Abstract
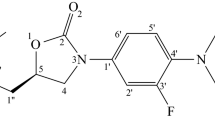
Macrolides are clinically important antibiotics that inhibit protein biosynthesis on ribosomes by binding to ribosomal tunnel. Tylosin belongs to the group of 16-membered macrolides. It is a potent inhibitor of translation whose activity is largely due to reversible covalent binding of its aldehyde group with the base of A2062 in 23S ribosomal RNA. It is known that the conversion of the aldehyde group of tylosin to methyl or carbinol groups dramatically reduces its inhibitory activity. However, earlier we obtained several derivatives of tylosin having comparable activity in spite of the fact that the aldehyde group of tylosin in these compounds was substituted with an amino acid or a peptide residue. Details of the interaction of these compounds with the ribosome that underlies their high inhibitory activity were not known. In the present work, the structure of the complex of tylosin derivative containing in position 20 the residue of ethyl ester of 2-imino(oxy)acetylphenylalanine with the tunnel of the E. coli ribosome was identified by means of molecular dynamics simulations, which could explain high biological activity of this compound.
Similar content being viewed by others
Abbreviations
- MD:
-
molecular dynamics
- NPET:
-
nascent peptide exit tunnel
- OMT:
-
5-O-mycaminosyltylonolide
- PTC:
-
peptidyltransferase center
- TylPhe:
-
phenylalanyl derivative of tylosin
References
Omura, S. (ed.) (2003) Macrolide Antibiotics: Chemistry, Biology and Practice, 2nd Edn., Academic Press, N.Y.
Dunkle, J. A., Xiong, L., Mankin, A. S., and Cate, J. H. D. (2010) Structures of the Escherichia coli ribosome with antibiotics bound near the peptidyl transferase center explain spectra of drug action, Proc. Natl. Acad. Sci. USA, 107, 17152–17157.
Hansen, J., Ippolito, J., Ban, N., Nissen P., Moore, P., and Steitz, T. (2002) The structures of four macrolide antibiotics bound to the large ribosomal subunit, Mol. Cell, 10, 117–128.
Wilson, D. N. (2009) The A-Z of bacterial translation inhibitors, Crit. Rev. Biochem. Mol. Biol., 44, 393–433.
Ruan, Z.-X., Huangfu, D.-S., Xu, X.-J., Sun, P.-H., and Chen, W.-M. (2013) 3D-QSAR and molecular docking for the discovery of ketolide derivatives, Exp. Opin. Drug Discov., 8, 427–444.
Korshunova, G. A., Sumbatyan, N. V., Fedorova, N. V., Kuznetsova, I. V., Shishkina, A. V., and Bogdanov, A. A. (2007) Peptide derivatives of tylosin-related macrolides, Russ. J. Bioorg. Chem., 33, 218–226.
Sumbatyan, N. V., Kuznetsova, I. V., Karpenko, V. V., Fedorova, N. V., Chertkov, V. A., Korshunova, G. A., and Bogdanov, A. A. (2010) Amino acid and peptide derivatives of the tylosin family of antibiotics modified by aldehyde function, Russ. J. Bioorg. Chem., 36, 245–256.
Starosta, A. L., Karpenko, V. V., Shishkina, A. V., Mikolajka, A., Sumbatyan, N. V., Schluenzen, F., Korshunova, G. A., Bogdanov, A. A., and Wilson, D. N. (2010) Interplay between the ribosomal tunnel, nascent chain, and macrolides influences drug inhibition, Chem. Biol., 17, 504–514.
Makarov, G. I., Makarova, T. M., Sumbatyan, N. V., and Bogdanov, A. A. (2016) Investigation of ribosomes using molecular dynamics simulation methods, Biochemistry (Moscow), 81, 1579–1588.
Shishkina, A., Makarov, G., Tereshchenkov, A., Korshunova, G., Sumbatyan, N., Golovin, A., Svetlov, M., and Bogdanov, A. (2013) Conjugates of amino acids and peptides with 5-O-mycaminosyltylonolide and their interaction with the ribosomal exit tunnel, Bioconjug. Chem., 24, 1861–1869.
Cannone, J. J., Subramanian, S., Schnare, M. N., Collett, J. R., D’Souza, L. M., Du, Y., Feng, B., Lin, N., Madabusi, L. V., Muller, K. M., Pande, N., Shang, Z., Yu, N., and Gutell, R. R. (2002) The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs, BMC Bioinformatics, 3, 1–31.
Byrd, R., Lu, P., and Nocedal, J. (1995) A limited memory algorithm for bound constrained optimization, SIAM J. Sci. Statist. Comput., 16, 1190–1208.
Petrone, P., Snow, C., Lucent, D., and Pande, V. (2008) Side-chain recognition and gating in the ribosome exit tunnel, Proc. Natl. Acad. Sci. USA, 105, 16549–16554.
Lucent, D., Snow, C., Aitken, C., and Pande, V. (2010) Non-bulk-like solvent behavior in the ribosome exit tunnel, PLoS Comput. Biol., 6, e1000963.
