-
PDF
- Split View
-
Views
-
Cite
Cite
Catherine R. Linnen, Brian D. Farrell, MITONUCLEAR DISCORDANCE IS CAUSED BY RAMPANT MITOCHONDRIAL INTROGRESSION IN NEODIPRION (HYMENOPTERA: DIPRIONIDAE) SAWFLIES, Evolution, Volume 61, Issue 6, 1 June 2007, Pages 1417–1438, https://doi.org/10.1111/j.1558-5646.2007.00114.x
Close - Share Icon Share
Abstract
We investigate the pervasiveness of hybridization and mitochondrial introgression in Neodiprion Rohwer (Hymenoptera; Diprionidae), a Holarctic genus of conifer-feeding sawflies. A phylogenetic analysis of the lecontei species group revealed extensive discordance between a contiguous mitochondrial region spanning three genes (COI, tRNA-leucine, and COII) and three nuclear loci (EF1α, CAD, and an anonymous nuclear locus). Bayesian tests of monophyly and Shimodaira–Hasegawa (SH) tests of topological congruence were consistent with mitochondrial introgression; however, these patterns could also be explained by lineage sorting (i.e., deep coalescence). Therefore, to explicitly test the mitochondrial introgression hypothesis, we used a novel application of coalescent-based isolation with migration (IM) models to measure interspecific gene flow at each locus. In support of our hypothesis, mitochondrial gene flow was consistently higher than nuclear gene flow across 120 pairwise species comparisons (P < 1 × 10−12). We combine phylogenetic and coalescent evidence to identify likely cases of recent and ancient introgression in Neodiprion, and based on these observations, we hypothesize that shared hosts and/or pheromones facilitate hybridization, whereas disparate abundances between hybridizing species promote mitochondrial introgression. Our results carry implications for phylogenetic analysis, and we advocate the separation of high and low gene flow regions to inform analyses of hybridization and speciational history, respectively.
Debate over the importance of hybridization (i.e., interbreeding between genetically divergent forms, Avise 2004) in evolution has largely followed taxonomic lines. Whereas zoologists have traditionally viewed hybridization as rare and evolutionarily unimportant (Arnold 1997, 2006; Dowling and Secor 1997), botanists have long appreciated its role in plant evolution (e.g., Anderson and Stebbins 1954; Grant 1981). This dichotomy has changed in recent years, and hybridization is now regarded as a significant evolutionary force in both plants and animals (Arnold 2004). This shift can be attributed to surveys of the amount of hybridization in various groups (Grant and Grant 1992; Arnold 1997, 2006) and to molecular and analytical advances that have facilitated the detection of cryptic hybridization events (Avise 2004). These studies have revealed variation in both the frequencies and outcomes of hybridization in different groups of organisms (reviewed in Arnold 1997, 2006).
One particularly common outcome of hybridization is introgression of mitochondrial DNA with comparatively low levels of nuclear introgression (reviewed in Avise 2004; Chan and Levin 2005). Several hypotheses have been advanced to explain this pattern (e.g., Turelli et al. 1992; Martinsen et al. 2001; Funk and Omland 2003; Ballard and Whitlock 2004; Hurst and Jiggins 2005). Most recently, Chan and Levin (2005) provide theoretical and empirical support for their hypothesis that frequency-dependent assortative mating leads to biased cytoplasmic (mitochondrial and chloroplast) introgression. Testing this and other hypotheses will require several complementary approaches: additional theoretical work, detailed mitochondrial introgression case studies, and surveys of the prevalence of differential mitochondrial introgression within and between groups of organisms.
If mitochondrial introgression has been prevalent throughout a group's history, two phylogenetic patterns are expected: (1) nonmonophyly of species in the mitochondrial gene tree and (2) discordance between mitochondrial and nuclear topologies. Unfortunately, these two patterns can also arise as ancestral polymorphism sorts randomly into descendent lineages. Therefore, additional (nonphylogenetic) evidence is required to distinguish between introgression and lineage sorting (i.e., deep coalescence) as explanations for these patterns (Maddison 1997; Funk and Omland 2003). There is a growing body of literature documenting nonphylogenetic evidence for recent (i.e., mitochondrial gene trees have not become reciprocally monophyletic) introgression events (e.g., Hey and Nielsen 2004; Buckley et al. 2006). In contrast, few studies have considered ancient (i.e., mitochondrial gene trees have become reciprocally monophyletic) introgression as a potential explanation for mitonuclear discordance (however, see Sota and Vogler 2001; Shaw 2002; Gomez-Zurita and Vogler 2006). One reason for this asymmetry is that introgression events become progressively more difficult to detect as time since hybridization accumulates and geographic signals of introgression (e.g., shared haplotypes in areas of sympatry) are eroded by range changes and mutation (Funk and Omland 2003). Fortunately, ancient introgression is expected to leave its mark in the gene trees themselves—mitochondrial introgression will cause species to share a mitochondrial ancestor (i.e., an interspecific coalescence event) more recently than ancestors at nuclear loci. Also, if the hybridization event was between nonsister taxa, differences in coalescence times will be accompanied by incongruence between mitochondrial and nuclear topologies. Coalescent-based analyses of multilocus datasets should therefore be able to distinguish between introgression and lineage sorting at all levels of divergence.
In this paper, we investigate the pervasiveness of hybridization and mitochondrial introgression in the sawfly genus Neodiprion Rohwer. We document extensive discordance between mitochondrial and nuclear datasets and describe two phylogenetic patterns (nonmonophyly of species and incongruent topologies) that are consistent with a hypothesis of rampant mitochondrial introgression. We distinguish between lineage sorting and mitochondrial introgression as explanations for observed patterns of discordance using a novel application of coalescent-based divergence with gene flow models (Nielsen and Wakeley 2001; Hey and Nielsen 2004). Specifically, we estimate interspecific gene flow at nuclear and mitochondrial loci—the mitochondrial introgression hypothesis predicts that mitochondrial gene flow has been consistently higher than nuclear gene flow throughout Neodiprion's evolutionary history.
Study System: Neodiprion Rohwer
Neodiprion Rohwer (Hymenoptera: Diprionidae) is a Holarctic genus of conifer-feeding sawflies containing approximately 51 described species and subspecies (Smith 1979, 1988; Wallace and Cunningham 1995). Because several members of the genus are economically important pests (Arnett 1993), the life histories of many Neodiprion species have been studied in great detail, yielding much information on host use, behavior, and development (reviewed in Ross 1955; Coppel and Benjamin 1965; Knerer and Atwood 1973; Knerer 1993). Neodiprion species are host specialists and feed exclusively on plants in the family Pinaceae. Most species further restrict their feeding to hosts in the genus Pinus, and many species specialize on one or two Pinus species. Neodiprion females attract males via sex pheromones and mating occurs on the host plant. Using their saw-like ovipositors, females deposit their eggs into the host plant needles. Larvae of some species feed gregariously and have conspicuous defensive displays, whereas larvae of other species are more solitary. Once feeding is completed, mature larvae spin tough, papery cocoons in the soil or on their host plant. Many species pass the winter as mature larvae in cocoons, others overwinter as eggs in the needles of their host plant, and one species (Neodiprion maurus) overwinters as fully formed adults in cocoons (Knerer 1983, 1990, 1991).
In his 1955 revision of the genus, Ross divided Neodiprion into two species groups based on morphology and geography—the lecontei group has large, distinct punctures on the mesoscutellum and are found in eastern North America; the sertifer group has no punctures on the mesoscutellum, and is found throughout the Holarctic region. Ross (1955) and subsequent authors (e.g., Coppel and Benjamin 1965; Knerer and Atwood 1973; Smith 1979) also noted the existence of multiple species complexes and numerous geographic and host plant races within species. A substantial amount of work on the lecontei group has helped untangle some of the more perplexing species complexes (e.g., Ross 1961; Becker et al. 1966; Becker and Benjamin 1967; Knerer 1984; Knerer and Wilkinson 1990), and species in this group can now be identified with some confidence. Progress has also been made in the sertifer group (e.g., Sheehan and Dahlsten 1985; Smith and Wagner 1986), but this group is still poorly known in comparison to the lecontei group. Because our study requires confidence in a priori taxonomic designations, we focus on the much more intensively studied lecontei group. In particular we (1) estimate the phylogeny of the lecontei group using DNA sequence data from nuclear and mitochondrial loci, (2) compare the number of species for which monophyly can be statistically rejected for each locus, (3) examine patterns of topological concordance between different data partitions, and (4) measure interspecific gene flow at each locus for all species pairs.
