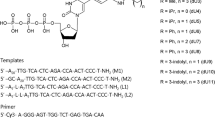
Abstract
(CA/TG)n repeats belong to microsatellite DNA. They are the most abundant among the other dinucleotide repeats in mammals, constituting approximately 0.25% of the entire genome. These repeats are recombination hot spots; however, the corresponding mechanisms are yet vague. We postulated that one of the reasons underlying an increase in the recombination frequency in the repetitive region could be the con-formational characteristics of duplex resulting from a specific geometry of base-stacking contacts, providing for initiation of a single-stranded DNA invasion in th e duplex homologous regions. This work for the first time demonstrates a DNA-DNA interaction of the d(CA)10 and d(TG)10 oligonucleotides with linear and circular duplexes containing (CA/TG)31 repeats during their coincubation in a protein-free water solution at 37°C. Using radioactively labeled oligonucleotides, we demonstrated that the duplex—oligonucleotide interaction intensity depended on the molar ratio of duplex-to-oligonucleotide at a duplex concentration of 30 nM. A decrease in this concentration to 3 nM had no effect on the intensity of oligonucleotide invasion. It was demonstrated that over 1% of the duplexes yet much less than 10% were involved in the interaction with oligonucleotides assuming that one oligonucleotide molecule interacted with one molecule of the duplex. Analysis of the kinetics showed that d(CA)10 invasion commenced from the first minute of incubation with duplexes, while d(TG)10 interacted with the duplex even at a higher rate. The role of conformational plasticity of CA/TG repeats in the discovered interaction is discussed as well as its biological significance, in particular, the role of CA microsatellites in the initiation of homologous recombination.
Similar content being viewed by others
Abbreviations
- PCR:
-
polymerase chain reaction
- NMR:
-
nuclear magnetic resonance
References
Lander E.S., Linton L.M., Birren B., et al. 2001. Initial sequencing and analysis of the human genome. Nature. 409, 860–921.
Treco D., Thomas B., Arnheim N. 1985. Recombination hot spot in the human beta-globin gene cluster: Meiotic recombination of human DNA fragments in Saccharomyces cerevisiae. Mol. Cell Biol. 5, 2029–2038.
Gendrel C-G., Boulet A., Dutreix M. 2000. (CA/GT)n microsatellites affect homologous recombination during yeast meiosis. Genet. Dev. 14, 1261–1268.
Murphy K.E., Stringer R. 1986. RecA independent recombination of poly d(GT)-d(CA) in pBR322. Nucleic Acids Res. 14, 7325–7340.
Dutriex M. 1997. (GT)n repetitive tracts affect several stages of RecA-promoted recombination. J. Mol. Biol. 273, 105–113.
Majewski J., Ott J. 2000. GT repeats are associated with recombination on human chromosome 22. Genet. Res. 10, 1108–1114.
Adachi M., Tsujimoto Y. 1990. Potential Z-DNA elements surround the breakpoints of chromosome trans-location within the 5’ flanking region of bcl-2 gene. Oncogene. 5, 1653–1657.
Ford A.M., Bennet C.A., Price C.M., Bruin M.C.A., van Wering E.R., Greaves M. 1998. Fetal origins of the TEL-AML fusion gene in identical twins with leukemia. Proc. Natl. Acad. Sci. USA. 95, 4584.
Thandla S.P., Ploski J.E., Razaigilmez S.Z., Chhal-liyil P.P., Block A.M.W., de Jong P.J., Aplan P.D. 1999. ETV6-AML1 translocation breakpoints cluster near a purine/pyrimidine repeat region in the ETV6 gene. Blood. 93, 293–299.
Naylor L.H., Clark E.M. 1990. d(TG)n d(CA)n sequences upstream of the rat prolactin gene form Z-DNA and inhibit gene transcription. Nucleic Acids Res. 18, 1595–1601.
Shimajiri S., Arima N., Tanimoto A., Murata Y., Hamada T., Wang K.Y., Sasaguri Y. 1999. Shortened microsatellite d(CA)21 sequence down-regulates promoter activity of matrix metalloproteinase 9 gene. FEBS Lett. 455, 70–74.
Hui J., Hung L.H., Heiner M., Schreiner S., Neumüller N., Reither G., Haas S.A., Bindereif A. 2005. Intronic CA-repeat and CA-rich elements: A new class of regulators of mammalian alternative splicing. EMBOJ. 24, 1988–1998.
Lorenz M., Hewing B., Hui J., Zepp A., Baumann G., Bindereif A., Stangl V., Stangl K. 2007. Alternative splicing in intron 13 of the human eNOS gene: A potential mechanism for regulating eNOS activity. FASEB J. 21, 1556–1564.
Yu J., Hai Y., Liu G., Fang T., Kung S.K., Xie J. 2009. The heterogeneous nuclear ribonucleoprotein L is an essential component in the Ca2+/calmodulin-depen-dent protein kinase IV-regulated alternative splicing through cytidine-adenosine repeats. J. Biol. Chem. 284, 1505–15013.
Rossbach O., Hung L.H., Schreiner S., Grishina I., Heiner M., Hui J., Bindereif A. 2009. Auto-and cross-regulation of the hnRNP L proteins by alternative splicing. Mol. Cell. Biol. 29, 1442–1451.
Benet A., Azorin F. 1999. The formation of triple-stranded DNA prevents spontaneous branch-migra-tion. J. Mol. Biol. 294, 851–857.
Wang G., Vasquez K.M. 2004. Naturally occurring H-DNA-forming sequences are mutagenic in mammalian cells. Proc. Natl. Acad. Sci. USA. 101, 13448–13453.
Wang G., Vasquez K.M. 2006. Non-B DNA structure induced genetic instability. Mutat. Res. 598, 103–119.