Ruiz-Carmona, S., Alvarez-Garcia, D., Foloppe, N., Garmendia-Doval, A. B., Juhos, S., Schmidtke, P., Barril, X., Hubbard, R. E., and Morley, S. D. (2014) rDock: a fast, versatile and open source program for docking ligands to proteins and nucleic acids, PLoS Comput. Biol., 10, e1003571.
Van der Spoel, D., Lindahl, E., Hess, B., Groenhof, G., Mark, A., and Berendsen, H. (2005) GROMACS: fast, flexible, free, J. Comput. Chem., 26, 1701–1718.
Van der Spoel, D., Lindahl, E., Hess, B., and Kutzner, C. (2008) GROMACS 4: algorithms for highly efficient, loadbalanced, and scalable molecular simulation, J. Chem. Theory Comp., 4, 435–447.
Hornak, V., Abel, R., Okur, A., Strockbine, B., Roitberg, A., and Simmerling, C. (2006) Comparison of multiple Amber force fields and development of improved protein backbone parameters, Prot. Struct. Funct. Bioinform., 65, 712–725.
Wang, J., Wolf, R. M., Caldwell, J. W., Kollman, P. A., and Case, D. A. (2004) Development and testing of a general Amber force field, J. Comput. Chem., 25, 1157–1174.
Bayly, C. I., Cieplak, P., Cornell, W., and Kollman, P. A. (1993) A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges: the RESP model, J. Phys. Chem., 97, 10269–10280.
Bussi, G., Donadio, D., and Parrinello, M. (2007) Canonical sampling through velocity rescaling, J. Chem. Phys., 126, 014107–014106.
Berendsen, H., Postma, J., van Gunsteren, W., DiNola, A., and Haak, J. (1984) Molecular dynamics with coupling to an external bath, J. Chem. Phys., 81, 3684–3690.
Darden, T., York, D., and Pedersen, L. (1993) Particle mesh Ewald: an Nlog(N) method for Ewald sums in large systems, J. Chem. Phys., 98, 10089–10092.
Horn, H. W., Swope, W. C., Pitera, J. W., Madura, J. D., Dick, T. J., Hura, G. L., and Head-Gordon, T. (2004) Development of an improved four-site water model for biomolecular simulations: TIP4P-EW, J. Chem. Phys., 120, 9665–9678.
Joung, I. S., and Cheatham, T. E. (2008) Determination of alkali and halide monovalent ion parameters for use in explicitly solvated biomolecular simulations, J. Phys. Chem. B, 112, 9020–9041.
Athavale, S., Petrov, A., Hsiao, C., Watkins, D., Prickett, C., Gossett, J., Lie, L., Bowman, J., O’Neill, E., Hud, C. B. N., Wartell, R., Harvey, S., and Williams, L. (2012) RNA folding and catalysis mediated by iron (II), PLoS One, 7, 1–7.
Hess, B., Bekker, H., Berendsen, H. J., and Fraaije, J. G. (1997) LINCS: a linear constraint solver for molecular simulations, J. Comput. Chem., 18, 1463–1472.
Tribello, G. A., Bonomi, M., Branduardi, D., Camilloni, C., and Bussi, G. (2014) PLUMED 2: new feathers for an old bird, Comp. Phys. Commun., 185, 604–613.
Daura, X., Gademann, K., Jaun, B., Seebach, D., Gunsteren, W. F., and Mark, A. E. (1999) Peptide folding: when simulation meets experiment, Ang. Chem. Inter. Ed., 38, 236–240.
Wilson, D., Harms, J., Nierhaus, K., Schlunzen, F., and Fucini, P. (2005) Species-specific antibiotic–ribosome interactions: implications for drug development, Biol. Chem., 386, 1239–1252.
Marks, J., Kannan, K., Roncase, E. J., Klepacki, D., Kefi, A., Orelle, C., Vázquez-Laslop, N., and Mankin, A. S. (2016) Context-specific inhibition of translation by ribosomal antibiotics targeting the peptidyl transferase center, Proc. Natl. Acad. Sci. USA, 113, 12150–12155.
Sothiselvam, S., Neuner, S., Rigger, L., Klepacki, D., Micura, R., Vázquez-Laslop, N., and Mankin, A. S. (2017) Binding of macrolide antibiotics leads to ribosomal selection against specific substrates based on their charge and size, Cell Rep., 16, 1789–1799.
Author information
Authors and Affiliations
Corresponding author
Additional information
Published in Russian in Biokhimiya, 2017, Vol. 82, No. 8, pp. 1199-1208.
Originally published in Biochemistry (Moscow) On-Line Papers in Press, as Manuscript BM17-179, July 3, 2017.
Rights and permissions
About this article
Cite this article
Makarov, G.I., Sumbatyan, N.V. & Bogdanov, A.A. Structural insight into interaction between C20 phenylalanyl derivative of tylosin and ribosomal tunnel. Biochemistry Moscow 82, 925–932 (2017). https://doi.org/10.1134/S0006297917080077
Received:
Revised:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0006297917080077