Abstract
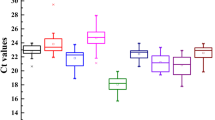
qRT-PCR is becoming a routine tool in molecular biology to study gene expression. It is necessary to find stable reference genes when performing qRT-PCR. The expression of genes cloned in oil-tea camellia currently cannot be accurately analyzed due to a lack of suitable reference genes. We collected different tissues (including roots, stems, leaves, flowers and seeds) from six oil-tea camellia species to determine stable reference genes. Five novel and ten traditional reference gene sequences were selected from the RNA-seq database of Camellia oleifera Abel seeds and specific PCR Primers were designed for each. Cycle threshold (C t) data were obtained from each reaction for all samples. Three different software tools, geNorm, NormFinder and Best-Keeper were applied to calculate the expression stability of the candidate reference genes according to the C t values. The results were similar between the three software packages, and indicated that the traditional genes TUBα-3, ACT7α and the novel gene CESA were relatively stable in all species and tissues. However, no genes were sufficiently stable across all species and tissues, thus the optimal number of reference genes required for accurate normalization varied from 2 to 6. Finally, the relative expression of squalene synthase (SQS) and squalene epoxidase (SQE) genes related to important ingredients squalene and tea saponin in oil-tea camellia seeds were compared by using stable to less stable reference genes. The comparison results validated the selection of reference genes in the current study. In summary, for the different tissues of six oil-tea camellia species different optimal numbers of suitable reference genes were found.
Similar content being viewed by others
References
Chang Z., Ling C., Yamashita M., et al. 2010. Microarray-driven validation of reference genes for quantitative real-time polymerase chain reaction in a rat vocal fold model of mucosal injury. Anal. Biochem. 406(2), 214–221.
Umenishi F., Verkman A.S., Gropper M.A. 1996. Quantitative analysis of aquaporin mRNA expression in rat tissues by RNase protection assay. DNA Cell Biol. 15, 475–480.
Zhang L., Zhou W., Velculescu V.E., et al. 1997. Gene expression profiles in normal and cancer cells. Science. 276(5316), 1268–1272.
Demidenko N.V., Logacheva M.D., Penin A.A. 2011. Selection and validation of reference genes for quantitative real-time PCR in Buckwheat (Fagopyrum esculentum) based on transcriptome sequence data. PloS ONE. 6(5), e19434.
Huggett J., Dheda K., Bustin S., et al. 2005. Real-time RT-PCR normalization: Strategies and considerations. Genes Immun. 6(4), 279–284.
Paolacci A.R., Tanzarella O.A., Porceddu E., Ciaffi M. 2009. Identification and validation of reference genes for quantitative RT-PCR normalization in wheat. BMC Mol. Biol. 10(1), 11.
Maccoux L.J., Clements D.N., Salway F., Day P.J. 2007. Identification of new reference genes for the normalization of canine osteoarthritic joint tissue transcripts from microarray data. BMC Mol. Biol. 8(1), 62.
Migocka M., Papierniak A. 2011. Identification of suitable reference genes for studying gene expression in cucumber plants subjected to abiotic stress and growth regulators. Mol. Breeding. 28(3), 343–357.
Dheda K., Huggett J.F., Chang J.S., et al. 2005. The implications of using an inappropriate reference gene for real-time reverse transcription PCR data normalization. Anal. Biochem. 344(1), 141–143.
Andersen C.L., Jensen J.L., Ørntoft T.F. 2004. Normalization of real-time quantitative reverse transcription-PCR data, a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 64(15), 5245.
Qi J., Yu S., Zhang F., Shen X., Zhao X., Yu Y., Zhang D. 2010. Reference gene selection for real-time quantitative polymerase chain reaction of mRNA transcript levels in Chinese cabbage (Brassica rapa L. ss. pekinensis). Plant Mol. Biol. Rep. 28(4), 597–604.
Hruz T., Wyss M., Docquier M., et al. 2011. RefGenes, identification of reliable and condition specific reference genes for RT-qPCR data normalization. BMC Genomics. 12(1), 156.
Zhuang R.L., Yao X.H. 2008. Oil-Tea Camellia of China, 2nd ed. Beijing: Chinese Forestry Publ., pp. 1–10 (in Chinese).
Vijayan K., Zhang W.J., Tsou C.H. 2009. Molecular taxonomy of Camellia (Theaceae) inferred from nrITS sequences. Am. J. Bot. 96(7), 1348–1360.
Tan X.F., Hu F.M., Xie L.S., et al. 2006. Construction of EST library and analysis of main expressed genes of Camellia oleifera seeds. Sci. Silvae Sinicae (China). 42(1), 43–48.
Lin P., Cao Y.Q., Yao X.H., et al. 2011. Transcriptome analysis of Camellia oleifera Abel seed in four development stages. Mol. Plant Breeding (China). 19(4), 498–505.
Hu X., Tan X., Tian X., et al. 2008. cDNA cloning, sequence analysis and physiological role speculation of a dehydrin-like protein from Camellia oleifera. Acta Bot. Boreali-Occidentalia Sinica (China). 28(8), 1541–1548.