Methods
SPECIMENS
Except where noted, specimens were collected by C. Linnen as feeding larvae on multiple trips throughout the United States and Canada in 2001–2004 (Appendix). For each collection, a subset of larvae were stored in 100% ethanol for molecular work, and remaining larvae were reared to adults and frozen at –80°C upon emergence. Initial species identifications were based on larval morphology and an extensive literature on larval forms (e.g., Atwood and Peck 1943; Ross 1955; Becker et al. 1966; Becker and Benjamin 1967; Wilson 1977; Knerer 1984; Dixon 2004; an unpublished key to Ontario larvae by Lindquist, Miller, and Nystrom of the Great Lakes Forest Research Center in Sault Ste. Marie, Ontario; and an unpublished key to Florida larvae by H. Greenbaum 1972). When possible, larval identifications were also confirmed with reared females using the key in Ross 1955 and additional species descriptions (Ross 1961; Becker et al. 1966; Smith and Wagner 1986). Specimens included in this study were chosen to maximize the geographical and ecological range sampled for each species. In all analyses where a priori taxonomic designations are required for hypothesis testing, we follow the recommendation of Knerer (1984) that N. pratti subspecies be treated as a single entity. Multiple outgroup taxa belonging to the sertifer group were included to root trees in phylogenetic analyses. Larval and adult voucher specimens are located at the Museum of Comparative Zoology at Harvard University.
DNA SEQUENCE DATA
Genomic DNA was extracted from prolegs of larvae or legs of adults using either the Qiagen Dneasy Tissue kit (Qiagen, Inc., Valencia, CA) or the “salting out” protocol of Sunnucks and Hales (1996). The polymerase chain reaction (PCR) was used to amplify the following regions (primers are available online in Supplementary Materials Table S1): a large region spanning the mitochondrial genes cytochrome c oxidase I, tRNA-leucine, and cytochrome c oxidase II (COI/COII); a region of the F2 copy of elongation factor-1α (EF1α) that spanned portions of two exons and a large intervening intron (Danforth and Ji 1998; Danforth et al. 1999; Nyman et al. 2006); a region of rudimentary (CAD) that spanned portions of two exons and two introns; and an anonymous (i.e., a BLAST search in GenBank failed to return a homologous match) nuclear locus (ANL43) that was developed for this study using a TOPO Shotgun Subcloning Kit (Invitrogen, Carlsbad, CA). PCR reactions (25 or 50 μL) typically consisted of: 0.5–5.0 μL template DNA, 2 μM each primer, 0.15mM each dNTP, 2.5 μM MgCl2, 1× Qiagen reaction buffer, and one unit of Taq DNA polymerase (Qiagen). Typical PCR temperature profiles consisted of 40 cycles of 30 sec at 95°C, 30 sec at 49–56°C, and 1.5–2 min at 72°C, followed by a 5 min extension step at 72°C. Double-stranded PCR products were purified enzymatically using shrimp alkaline phosphatase and exonuclease I (GE Healthcare, Piscataway, NJ) or were purified with or without a gel extraction step using QIAquick PCR purification kits (Qiagen). Purified PCR products were sequenced in both directions with the sequencing primers listed in Supplementary Materials Table S1 (available online), BigDye Terminator version 3.0 or 3.1 Cycle Sequencing Kits (Applied Biosystems, Foster City, CA), and an ABI 3100 automated sequencer (Applied Biosystems).
Contigs for all loci were assembled and edited in Sequencher version 4.1 (GeneCodes, Ann Arbor, MI) and the entire length of each sequence was examined by eye to confirm base calls and to identify heterozygous sites. Sites with two clear, same-sized peaks were considered heterozygous and were coded as Ns for phylogenetic analysis. Some noncoding regions of EF1α, CAD, and ANL43 contained length heterozygosities. Generally, these consisted of small indels and were easily identified. Regions heterozygous for an indel were coded using Ns. This should not adversely affect subsequent analysis because gaps were treated as missing data in all analyses. Finally, protein-coding regions were checked in Sequencher 4.1 for the presence of stop codons—all protein-coding regions were confirmed to have open reading frames.
PHYLOGENETIC ANALYSIS
The mitochondrial gene region (COI/COII) was aligned by eye, and the three nuclear regions were aligned using default settings in Clustal X version 1.83 (Thompson et al. 1997), followed by a manual adjustment. Each partition (COI/COII, EF1α, CAD, and ANL43) was analyzed separately using maximum parsimony, maximum likelihood, and Bayesian approaches. Because each gene is expected to have an independent genealogical history, estimates of the underlying species tree should consider evidence from all genes that share that history (Maddison 1997). Therefore, a combined analysis of the three nuclear genes (NUC) was carried out to obtain a “best estimate” of Neodiprion relationships implied by nuclear genes. Nuclear and mitochondrial datasets were not combined because preliminary analysis suggested that they were recovering very different histories. Finally, for the combined analysis, nuclear genes were concatenated because this method is expected to maximize the signal contained within each dataset compared to consensus approaches (Baker and DeSalle 1997; Wiens 1998).
Maximum parsimony searches were performed in PAUP* 4.0b10 (Swofford 2000) in conjunction with PAUPRat (Sikes and Lewis 2000), which implements the parsimony ratchet method (Nixon 1999). For each of the five data partitions (COI/COII, EF1α, CAD, ANL43, and NUC), 10 consecutive ratchet searches (each with 15% reweighted characters and 200 iterations) were performed and trees found in separate searches were combined and filtered in PAUP* to find the shortest trees. Parsimony bootstrap analyses for each data partition consisted of 1000 replicates, each with 10 random addition sequences, TBR branch swapping, and no more than 10 trees were saved per replicate.
Maximum likelihood analyses were performed in PHYML version 2.4.4 (Guindon and Gascuel 2003). Models for each data partition were chosen according to the Akaike information criterion (AIC) using Modeltest version 3.7 (Posada and Crandall 1998), and model parameters were estimated in PHYML. Maximum likelihood bootstrap analyses were also conducted in PHMYL and consisted of 1000 replicates per partition.
Bayesian analyses of the five data partitions were performed in MrBayes version 3.1 (Ronquist and Huelsenbeck 2003) with models chosen according to the AIC and MrModeltest version 2.2 (Nylander et al. 2004). Model parameters were estimated separately for each locus in the combined nuclear analysis. Analyses for all but the NUC partition consisted of two concurrent runs (each with four Markov chains and a temperature of 0.2), 10 million generations, a sampling frequency of 1000 generations, and a burn-in of 2500 (25%) trees. Analysis of the NUC partition consisted of two runs (each with eight Markov chains and a temperature of 0.4 to improve mixing), approximately 11 million generations, a sampling frequency of 1000 generations, and a burn-in of 25%. Bayesian support in all runs was assessed with posterior probabilities.
BAYESIAN TESTS OF MONOPHYLY
Mitochondrial introgression predicts that mitochondrial gene trees, despite having a smaller effective population size on average than nuclear loci (Palumbi et al. 2001; Ballard and Whitlock 2004), will contain as many or more nonmonophyletic species as do nuclear gene trees. The Bayesian framework provides a straightforward method for evaluating the monophyly of each species because, assuming correct model specification, posterior probabilities of trees can be interpreted as the probability that those trees are correct (Huelsenbeck and Rannala 2004). For each of the five data partitions (COI/COII, EF1α, CAD, ANL43, and NUC), 16 constraint trees were constructed in MacClade version 4.05 (Maddison and Maddison 2000) to correspond to hypotheses of monophyly for each of the 16 species in which multiple populations had been sampled. These monophyly constraints were then imported into PAUP* and used to filter the post-burn-in set of trees obtained for each data partition. If less than 5% (0.31% after Bonferroni correction for multiple comparisons in each data partition) of the trees were retained after filtering with a given constraint tree, the null hypothesis of monophyly for that species and data partition was rejected (Miller et al. 2002; Buschbom and Barker 2006).
