., Abedi H., Schreiber V., Ménissier-de Murcia J., Collins M.K., Shall S., Farzaneh F. 1996. Efficient retroviral infection of mammalian cells is blocked by inhibition of poly(ADP-ribose) polymerase activity. J. Virol. 70, 3992–4000.
Ha H.C., Juluri K., Zhou Y., Leung S., Hermankova M., Snyder S.H. 2001. Poly(ADP-ribose) polymerase-1 is required for efficient HIV-1 integration. Proc. Natl. Acad. Sci. U. S. A. 98, 3364–3368.
Siva A.C., Bushman F. 2002. Poly(ADP-ribose) polymerase 1 is not strictly required for infection of murine cells by retroviruses. J. Virol. 76, 11904–11910.
Rom S., Reichenbach N.L., Dykstra H., Persidsky Y. 2015. The dual action of poly(ADP-ribose) polymerase-1 (PARP-1) inhibition in HIV-1 infection: HIV-1 LTR inhibition and diminution in Rho GTPase activity. Front. Microbiol. 6, 878.
Bueno M.T., Reyes D., Valdes L., Saheba A., Urias E., Mendoza C., Fregoso O.I., Llano M. 2013. Poly(ADP-ribose) polymerase 1 promotes transcriptional repression of integrated retroviruses. J. Virol. 87, 2496–2507.
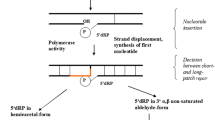
Yoder K.E., Bushman F.D. 2000. Repair of gaps in retroviral DNA integration intermediates. J. Virol. 74, 11191–11200.
Yoder K.E., Espeseth A., Wang X.H., Fang Q., Russo M.T., Lloyd R.S., Hazuda D., Sobol R.W., Fishel R. 2011. The base excision repair pathway is required for efficient lentivirus integration. PLoS One. 6, e17862.
Goetze R.W., Kim D.H., Schinazi R.F., Kim B. 2017. A CRISPR/Cas9 approach reveals that the polymerase activity of DNA polymerase β is dispensable for HIV-1 infection in dividing and nondividing cells. J. Biol. Chem. 292, 14016–14025.
Bennett G.R., Peters R., Wang X.H., Hanne J., Sobol R.W., Bundschuh R., Fishel R., Yoder K.E. 2014. Repair of oxidative DNA base damage in the host genome influences the HIV integration site sequence preference. PLoS One. 9, e103164.
Anisenko A.N., Knyazhanskaya E.S., Isaguliants M.G., Gottikh M.B. 2018. A qPCR assay for measuring the post-integrational DNA repair in HIV-1 replication. J. Virol. Methods. 262, 12–19.
Author information
Authors and Affiliations
Corresponding author
Additional information
Translated by D. Timchenko
Abbreviations: PIC, pre-integration complex; PI3K, phosphoinositide-3-kinase; DNA-PK, DNA-dependent protein kinase; ATM, аtaxia telangiectasia mutated protein; ATR, serine–threonine protein kinase, or ataxia-telangiectasia and Rad3-related protein; DNA-PKcs, catalytic subunit of DNA-PK; PARP, poly(АDP-ribose)-polymerase; NHEJ, non-homologous end joining repair mechanism; HR, homologous recombination; LigIV, ligase IV; RPA, replication protein A; AP site, apurinic/apyrimidinic site; BER, base excision repair; NER, nucleotide excision repair; RT, HIV-1 reverse transcriptase; IN, HIV-1 integrase; VSV-G, surface glycoprotein G of vesicular stomatitis virus; RSV, Rous sarcoma virus; MMLV, Moloney murine leukemia virus; A-MuLV, Abelson murine leukemia virus; MEF, mouse embryonic fibroblasts; MOI, multiplicity of infection.
Rights and permissions
About this article
Cite this article
Anisenko, A.N., Gottikh, M.B. Role of Cellular DNA Repair Systems in HIV-1 Replication. Mol Biol 53, 313–322 (2019). https://doi.org/10.1134/S0026893319030026
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0026893319030026