Gasanova V.K., Neschastnova A.A., Belitskii G.A., Yakubovskaya M.G. 2006. Specific oligonucleotide invasion into an end of a DNA duplex. Mol. Biol. 40, 132–138.
Saenger W. 1984. Principles of Nucleic Acid Structure. N.Y.: Springer.
Gaillard C., Strauss F. 1994. Association of poly(CA).poly(TG) DNA fragments into four-stranded complexes bound by HMG1 and 2. Science. 264, 433–436.
Sambrook J., Russell D.W. 2001. Molecular Cloning: A Laboratory Manual, 3rd ed., Cold Spring Harbor, NY: Cold Spring Harbor Lab. Press, vol. 1.
Yakubovskaya M. G., Neschastnova A. A., Popenko V.I., Lipatova Zh.V., Belitsky G.A. 1999. Holliday junctions are formed in concentrated solutions of purified products of DNA amplification. Biokhimiya. 64, 1550–1554.
Kaluzhny D., Shchyolkina A., Livshits M., Lysov Y., Borisova O. 2009. A novel intramolecular G-quartet-containing fold of single-stranded d(GT)(8) and d(GT)(16) oligonucleotides. Biophys. Chem. 143, 161–165.
Hite J.M., Eckert K.A., Cheng K.C. 1996. Factors affecting fidelity of DNA synthesis during PCR amplification of d(C-A)n d(G-T) n microsatellite repeats. Nucleic Acids Res. 24, 2429–2434.
Eckert K., Mowery A., Hile S. 2002. Misalignment-mediated DNA polymerase beta mutations: Comparison of microsatellite and frame-shift error rates using a forward mutation assay. Biochemistry. 41, 10490–10498.
Shinde D., Lai Y., Sun F., Arnheim N. 2003. Taq DNA polymerase slippage mutation rates measured by PCR and quasi-likelihood analysis: (CA/GT)n and (A/T)n microsatellites. Nucleic Acids Res. 31, 974–980.
Debrauwere H., Gendrel C.G., Lechat S., Dutreix M. 1997. Differences and similarities between various tandem repeat sequences: Minisatellites and microsatel-lites. Biochimie. 79, 577–586.
Yakubovskaya M.G., Neschastnova A A., Humphrey K.E., et al. 2001. Interaction of linear homologous DNA duplexes via Holliday junction formation. Eur. J. Bio-chem. 268, 7–14.
Markina V.K., Danilova O.A., Neschastnova A.A., Belitskii G.A., Yakubovskaya M.G. 2002. The role of duplex ends in the spontaneous interaction of homologous linear DNA fragments. Mol. Biol. 36, 693–697.
Bichara M., Pinet I., Schumacher S., Fuchs R.P. 2000. Mechanisms of dinucleotide repeat instability in Escherichia coli. Genetics. 154, 533–542.
Gaillard C., Shlyakhtenko L.S., Lyubchenko Y.L., Strauss F. 2002. Structural analysis of hemicatenated DNA loops. BMC Struct. Biol. 2, 7.
Kladde M.R, Kohwi Y, Kohwi-Shigematsu T., Gorski J. 1994. The non-B-DNA structure of d(CA/TG)n differs from that of Z-DNA. Proc. Natl. Acad. Sci. USA. 91, 1898–1902.
Ha S.C., Lowenhaupt K., Rich A., Kim Y.G., Kim K.K. 2005. Crystal structure of a junction between B-DNA and Z-DNA reveals two extruded bases. Nature. 437, 1183–1186.
Singleton C.K., Klysik J., Stirdivant S.M., Wells R.D. 1982. Left-handed Z-DNA is induced by supercoiling in physiological ionic conditions. Nature. 299, 312–316.
Suzuki M., Yagi N. 1995. Stereochemical basis of DNA bending by transcription factors. Nucleic Acids Res. 23, 2083–2091
Lankas F., Sponer J., Langowski J., Cheatham T.E. 3rd. 2003. DNA basepair step deformability inferred from molecular dynamics simulations. Biophys. J. 85, 2872–2883.
Cheung S., Arndt K., Lu P. 1984. Correlation of lac operator DNA imino proton exchange kinetics with its function. Proc. Natl. Acad. Sci. USA. 81, 3665–3669.
Kowalczykowski S.C., Dixon D.A., Eggleston A.K., Lauder S.D., Rehraurer W.M. 1994. Biochemistry of homologous recombination in Escherichia coli. Micro-biol.Rev. 58, 401–465.
Hunter N., Kleckner N. 2001. The single-end invasion: An asymmetric intermediate at the double-strand break to double-Holliday junction transition of meiotic recombination. Cell. 106, 59–70.
Wahls W.P. 1998. Meiotic recombination hotspots: Shaping the genome and insights into hypervariable minisatellite DNA change. Curr. Top. Dev. Biol. 37, 37–75.
Wang G., Carbajal S., Vijg J., DiGiovanni J., Vasquez K.M. 2008. DNA structure4nduced genomic instability itin vivo. J. Natl. Cancer Inst. 100, 1815–1817.
Author information
Authors and Affiliations
Corresponding author
Additional information
Original Russian Text © V.K. Gasanova, N V. Ryadninskaya, C. Gaillard, F. Strauss, G.A. Belitsky, M.G. Yakubovskaya, 2010, published in Molekulyarnaya Biologiya, 2010, Vol. 44, No. 3, pp. 520–528.
Rights and permissions
About this article
Cite this article
Gasanova, V.K., Ryadninskaya, N.V., Gaillard, C. et al. Invasion of complementary oligonucleotides into (CA/TG)31 repetitive region of linear and circular DNA duplexes. Mol Biol 44, 458–465 (2010). https://doi.org/10.1134/S0026893310030155
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0026893310030155