SHIMODAIRA–HASEGAWA TESTS OF CONGRUENCE
If mitochondrial introgression between nonsister taxa has been high relative to nuclear introgression, the mitochondrial tree is expected to conflict with nuclear gene trees more than nuclear gene trees conflict with one another—these predictions were tested using multiple Shimodaira and Hasegawa (1999; SH) tests. Because the phylogenetic relationships relevant to this prediction are interspecific rather than intraspecific ones, congruence tests were performed on a subsample of the data that consisted of a single individual per species. For each species, the individual with the lowest collection ID number that was homozygous across all three nuclear loci was chosen to include in the analysis (if no individuals were homozygous across all three genes for a particular species, we chose the individual with the fewest heterozygosities). This approach was taken because it allowed individuals to be chosen without respect to the phylogenetic relationships recovered in analyses of the full dataset and maximized the information content of each gene, as homozygous individuals did not contain ambiguities that were treated as missing data. Although taxon selection was nonrandom with respect to zygosity, this should not systematically bias our results with respect to congruence between partitions. The resulting subsample contained 18 ingroup and three outgroup species.
Six data partitions (COI/COII, EF1α, CAD, ANL43, NUC, and all genes combined) were analyzed using maximum likelihood in PAUP*4.0b10 with models chosen according to the AIC in Modeltest version 3.7 (Posada and Crandall 1998). Maximum likelihood analyses consisted of 1000 random addition sequences, TBR branch swapping, and the “MulTrees” option. ML bootstrap analyses were also performed for each partition, with 1000 replicates each consisting of 10 random addition sequences, TBR branch swapping, and the “MulTrees” option. ML trees obtained in these analyses were then used as constraints in an additional set of analyses. Five constrained ML searches were performed for each data partition, one search corresponding to each of five ML topologies obtained for the other data partitions. A total of 30 constrained searches (representing all possible partition/topological constraint combinations) were performed in PAUP*, each with the same settings as in the unconstrained searches (1000 RAS, TBR, MulTrees). For each partition, concordance with each of the other data partitions was assessed using SH tests to compare the likelihood scores of unconstrained and constrained searches. SH tests were performed in PAUP* using the RELL approximation with 10,000 bootstrap replicates.
HYPOTHESIS TESTING IN IM
If mitochondrial introgression has exceeded nuclear introgression in Neodiprion, interspecific mitochondrial gene flow should be consistently higher than interspecific nuclear gene flow. Because this prediction deals with species that have exchanged genes following divergence, an appropriate framework for measuring gene flow is provided by the isolation with migration (IM) model (Nielsen and Wakeley 2001; Hey and Nielsen 2004). In this model, an ancestral taxon with an effective population size NA splits into two descendant taxa (with effective sizes N1 and N2) at time t, after which populations 1 and 2 exchange genes at rates m1 and m2 (rates are per gene copy per generation). Nielsen and Wakeley (2001) developed a likelihood/Bayesian framework that uses a Markov chain Monte Carlo (MCMC) approach to fit single locus datasets to the IM model, and Hey and Nielsen (2004) extended this method to multiple loci in their program IM. The program IM outputs the following parameter estimates scaled by the neutral mutation rate u (Hey and Nielsen 2004): θ1 (= 4N1u), θ2 (= 4N2u),θA (= 4NAu), m1/u, m2/u, and tu. Under uniform parameter priors and after a sufficient “burn-in” to ensure the Markov chain is sampling from the posterior distribution, the mode of the posterior distribution for each parameter provides a maximum likelihood estimate of that parameter (Nielsen and Wakeley 2001; Hey and Nielsen 2004). In our analysis, we allowed separate migration rates for each of the four loci (COI/COII, EF1α, CAD, and ANL43). We assumed that migration rate was the same in both directions for each locus to reduce the number of parameters in the model. As the program IM can only accommodate pairs of taxa, estimates for the parameters in the four-locus model were obtained for each possible pairwise species comparison (120 total).
DATA FILE PREPARATION
Several steps were taken to prepare the sequence data for analysis in IM. First, a modification of Clark's (1990) method was used to reconstruct haplotypes from heterozygous sequence data. The majority of haplotype assignments were straightforward and previous work has indicated that IM analyses are not highly sensitive to the method of haplotype inference when the number of polymorphic sites is small (Won and Hey 2005), as was the case for this analysis (for each gene, 70–80% of the individuals sampled harbored unambiguous haplotypes and 86–93% had three or fewer heterozygous sites). Nevertheless, reconstructed haplotypes that resulted in the inference of additional recombination events (as inferred by the four-gamete test of Hudson and Kaplan 1985 implemented in DnaSP ver. 4.10, Rozas et al. 2003) were considered spurious and removed from analysis. The resulting dataset (all unambiguous haplotypes and reconstructed haplotypes that did not increase the number of inferred recombination events) is referred to as “IMset” and was used in all subsequent analyses. Second, because IM assumes no recombination within loci, the largest nonrecombined block was identified for each nuclear locus for each pairwise species comparison using the four-gamete test and DnaSP 4.10. This is a conservative approach because the four-gamete method assumes an infinite sites model and is therefore likely to overestimate the number of recombination events (Bull et al. 2006). Third, once haplotypes were reconstructed, spurious haplotypes discarded, and largest nonrecombined blocks identified, 120 IMset input files (one file for each pairwise species comparison) were constructed using PAUP* version 4.0b10 (Swofford 2000) as a data editor. And finally, to check for potential bias introduced by haplotype reconstruction method and/or exclusion of spurious haplotypes, two additional sets of 120 input files were prepared. “ALLset” contained all haplotypes for all species and “UNAMBset” contained only unambiguous haplotypes (no more than one heterozygous site). To check for potential bias introduced by choosing the largest nonrecombined block for inclusion in analyses (Won and Hey 2005), these two additional sets of data files contained the entire sequence for each locus. Numbers of haplotypes included for each species and data file are given in Table 1. Additional details regarding data file preparation and all three sets of input files are available on request from the authors.
Number of individuals and haplotypes for each species included in IM analysis. The three numbers given for each gene region correspond to the number of haplotypes included for that species in each of the three different types of IM datasets: ALLset/IMset/UNAMBset (see text for explanation of each IM dataset).
