Abstract
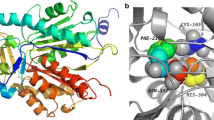
Prediction and analysis of molecular structure and biochemical function are of theoretical guiding significance for gene discovery and application, and considered as one of the central problem of computational biology. Here, some characteristic features of chalcone synthase (CHS) family from Scutellaria baicalensis were described via bioinformatic analysis, and showed as following: the nucleic acid sequences and amino acid sequences of three chs member genes shared high similarity in the molecular structures and physicochemical properties; SbCHS proteins were localized to the cytosol, and possessed a good hydrophobic nature, with lacking any transmembrane topological structure. The phylogram analysis suggested that they were a group genes with significant functional association and genetic conservation. The secondary structures of the SbCHSs were mainly composed of α-helixes and random coils, and the tertiary structures contained malonyl CoA linkers, besides, each of CHS-A and CHS-B with N-glycosylation motif included. Taken together, these results demonstrate that CHS family from S. baicalensis has the typical molecular structure and function of chalcone synthase, compared with the experimental data for Medicago sativa CHS protein.
Similar content being viewed by others
References
Stefan M., Axel M. 2005. Flavones and flavone synthases. Phytochemistry. 66, 2399–2407.
McKhann H.I., Hirsch A.M. 1994. Isolation of chalcone synthase and chalcone isomerase cDNAs from alfalfa (Medicago sativa L.): Highest transcript levels occur in young roots and root tips. Plant Mol. Biol. 5, 767–777.
Junghans H., Dalkin K., Dixon R.A. 1993. Stress responses in alfalfa ( Medicago sativa L.): 15. Characterization and expression patterns of members of a subset of the chalcone synthase multigene family. Plant Mol. Biol. 2, 239–253.
Jez J.M., Noel J.P. 2000. Mechanism of chalcone synthase: PKa of the catalytic cysteine and the role of the conserved histidine in a plant polyketide synthase. Biol. Chem. 275, 39640–39643.
Ferrer J.L., Jez J.M., Bowman M.E., Dixon R.A., Noel J.P. 1999. Structure of chalcone synthase and the molecular basis of plant polyketide biosynthesis. Nature. 8, 775–840.
Zhang D.Q., Tan X.F., Wang X.H. 2007. Gene characteristics and transgenic application of chalcone synthase and chalcone isomerase. J. Central South Univer. Forestry Technol. 2, 87–91.
Yamamoto H. 1991. In: Biotechnology in Agriculture and Forestry: Medicinal and Aromatic Plants III. Ed. Bajaj Y.P.S. Berlin: Springer, vol. 15, pp. 398–418.
Olof E., Henrik N., Soren B. Gunnar H. 2000. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300, 1005–1016.
Jensen J.L., Gupta R., Blom N., Devos D., Tamames J., Kesmir C., Nielsen H., Staerfeldt H., Rapacki K., Workman C. 2002. Ab initio prediction of human orphan protein function from post-translational modifications and localization features. J. Mol. Biol. 319, 1257–1265.
Jensen J.L., Sterfeldt H.H., Brunak S. 2003. Prediction of human protein function according to gene ontology categories. Bioinformatics. 19, 635–642.
Ikeda M., Arai M., Lao D.M. 2002. Transmembrane topology prediction methods: A reassessment and improvement by a consensus method using a dataset of experimentally characterized transmembrance topologies. In Silico Biol. 2, 19–33.
Kyte J., Doolittle R.F. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157, 105–132.
Combet C., Blanchet C., Geourjon C., Deléage G. 2000. NPS: Network protein sequence analysis. Trends Biochem. Sci. 25, 147–150.
Marchler B.A., Bryant S.H. 2004. CD-Search: Protein domain annotations on the fly. Nucleic Acids Res. 32, w327–w331.
Thompson J.D., Gibson T.J., Plewniaki F., Jeanmougin F., Higgins D.G. 1997. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882.
Saito N., Nei M. 1987. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425.
Kumar S.K., Tmamura K., Jakobseni I.B., Nei M. 2001. MEGA2: Molecular evolutionary genetics analysis software. Bioinformatics. 17, 1244–1245.
Felsenstein J. 1989. PHYL IP-Phylogeny Inference Package (Version 3.2). Cladistics. 5, 164–166.
Guex N., Peitsch M.C. 1997. SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modeling. Electrophoresis. 18, 2714–2723.
Schwede T., Kopp J., Guex N., Peitsch M.C. 2003. SWISS-MODEL: An automated protein homologymodeling server. Nucleic Acids Res. 31, 3381–3385.
Arnold K., Bordoli L., Kopp J., Schwede T. 2006. The SWISS-MODEL Workspace: A web-based environment for protein structure homology modelling. Bioinformatics. 22, 195–201.
WebLab ViewerLite, version 4.2. 2000. Molecular Simulations, Inc. San Diego, CA.
Hrazdina G. 1992. Compartmentation in aromatic metabolism, In: Recent Advances in Phytochemistry. Eds Stafford H.A., Ibrahim R.K. N.Y.: Plenum, pp. 1–23.
Saslowsky D., Winkel S.B. 2001. Localization of flavonoid enzymes in Arabidopsis roots. Plant J. 27, 37–48.
Stafford H.A. 1991. Flavonoid evolution: An enzymic approach. Plant Physiol. 96, 680–685.
Jez J.M., Bowman M.E., Noel J.P. 2002. Expanding the biosynthetic repertoire of plant type III polyketide synthases by altering starter molecule specificity. Proc. Natl. Acad. Sci. USA. 99, 5319–5324.
Kim S.H., Mizuno K., Fujimura T. 2002. Regulated expression of ADP-glucose pyrophosphorylase and chalcone synthase during root development in sweet potato. Plant Growth Regulation. 38, 173–179.
Jiang M., Cao J.S. 2007. Chalcone synthase gene. Chinese J. Cell Biol. 29, 525–529.
Author information
Authors and Affiliations
Corresponding author
Additional information
The article is published in the original.
Rights and permissions
About this article
Cite this article
Lei, W., Sun, M., Luo, K.M. et al. Compute simulation to characterize structure and function of chalcone synthase from Scutellaria baicalensis georgi. Mol Biol 43, 1008–1013 (2009). https://doi.org/10.1134/S0026893309060144
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0026893309060144