Jiang Y., Tan X.F., Zhang D.Q., et al. 2009. Cloning and sequence analysis of a metallothionein gene from Camellia oleifera. Acta Agric. Univ. Jiangxiensis (China). 31(4), 699–705.
Luo Q., Xie L.S., Tan X.F., et al. 2008. Cloning of full-length cDNA of FAD2 gene from Camellia oleifera. Sci. Silvae Sinicae (China). 44(3), 70–75.
Zhang D., Tan X., Chen H., et al. 2008. Full-length cDNA cloning and bioinformatic analysis of Camellia oleifera SAD. Sci. Silvae Sinicae. 44(2), 155–159.
Wang B., Tan X.F., Chen Y., et al. 2012. Molecular cloning and expression analysis of two calmodulin genes encoding an identical protein from Camellia olerfera. Pak. J. Bot. 44(3), 961–968.
Shao G., Tan X.F., Chen H., et al. 2012. Isolation and characterization of an aldo-keto reductase cDNA from Camellia oleifera seed. Adv. Sci. Lett. (China). 10(1), 153–157.
Han X.J., Lu M., Chen Y., et al. 2012. Selection of reliable reference genes for gene expression studies using real-time PCR in tung tree during seed development. PloS ONE. 7(8), e43084.
Li R., Zhu H., Ruan J., et al. 2010. De novo assembly of human genomes with massively parallel short read sequencing. Genome Res. 20, 265–272.
Mortazavi A., Williams B.A., McCue K., Schaeffer L., Wold B. 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nature Methods. 5(7), 621–628.
Czechowski T., Stitt M., Altmann T., et al. 2005. Genome-wide identification and testing of superior referencegenes for transcript normalization in Arabidopsis. Plant Physiol. 139, 5–17.
Expósito-Rodríguez M., Borges A.A., Borges-Pérez A., et al. 2008. Selection of internal control genes for quantitative real-time RT-PCR studies during tomato development process. BMC Plant Biol. 8, 131.
Chang E., Shi S., Liu J., et al. 2012. Selection of reference genes for quantitative gene expression studies in Platycladus orientalis (Cupressaceae) using real-time PCR. PloS ONE. 7(3), e33278.
Do R., Kiss R.S., Gaudet D., et al. 2009. Squalene synthase, a critical enzyme in the cholesterol biosynthesis pathway. Clin. Genet. 75(1), 19–29.
Beytia E., Qureshi A.A., Porter J.W. 1973. Squalene synthetase III. Mechanism of the reacion. J. Biol. Chem. 248(5), 1856–1867.
M’Baya B., Fegueur M., Servouse M., et al. 1989. Regulation of squalene synthetase and squalene epoxidase activities in Saccharomyces cerevisiae. Lipids. 24(12), 1020–1023.
Vandesompele J., De Preter K., Pattyn F., et al. 2002. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3(7), res. 0034.
Pfaffl M.W., Tichopad A., Prgomet C., et al. 2004. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper-Excel-based tool using pair-wise correlations. Biotechnol. Lett. 26(6), 509–515.
Livak K.J., Schmittgen T.D. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔCt method. Methods. 25(4), 402–408.
Benn C.L., Fox H., Bates G. 2008. Optimisation of region-specific reference gene selection and relative gene expression analysis methods for pre-clinical trials of Huntington’s disease. Mol. Neurodegener. 3(1), 17.
Yan J., Yuan F., Long G., et al. 2011. Selection of reference genes for quantitative real-time RT-PCR analysis in citrus. Mol. Biol. Rep. 39(2), 1831–1838.
Tong Z., Gao Z., Wang F., et al. 2009. Selection of reliable reference genes for gene expression studies in peach using real-time PCR. BMC Mol. Biol. 10(1), 71.
Zhu G.P., Li J.Y., Fan Z.Q., et al. 2011. Isolation of B function CjDEF-1 gene involved in floral development in Camellia japonica Hongshibaxueshi and its expression analysis. J. Agric. Biotechnol. 19(3), 442–448.
Zhou X.W., Li J.Y., Fan Z.Q. 2012. Cloning and expression analysis of chalcone isomerase gene cDNA from Camellia nitidissima. Forest Res. (China). 25(1), 93–99.
Radonić A., Thulke S., Mackay I.M., Landt O., Siegert W., Nitsche A. 2004. Guideline to reference gene selection for quantitative real-time PCR. Biochem. Biophys. Res. Commun. 313(4), 856–862.
Raaijmakers M.H., van Emst L., de Witte T., Mensink E., Raymakers R.A. 2002. Quantitative assessment of gene expression in highly purified hematopoietic cells using real-time reverse transcriptase polymerase chain reaction. Exp. Hematol. 30(5), 481–487.
Author information
Authors and Affiliations
Corresponding author
Additional information
Published in Russian in Molekulyarnaya Biologiya, 2013, Vol. 47, No. 6, pp. 959–975.
The article is published in the original.
Zhou C.F. and Lin P. made an equal contribution to the study, and should be regarded as joint first authors.
Rights and permissions
About this article
Cite this article
Zhou, C.F., Lin, P., Yao, X.H. et al. Selection of reference genes for quantitative real-time PCR in six oil-tea camellia based on RNA-seq. Mol Biol 47, 836–851 (2013). https://doi.org/10.1134/S0026893313060198
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0026893313060198