| Species | Individuals | COI/COII | EF1α | CAD | ANL43 |
| N. abbotii | 9 | 9/9/9 | 14/8/8 | 14/12/6 | 12/6/6 |
| N. compar | 12 | 12/12/12 | 16/14/8 | 15/13/11 | 15/11/11 |
| N. dubiosus | 6 | 6/6/6 | 7/7/7 | 7/7/7 | 8/6/6 |
| N. excitans | 11 | 11/11/11 | 14/14/12 | 18/10/6 | 19/11/5 |
| N. hetricki | 4 | 4/4/4 | 4/4/4 | 4/4/4 | 5/5/5 |
| N. lecontei | 14 | 14/14/14 | 17/17/15 | 21/17/13 | 23/17/13 |
| N. nigroscutum | 3 | 3/3/3 | 4/4/2 | 4/2/2 | 4/2/2 |
| N. pinetum | 5 | 5/5/5 | 10/10/2 | 6/6/4 | 6/6/6 |
| N. pinusrigidae | 5 | 5/5/5 | 5/5/5 | 9/7/5 | 6/4/4 |
| N. pratti | 14 | 14/14/14 | 22/22/12 | 19/19/15 | 23/17/7 |
| N. rugifrons | 10 | 10/10/10 | 11/11/11 | 16/14/4 | 14/14/14 |
| N. species 1 | 11 | 11/11/11 | 17/15/7 | 18/18/6 | 15/9/9 |
| N. taedae linearis | 3 | 3/3/3 | 6/6/6 | 6/6/6 | 4/4/4 |
| N. virginiana | 5 | 5/5/5 | 6/6/6 | 6/6/4 | 8/4/4 |
| N. warreni | 4 | 4/4/4 | 6/4/4 | 6/4/2 | 6/2/2 |
| N. swainei | 7 | 7/7/7 | 7/7/7 | 8/8/6 | 7/7/7 |
| Species | Individuals | COI/COII | EF1α | CAD | ANL43 |
| N. abbotii | 9 | 9/9/9 | 14/8/8 | 14/12/6 | 12/6/6 |
| N. compar | 12 | 12/12/12 | 16/14/8 | 15/13/11 | 15/11/11 |
| N. dubiosus | 6 | 6/6/6 | 7/7/7 | 7/7/7 | 8/6/6 |
| N. excitans | 11 | 11/11/11 | 14/14/12 | 18/10/6 | 19/11/5 |
| N. hetricki | 4 | 4/4/4 | 4/4/4 | 4/4/4 | 5/5/5 |
| N. lecontei | 14 | 14/14/14 | 17/17/15 | 21/17/13 | 23/17/13 |
| N. nigroscutum | 3 | 3/3/3 | 4/4/2 | 4/2/2 | 4/2/2 |
| N. pinetum | 5 | 5/5/5 | 10/10/2 | 6/6/4 | 6/6/6 |
| N. pinusrigidae | 5 | 5/5/5 | 5/5/5 | 9/7/5 | 6/4/4 |
| N. pratti | 14 | 14/14/14 | 22/22/12 | 19/19/15 | 23/17/7 |
| N. rugifrons | 10 | 10/10/10 | 11/11/11 | 16/14/4 | 14/14/14 |
| N. species 1 | 11 | 11/11/11 | 17/15/7 | 18/18/6 | 15/9/9 |
| N. taedae linearis | 3 | 3/3/3 | 6/6/6 | 6/6/6 | 4/4/4 |
| N. virginiana | 5 | 5/5/5 | 6/6/6 | 6/6/4 | 8/4/4 |
| N. warreni | 4 | 4/4/4 | 6/4/4 | 6/4/2 | 6/2/2 |
| N. swainei | 7 | 7/7/7 | 7/7/7 | 8/8/6 | 7/7/7 |
Number of individuals and haplotypes for each species included in IM analysis. The three numbers given for each gene region correspond to the number of haplotypes included for that species in each of the three different types of IM datasets: ALLset/IMset/UNAMBset (see text for explanation of each IM dataset).
| Species | Individuals | COI/COII | EF1α | CAD | ANL43 |
| N. abbotii | 9 | 9/9/9 | 14/8/8 | 14/12/6 | 12/6/6 |
| N. compar | 12 | 12/12/12 | 16/14/8 | 15/13/11 | 15/11/11 |
| N. dubiosus | 6 | 6/6/6 | 7/7/7 | 7/7/7 | 8/6/6 |
| N. excitans | 11 | 11/11/11 | 14/14/12 | 18/10/6 | 19/11/5 |
| N. hetricki | 4 | 4/4/4 | 4/4/4 | 4/4/4 | 5/5/5 |
| N. lecontei | 14 | 14/14/14 | 17/17/15 | 21/17/13 | 23/17/13 |
| N. nigroscutum | 3 | 3/3/3 | 4/4/2 | 4/2/2 | 4/2/2 |
| N. pinetum | 5 | 5/5/5 | 10/10/2 | 6/6/4 | 6/6/6 |
| N. pinusrigidae | 5 | 5/5/5 | 5/5/5 | 9/7/5 | 6/4/4 |
| N. pratti | 14 | 14/14/14 | 22/22/12 | 19/19/15 | 23/17/7 |
| N. rugifrons | 10 | 10/10/10 | 11/11/11 | 16/14/4 | 14/14/14 |
| N. species 1 | 11 | 11/11/11 | 17/15/7 | 18/18/6 | 15/9/9 |
| N. taedae linearis | 3 | 3/3/3 | 6/6/6 | 6/6/6 | 4/4/4 |
| N. virginiana | 5 | 5/5/5 | 6/6/6 | 6/6/4 | 8/4/4 |
| N. warreni | 4 | 4/4/4 | 6/4/4 | 6/4/2 | 6/2/2 |
| N. swainei | 7 | 7/7/7 | 7/7/7 | 8/8/6 | 7/7/7 |
| Species | Individuals | COI/COII | EF1α | CAD | ANL43 |
| N. abbotii | 9 | 9/9/9 | 14/8/8 | 14/12/6 | 12/6/6 |
| N. compar | 12 | 12/12/12 | 16/14/8 | 15/13/11 | 15/11/11 |
| N. dubiosus | 6 | 6/6/6 | 7/7/7 | 7/7/7 | 8/6/6 |
| N. excitans | 11 | 11/11/11 | 14/14/12 | 18/10/6 | 19/11/5 |
| N. hetricki | 4 | 4/4/4 | 4/4/4 | 4/4/4 | 5/5/5 |
| N. lecontei | 14 | 14/14/14 | 17/17/15 | 21/17/13 | 23/17/13 |
| N. nigroscutum | 3 | 3/3/3 | 4/4/2 | 4/2/2 | 4/2/2 |
| N. pinetum | 5 | 5/5/5 | 10/10/2 | 6/6/4 | 6/6/6 |
| N. pinusrigidae | 5 | 5/5/5 | 5/5/5 | 9/7/5 | 6/4/4 |
| N. pratti | 14 | 14/14/14 | 22/22/12 | 19/19/15 | 23/17/7 |
| N. rugifrons | 10 | 10/10/10 | 11/11/11 | 16/14/4 | 14/14/14 |
| N. species 1 | 11 | 11/11/11 | 17/15/7 | 18/18/6 | 15/9/9 |
| N. taedae linearis | 3 | 3/3/3 | 6/6/6 | 6/6/6 | 4/4/4 |
| N. virginiana | 5 | 5/5/5 | 6/6/6 | 6/6/4 | 8/4/4 |
| N. warreni | 4 | 4/4/4 | 6/4/4 | 6/4/2 | 6/2/2 |
| N. swainei | 7 | 7/7/7 | 7/7/7 | 8/8/6 | 7/7/7 |
IM ANALYSES
For each of 120 IMset input files, an initial run of one million or more steps (following a burn-in of 100,000 steps) was performed in the program IM (Hey and Nielsen 2004, 2006). Each run implemented Metropolis-coupling with heated chains to improve mixing (Geyer 1991; IM options: –f1 –n 6 –g1 0.05). Each locus had its own pair of migration rates, which were set to be equal in both directions (options: –j 5 and –j 6). An HKY substitution model was chosen for each locus because it is the most complex model currently available in IM that is applicable to our data. Inheritance scalars were included in the input files to account for expected differences in effective population size due to inheritance mode (because sawflies are haplodiploid, these values were 0.75 for each nuclear locus and 0.25 for the mitochondrial locus). Wide, noninformative priors for thetas and migration rates (options: –q1 10 –m1 10 –m2 10) were used in initial analyses because no prior information was available for these parameters. Divergence time priors were initially chosen to correspond to a maximum of roughly 20 million years (tu= 30; option –t 30) based on biogeographic information (Graham 1999; Sanmartin et al. 2001) and genetic divergence estimates for the lecontei group (C. Linnen, unpubl. data).
The results of the first set of runs (“A” runs) were used to individually adjust the run conditions for a second set of runs (“B” runs). The posterior distributions for the parameters for each comparison were examined and new parameter priors were chosen. In particular, because wide upper limits were initially chosen for each parameter, most distributions were fully contained within the prior bounds and had peaks that were well to the left of the upper bound. In such cases, maximum values were reduced in subsequent runs. When parameter distributions were not contained fully within the original upper bounds (i.e., flat or rising distributions), maxima were increased accordingly. In “B” and subsequent IMset runs, the option “–qu 1” was used to allow separate upper limits for each theta. Also, the number of chains and heating increments were adjusted for each run to achieve better mixing. B runs each consisted of two million steps following a burn-in of 100,000 steps. Convergence on the stationary distribution was assessed using parameter effective sample sizes (ESS), which estimate the extent to which model parameters are autocorrelated over the course of the run (Hey and Nielsen 2006). Following Hey and Nielsen (2006), runs were considered to have converged if the lowest ESS value among the parameters was at least 50. Comparisons that failed to meet this criterion and were rerun with further adjustments (more steps, more chains, different heating schemes). Run conditions for all IMset runs are available online (Supplementary Materials, Table S2).
To ensure that our results were not influenced by the methods used to construct the IMset data files, two additional sets of runs were performed. A total of 120 ALLset files (all haplotypes and the full sequence length included for each gene for all comparisons) were run with the following options: −l 2000000 −b 100000 −fl −n8 −g1 0.025 −q1 10 −m1 10 −m2 10 −t 40 −j 5 −j 6. Similarly, 120 UNAMBset files (unambiguous haplotypes only and full sequence lengths for each gene for all comparisons) were run with all of the same options as ALLset, except for the length (which was 2.5 million steps). All IM analyses were run on a Linux cluster housed at the Bauer Center for Genomics Research at Harvard University.
The program IM gives posterior probability distributions for each parameter in the IM model, and the peaks of these distributions provide maximum likelihood estimates for these parameters (Hey and Nielsen 2004, 2006). For each comparison, the parameters θ1, θ2, and migration rates for each of the four loci were used to calculate estimates of 2Nm, the population migration rate, for each locus. Briefly, for each population (or species) “i,” 2Nimi was obtained for each locus by multiplying θi (= 4Niu) by the locus-specific mi (=mi/u) and dividing the product by two (Hey and Nielsen 2006). Because migration rates were constrained to be equal in both directions for each locus, an average of the two population migration rates (2Nm) was calculated for each locus. For significance testing with the IMset results, the longest run for a given comparison that met our criterion for convergence (all ESS values > 50) and returned complete parameter distributions was used.
The significance of the difference between mitochondrial and nuclear gene flow was tested using nonparametric Wilcoxon matched-pairs signed-ranks tests. The null hypothesis in this case is that the median difference in gene flow between a pair of loci (e.g., COI/COII vs. EF1α) is zero, and matched pairs consist of two gene flow observations taken from a single-species comparison.
Results
SPECIMEN COLLECTION
In total, 18 lecontei group species were collected, and multiple populations were obtained for 16 of these species (Appendix). Absent are two species that are known only from Cuba: N. cubensis Hochmut and N. insularis (Cresson). Also missing are two subspecies: N. merkeli maestrensis Hochmut, which is known only from Cuba and N. taedae taedae Ross, from the southeastern United States. In addition, multiple populations of an undescribed Neodiprion species belonging to the lecontei group were collected throughout the southeastern United States. Larval and adult morphologies of this species were uniform across geographically separated populations and distinct from all other known species, even in tight sympatry (i.e., on the same tree). Thus, we are confident that this entity represents a new Neodiprion species, which we will refer to as N. species 1 throughout the remainder of the paper. This species will be formally described elsewhere.
PHYLOGENETIC ANALYSIS
Only a single base pair deletion was present in the tRNA-leucine gene of some taxa, therefore, the mitochondrial gene region (COI/COII) was easily and unambiguously aligned. EF1α and CAD exons did not contain any insertions or deletions, and the introns of these genes contained few gaps, all of which were easily aligned. In contrast, ANL43 had a large repetitive region that was highly polymorphic in length and could not be unambiguously aligned—this region was discarded before further analysis. The trimmed lengths and percentages of variable sites are given for each of the four loci in Table 2—COI/COII contained the most variation and parsimony-informative (PI) sites, whereas CAD had the least.
Final trimmed length, percentage of variable sites, and percentage of parsimony-informative (PI) sites for loci included in phylogenetic analysis.
| Locus | Length (bp) | Variable sites (%) | PI Sites (%) |
| COI/COII | 1752 | 23.3 | 19.5 |
| EF1α | 1094 | 14.9 | 11.1 |
| CAD | 916 | 8.8 | 5.2 |
| ANL43 | 776 | 18.4 | 12.1 |
| Locus | Length (bp) | Variable sites (%) | PI Sites (%) |
| COI/COII | 1752 | 23.3 | 19.5 |
| EF1α | 1094 | 14.9 | 11.1 |
| CAD | 916 | 8.8 | 5.2 |
| ANL43 | 776 | 18.4 | 12.1 |
Final trimmed length, percentage of variable sites, and percentage of parsimony-informative (PI) sites for loci included in phylogenetic analysis.
| Locus | Length (bp) | Variable sites (%) | PI Sites (%) |
| COI/COII | 1752 | 23.3 | 19.5 |
| EF1α | 1094 | 14.9 | 11.1 |
| CAD | 916 | 8.8 | 5.2 |
| ANL43 | 776 | 18.4 | 12.1 |
| Locus | Length (bp) | Variable sites (%) | PI Sites (%) |
| COI/COII | 1752 | 23.3 | 19.5 |
| EF1α | 1094 | 14.9 | 11.1 |
| CAD | 916 | 8.8 | 5.2 |
| ANL43 | 776 | 18.4 | 12.1 |
Models chosen by Modeltest 3.7 (Posada and Crandall 1998) and the AIC for each partition were: GTR + I +Γ (COI/COII), HKY +Γ (EF1α), TrN +Γ (CAD), TVM + I +Γ (ANL43), and TVM + I +Γ (NUC). Models chosen by MrModeltest version 2.2 (Nylander et al. 2004) and the AIC were: GTR + I +Γ (COI/COII), HKY +Γ (EF1α), GTR +Γ (CAD), and GTR + I +Γ (ANL43). All Bayesian searches showed evidence of sufficiently long burn-ins and convergence on the stationary distribution, as there were no obvious trends in the generation versus log-likelihood plots and the potential scale reduction factor (PSRF) values for all parameters were near 1.0 (Ronquist et al. 2005).
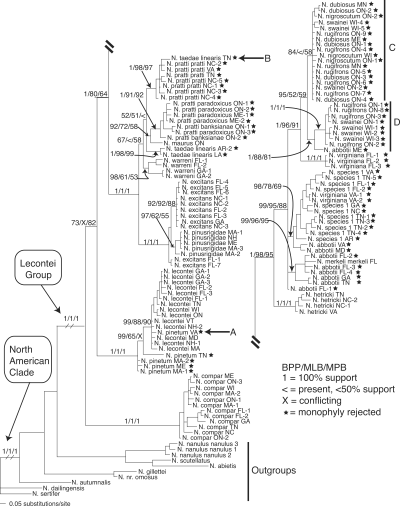
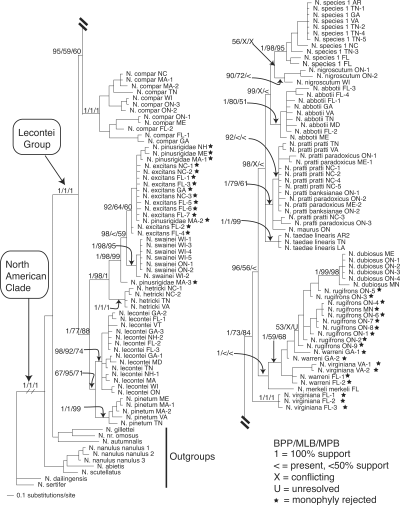
Figure 1 summarizes the results obtained from parsimony, Bayesian, and likelihood analyses of the mitochondrial (COI/COII) partition of the complete taxon set. Figure 2 summarizes the results obtained from analyses of the combined nuclear (EF1α, CAD, and ANL43) partition. Because they represent the best estimates currently available for Neodiprion relationships implied by nuclear genes and because individual nuclear genes were concordant with the combined nuclear partition (see the “SH Tests of Congruence” section below), only the results of the combined nuclear analyses are shown. Branch lengths obtained in Bayesian analyses are included in both figures and are intended only as rough guides to the amount of evolutionary change (in expected number of substitutions per site) that has occurred along each branch.
Bayesian phylogram with Bayesian, likelihood, and parsimony support values for the COI/COII data partition. Support is given for selected nodes in the following order: Bayesian posterior probabilities (BPP)/maximum likelihood bootstrap (MLB)/maximum parsimony bootstrap (MPB). Stars indicate species for which monophyly was rejected (see Bayesian Tests of Monophyly sections). The letters A, B, C, and D refer to species or clades that are discussed further in the text.
Bayesian phylogram with Bayesian, likelihood, and parsimony support values for the combined nuclear (NUC) data partition. Support is given for selected nodes in the following order: Bayesian posterior probabilities (BPP)/maximum likelihood bootstrap (MLB)/maximum parsimony bootstrap (MPB). Stars indicate species for which monophyly was rejected (see Bayesian Tests of Monophyly sections).
Both mitochondrial and nuclear genes strongly supported the monophyly of the North American Neodiprion and the monophyly of the lecontei group—these clades received 100% Bayesian, parsimony, and likelihood support in both data partitions. However, relationships at all levels of divergence within the lecontei group are strikingly different in the two topologies. For example, both data partitions recover several nonmonophyletic species, but the identities of these species differ in the two trees (see starred taxa in Figs. 1 and 2 and more detailed explanation in the “Bayesian Tests of Monophyly” section below). Also, the relationships recovered between species are dramatically different in the two gene trees. For example, the nuclear phylogeny strongly supports (98–100% support under all optimality criteria) the monophyly of the N. pinusrigidae species complex (N. hetricki, N. pinusrigidae, N. swainei, and N. excitans; Ross 1955); in contrast, these species are distributed in two divergent, well-supported clades in the mitochondrial phylogeny.
DNA sequences have been deposited in GenBank (accession nos. EF361837–EF362376), and DNA sequence alignments and trees have been submitted to TreeBASE (accession nos. M3107, M3108, and S1716).
BAYESIAN TESTS OF MONOPHYLY
Null hypotheses of monophyly were rejected in each of the five data partitions (Table 3). With the exception of CAD, each nuclear dataset rejected species monophyly in fewer instances than did the mitochondrial dataset (monophyly was rejected for three species for EF1α, eight species for ANL43, and five species for the combined nuclear dataset). For both CAD and COI/COII, monophyly was rejected for a majority of the species (11 of 16 and 10 of 16, respectively). Finally, there were several species for which monophyly was rejected by the COI/COII partition, but strongly supported by most or all of the nuclear genes (e.g., N. dubiosus, N. pinetum, and N. taedae linearis—see Table 3 and Fig. 2).
Bayesian tests of monophyly. A total of 16 constraint trees were created to correspond to the monophyly of each species. For each dataset, the number of trees in the posterior probability distribution that were consistent with each monophyly constraint, as well as the total number of post-burn-in trees sampled, is given. Asterisks indicate constraint/dataset combinations for which the null hypothesis of monophyly was rejected at a significance level of α=0.05 (after Bonferroni correction for multiple comparisons). The total number of species for which monophyly was rejected is also given for each dataset.
| Monophyly constraint | COI/COII | EF1α | CAD | ANL43 | Nuclear |
| N. abbotii | 0* | 12,073 | 0* | 0* | 16,600 |
| N. compar | 15,002 | 15,002 | 569 | 14,978 | 16,610 |
| N. dubiosus | 2* | 15,002 | 12,158 | 0* | 16,610 |
| N. excitans | 1051 | 0* | 0* | 0* | 0* |
| N. hetricki | 15,002 | 14,988 | 585 | 15,002 | 16,610 |
| N. lecontei | 1226 | 14,655 | 0* | 14,936 | 16,347 |
| N. nigroscutum | 8* | 1876 | 295 | 0* | 14,887 |
| N. pinetum | 0* | 14,629 | 2733 | 15,002 | 16,610 |
| N. pinusrigidae | 14,522 | 4* | 0* | 0* | 0* |
| N. pratti | 0* | 549 | 0* | 0* | 5378 |
| N. rugifrons | 0* | 88 | 0* | 0* | 0* |
| N. species 1 | 13* | 12,778 | 37* | 99 | 16,610 |
| N. swainei | 0* | 1120 | 0* | 15,002 | 16,610 |
| N. taedae linearis | 0* | 15,002 | 18* | 15,001 | 16,610 |
| N. virginiana | 0* | 0* | 0* | 4774 | 0* |
| N. warreni | 14,686 | 2079 | 0* | 0* | 0* |
| Total number of trees | 15,002 | 15,002 | 15,002 | 15,002 | 16,610 |
| Nonmonophyletic | 10 | 3 | 11 | 8 | 5 |
| Monophyly constraint | COI/COII | EF1α | CAD | ANL43 | Nuclear |
| N. abbotii | 0* | 12,073 | 0* | 0* | 16,600 |
| N. compar | 15,002 | 15,002 | 569 | 14,978 | 16,610 |
| N. dubiosus | 2* | 15,002 | 12,158 | 0* | 16,610 |
| N. excitans | 1051 | 0* | 0* | 0* | 0* |
| N. hetricki | 15,002 | 14,988 | 585 | 15,002 | 16,610 |
| N. lecontei | 1226 | 14,655 | 0* | 14,936 | 16,347 |
| N. nigroscutum | 8* | 1876 | 295 | 0* | 14,887 |
| N. pinetum | 0* | 14,629 | 2733 | 15,002 | 16,610 |
| N. pinusrigidae | 14,522 | 4* | 0* | 0* | 0* |
| N. pratti | 0* | 549 | 0* | 0* | 5378 |
| N. rugifrons | 0* | 88 | 0* | 0* | 0* |
| N. species 1 | 13* | 12,778 | 37* | 99 | 16,610 |
| N. swainei | 0* | 1120 | 0* | 15,002 | 16,610 |
| N. taedae linearis | 0* | 15,002 | 18* | 15,001 | 16,610 |
| N. virginiana | 0* | 0* | 0* | 4774 | 0* |
| N. warreni | 14,686 | 2079 | 0* | 0* | 0* |
| Total number of trees | 15,002 | 15,002 | 15,002 | 15,002 | 16,610 |
| Nonmonophyletic | 10 | 3 | 11 | 8 | 5 |
Bayesian tests of monophyly. A total of 16 constraint trees were created to correspond to the monophyly of each species. For each dataset, the number of trees in the posterior probability distribution that were consistent with each monophyly constraint, as well as the total number of post-burn-in trees sampled, is given. Asterisks indicate constraint/dataset combinations for which the null hypothesis of monophyly was rejected at a significance level of α=0.05 (after Bonferroni correction for multiple comparisons). The total number of species for which monophyly was rejected is also given for each dataset.
| Monophyly constraint | COI/COII | EF1α | CAD | ANL43 | Nuclear |
| N. abbotii | 0* | 12,073 | 0* | 0* | 16,600 |
| N. compar | 15,002 | 15,002 | 569 | 14,978 | 16,610 |
| N. dubiosus | 2* | 15,002 | 12,158 | 0* | 16,610 |
| N. excitans | 1051 | 0* | 0* | 0* | 0* |
| N. hetricki | 15,002 | 14,988 | 585 | 15,002 | 16,610 |
| N. lecontei | 1226 | 14,655 | 0* | 14,936 | 16,347 |
| N. nigroscutum | 8* | 1876 | 295 | 0* | 14,887 |
| N. pinetum | 0* | 14,629 | 2733 | 15,002 | 16,610 |
| N. pinusrigidae | 14,522 | 4* | 0* | 0* | 0* |
| N. pratti | 0* | 549 | 0* | 0* | 5378 |
| N. rugifrons | 0* | 88 | 0* | 0* | 0* |
| N. species 1 | 13* | 12,778 | 37* | 99 | 16,610 |
| N. swainei | 0* | 1120 | 0* | 15,002 | 16,610 |
| N. taedae linearis | 0* | 15,002 | 18* | 15,001 | 16,610 |
| N. virginiana | 0* | 0* | 0* | 4774 | 0* |
| N. warreni | 14,686 | 2079 | 0* | 0* | 0* |
| Total number of trees | 15,002 | 15,002 | 15,002 | 15,002 | 16,610 |
| Nonmonophyletic | 10 | 3 | 11 | 8 | 5 |
| Monophyly constraint | COI/COII | EF1α | CAD | ANL43 | Nuclear |
| N. abbotii | 0* | 12,073 | 0* | 0* | 16,600 |
| N. compar | 15,002 | 15,002 | 569 | 14,978 | 16,610 |
| N. dubiosus | 2* | 15,002 | 12,158 | 0* | 16,610 |
| N. excitans | 1051 | 0* | 0* | 0* | 0* |
| N. hetricki | 15,002 | 14,988 | 585 | 15,002 | 16,610 |
| N. lecontei | 1226 | 14,655 | 0* | 14,936 | 16,347 |
| N. nigroscutum | 8* | 1876 | 295 | 0* | 14,887 |
| N. pinetum | 0* | 14,629 | 2733 | 15,002 | 16,610 |
| N. pinusrigidae | 14,522 | 4* | 0* | 0* | 0* |
| N. pratti | 0* | 549 | 0* | 0* | 5378 |
| N. rugifrons | 0* | 88 | 0* | 0* | 0* |
| N. species 1 | 13* | 12,778 | 37* | 99 | 16,610 |
| N. swainei | 0* | 1120 | 0* | 15,002 | 16,610 |
| N. taedae linearis | 0* | 15,002 | 18* | 15,001 | 16,610 |
| N. virginiana | 0* | 0* | 0* | 4774 | 0* |
| N. warreni | 14,686 | 2079 | 0* | 0* | 0* |
| Total number of trees | 15,002 | 15,002 | 15,002 | 15,002 | 16,610 |
| Nonmonophyletic | 10 | 3 | 11 | 8 | 5 |
SH TESTS OF CONGRUENCE
For each of six data partitions (COI/COII, EF1α, CAD, ANL43, NUC, and ALL), Table 4 lists the differences in log-likelihood scores between the unconstrained ML topology and topologies constrained by each of the five ML trees for the remaining partitions. Differences that were found to be significant according to SH tests are also indicated. Several patterns are evident from these analyses. First, datasets from individual loci (COI/COII, EF1α, CAD, ANL43) each significantly reject all other single-locus topologies (e.g., the EF1α dataset significantly rejects topologies for COI/COII, CAD, and ANL43). Second, each nuclear locus dataset fails to reject the combined nuclear topology, yet strongly rejects the ALL topology; likewise the NUC dataset rejects ALL and COI/COII topologies, but not the topologies for EF1α, CAD, and ANL43 topologies. Third, COI/COII and ALL do not reject one another's topologies, but are incongruent with all other partitions. Finally, the largest differences in likelihood scores are observed in comparisons between mitochondrial and nuclear datasets. The ML phylogenies used in these analyses are available in Supplementary Materials Figure S1 (available online).
Shimodaira–Hasegawa tests of topological congruence. For each dataset, the difference in –ln L between the ML topology for that partition and the ML topology constrained by each of the trees listed in the top row is given. This difference corresponds to the test statistic used in the SH test, and its significance was assessed with 10,000 RELL bootstrap replicates. Significant P values are indicated as follows: *P<0.05 **P<0.001. Note that the only nonsignificant –ln L differences are observed between COI/COII versus ALL comparisons and NUC versus individual nuclear gene (EF1α, CAD, and ANL43) comparisons.
| COI/COII tree | EF1α tree | CAD tree | ANL43 tree | NUC tree | ALL tree | |
| COI/COII data | – | 399.08** | 455.04** | 455.56** | 418.05** | 38.28 |
| EF1α data | 199.58** | 60.25* | 73.94* | 23.62 | 141.45** | |
| CAD data | 108.46* | 39.11* | – | 54.06* | 37.27 | 83.46* |
| ANL43 data | 74.12* | 68.73* | 79.86** | 26.43 | 53.79* | |
| NUC data | 238.90** | 22.95 | 50.14 | 29.07 | – | 161.28** |
| ALL data | 29.98 | 328.20** | 430.58** | 406.30** | 312.50** |
| COI/COII tree | EF1α tree | CAD tree | ANL43 tree | NUC tree | ALL tree | |
| COI/COII data | – | 399.08** | 455.04** | 455.56** | 418.05** | 38.28 |
| EF1α data | 199.58** | 60.25* | 73.94* | 23.62 | 141.45** | |
| CAD data | 108.46* | 39.11* | – | 54.06* | 37.27 | 83.46* |
| ANL43 data | 74.12* | 68.73* | 79.86** | 26.43 | 53.79* | |
| NUC data | 238.90** | 22.95 | 50.14 | 29.07 | – | 161.28** |
| ALL data | 29.98 | 328.20** | 430.58** | 406.30** | 312.50** |
Shimodaira–Hasegawa tests of topological congruence. For each dataset, the difference in –ln L between the ML topology for that partition and the ML topology constrained by each of the trees listed in the top row is given. This difference corresponds to the test statistic used in the SH test, and its significance was assessed with 10,000 RELL bootstrap replicates. Significant P values are indicated as follows: *P<0.05 **P<0.001. Note that the only nonsignificant –ln L differences are observed between COI/COII versus ALL comparisons and NUC versus individual nuclear gene (EF1α, CAD, and ANL43) comparisons.
| COI/COII tree | EF1α tree | CAD tree | ANL43 tree | NUC tree | ALL tree | |
| COI/COII data | – | 399.08** | 455.04** | 455.56** | 418.05** | 38.28 |
| EF1α data | 199.58** | 60.25* | 73.94* | 23.62 | 141.45** | |
| CAD data | 108.46* | 39.11* | – | 54.06* | 37.27 | 83.46* |
| ANL43 data | 74.12* | 68.73* | 79.86** | 26.43 | 53.79* | |
| NUC data | 238.90** | 22.95 | 50.14 | 29.07 | – | 161.28** |
| ALL data | 29.98 | 328.20** | 430.58** | 406.30** | 312.50** |
| COI/COII tree | EF1α tree | CAD tree | ANL43 tree | NUC tree | ALL tree | |
| COI/COII data | – | 399.08** | 455.04** | 455.56** | 418.05** | 38.28 |
| EF1α data | 199.58** | 60.25* | 73.94* | 23.62 | 141.45** | |
| CAD data | 108.46* | 39.11* | – | 54.06* | 37.27 | 83.46* |
| ANL43 data | 74.12* | 68.73* | 79.86** | 26.43 | 53.79* | |
| NUC data | 238.90** | 22.95 | 50.14 | 29.07 | – | 161.28** |
| ALL data | 29.98 | 328.20** | 430.58** | 406.30** | 312.50** |
IM ANALYSES
Two to five IM runs (A–E, see Supplementary Materials Tables S2 and 3 available online) with varying conditions were performed for each of the 120 IMset species comparisons, and at least one run per comparison returned ESS values above 50 for all parameters. Several comparisons returned parameter distributions that were flat, rising, or otherwise incomplete for θA and/or t. These patterns likely result from insufficient data to infer these distributions and should not affect estimates of other parameters (Hey 2005); as expected, varying priors on these parameters had little impact on other parameter estimates (see Supplementary Materials Tables S2 and 3 available online). For eight comparisons, we were unable to obtain complete distributions for θ1, θ2, or locus-specific migration rates (m/u) after multiple runs with different parameter priors. Because these parameters are required to estimate 2Nm, these comparisons were excluded from further analysis.
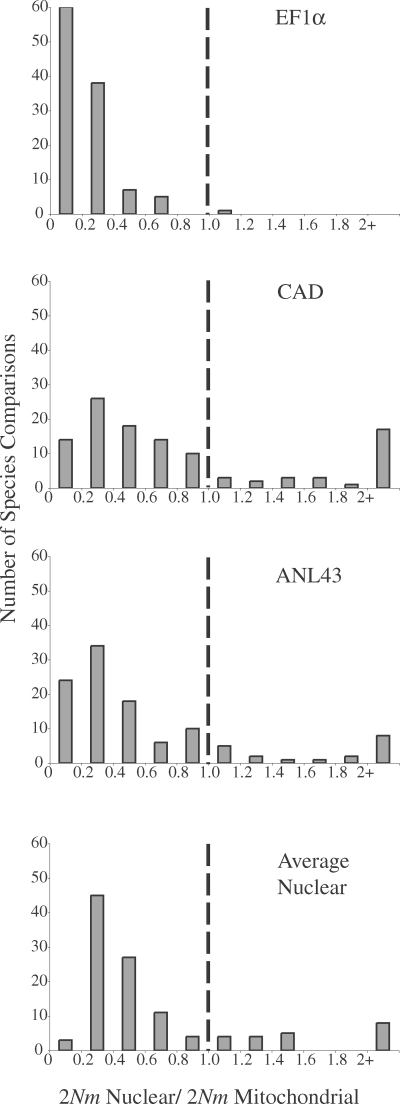
Tables 5 and 6 give the average population migration rate (2Nm) for every possible pairwise species comparison for each of the four loci examined (COI/COII, EF1α, CAD, and ANL43), and the locus with the highest migration rate is underlined for each comparison. Although mitochondrial gene flow generally exceeded gene flow at other loci, the three nuclear loci also appeared to differ in their gene flow rates (Fig. 3). CAD seems to have experienced the most gene flow and EF1α the least (Tables 5 and 6, Fig. 3).
Average population gene flow estimates for COI/COII (above diagonal) and EF1α (below diagonal). Each of 16 species is listed along the top row and first column, and gene flow estimates (2Nm) for each pair are given in the cells where they intersect. Underlined values indicated maximum 2Nm values observed across all loci for a given comparison. Asterisks indicate values where there was a tie for the highest 2Nm. Dashes indicate species comparisons for which one or more migration rate parameters could not be estimated after multiple independent analyses. Species names are abbreviated as follows: N. species 1 (Nsp1); N. abbotii (Nabb); N. nigroscutum(Nnig);N. rugifrons(Nrug);N. virginiana(Nvir);N. warreni(Nwar);N. dubiosus(Ndub);N. pratti(Npra);N. taedae linearis(Ntdl);N. excitans(Nexc);N. pinusrigidae(Npri);N. swainei(Nswa);N. hetricki(Nhet);N. lecontei(Nlec);N. pinetum(Npin); andN. compar (Ncom).
| Nsp1 | Nabb | Nnig | Nrug | Nvir | Nwar | Ndub | Npra | Ntdl | Nexc | Npri | Nswa | Nhet | Nlec | Npin | Ncom | |
| Nsp1 | 0.50 | 0.04 | 0.28 | 1.04 | 0.33 | 0.35 | 0.29 | 0.27 | 0.17 | 0.23 | 0.48 | 0.38 | 0.24 | 0.39 | 0.22 | |
| Nabb | 0.00 | 0.77 | 41.60 | 0.29 | 0.58 | 0.04 | 0.70 | 0.19 | 0.21 | 1.34 | 2.39 | 0.27 | 0.22 | 0.05 | ||
| Nnig | 0.04 | – | 12.55 | 0.53 | – | 7.69 | 0.25 | 0.29 | 0.03 | – | – | 0.18 | 0.26 | 0.33 | 0.17 | |
| Nrug | 0.00 | 0.00 | 0.14 | 0.46 | 0.13 | 2.34 | 0.23 | 0.15 | 0.09 | 0.08 | 6.58 | 0.15 | 0.22 | 0.19 | 0.19 | |
| Nvir | 0.00 | 9.30 | 0.21 | 0.09 | – | 0.19 | 0.22 | 0.22 | 0.06 | 0.09 | 0.55 | 0.43 | 0.25 | 0.28 | 0.21 | |
| Nwar | 0.04 | 0.00 | 0.02 | 0.00 | 0.54 | 0.71 | 0.16 | 0.27 | 0.18 | 0.28 | 0.25* | 0.13 | ||||
| Ndub | 0.02 | 0.00 | 0.00 | 0.00 | 0.03 | 0.00 | 0.16 | 0.09* | 0.02 | 0.02 | 1.08 | 0.09 | 0.16 | 0.16 | 0.12 | |
| Npra | 0.00 | 0.00 | 0.14 | 0.00 | 0.05 | 0.05 | 0.01 | 39.53 | 0.28 | 0.29 | 0.23 | 0.27 | 0.22 | 0.29 | 0.02 | |
| Ntdl | 0.01 | 0.00 | 0.05 | 0.00 | 0.10 | 0.00 | 0.00 | 0.00 | 0.18 | 0.27 | 0.21 | 0.21 | 0.22 | 0.33 | 0.22 | |
| Nexc | 0.00 | 0.00 | 0.02 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.04 | 0.13 | 0.10 | 0.08 | |||
| Npri | 0.02 | 0.00 | – | 0.00 | 0.00 | – | 0.00 | 0.01 | 0.00 | – | 0.07 | 0.05 | 0.12 | 0.11 | 0.07 | |
| Nswa | 0.03 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.01 | 0.00 | 0.00 | 0.11 | 0.21 | 0.33 | 0.22 | |||
| Nhet | 0.01 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.03 | 0.00 | 0.01 | 0.00 | 0.00 | 0.16 | 0.23 | 0.17 | |
| Nlec | 0.00 | 0.00 | 0.05 | 0.01 | 0.06 | 0.03 | 0.01 | 0.01 | 0.03 | 0.02 | 0.00 | 0.04 | 0.03 | 1.28 | 0.03 | |
| Npin | 0.00 | 0.01 | 0.03 | 0.01 | 0.05 | 0.00 | 0.00 | 0.02 | 0.01 | 0.01 | 0.00 | 0.00 | 0.00 | 0.00 | 0.15 | |
| Ncom | 0.02 | 0.00 | 0.02 | 0.03 | 0.09 | 0.07 | 0.02 | 0.00 | 0.08 | 0.00 | 0.00 | 0.00 | 0.02 | 0.00 | 0.04 |
| Nsp1 | Nabb | Nnig | Nrug | Nvir | Nwar | Ndub | Npra | Ntdl | Nexc | Npri | Nswa | Nhet | Nlec | Npin | Ncom | |
| Nsp1 | 0.50 | 0.04 | 0.28 | 1.04 | 0.33 | 0.35 | 0.29 | 0.27 | 0.17 | 0.23 | 0.48 | 0.38 | 0.24 | 0.39 | 0.22 | |
| Nabb | 0.00 | 0.77 | 41.60 | 0.29 | 0.58 | 0.04 | 0.70 | 0.19 | 0.21 | 1.34 | 2.39 | 0.27 | 0.22 | 0.05 | ||
| Nnig | 0.04 | – | 12.55 | 0.53 | – | 7.69 | 0.25 | 0.29 | 0.03 | – | – | 0.18 | 0.26 | 0.33 | 0.17 | |
| Nrug | 0.00 | 0.00 | 0.14 | 0.46 | 0.13 | 2.34 | 0.23 | 0.15 | 0.09 | 0.08 | 6.58 | 0.15 | 0.22 | 0.19 | 0.19 | |
| Nvir | 0.00 | 9.30 | 0.21 | 0.09 | – | 0.19 | 0.22 | 0.22 | 0.06 | 0.09 | 0.55 | 0.43 | 0.25 | 0.28 | 0.21 | |
| Nwar | 0.04 | 0.00 | 0.02 | 0.00 | 0.54 | 0.71 | 0.16 | 0.27 | 0.18 | 0.28 | 0.25* | 0.13 | ||||
| Ndub | 0.02 | 0.00 | 0.00 | 0.00 | 0.03 | 0.00 | 0.16 | 0.09* | 0.02 | 0.02 | 1.08 | 0.09 | 0.16 | 0.16 | 0.12 | |
| Npra | 0.00 | 0.00 | 0.14 | 0.00 | 0.05 | 0.05 | 0.01 | 39.53 | 0.28 | 0.29 | 0.23 | 0.27 | 0.22 | 0.29 | 0.02 | |
| Ntdl | 0.01 | 0.00 | 0.05 | 0.00 | 0.10 | 0.00 | 0.00 | 0.00 | 0.18 | 0.27 | 0.21 | 0.21 | 0.22 | 0.33 | 0.22 | |
| Nexc | 0.00 | 0.00 | 0.02 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.04 | 0.13 | 0.10 | 0.08 | |||
| Npri | 0.02 | 0.00 | – | 0.00 | 0.00 | – | 0.00 | 0.01 | 0.00 | – | 0.07 | 0.05 | 0.12 | 0.11 | 0.07 | |
| Nswa | 0.03 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.01 | 0.00 | 0.00 | 0.11 | 0.21 | 0.33 | 0.22 | |||
| Nhet | 0.01 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.03 | 0.00 | 0.01 | 0.00 | 0.00 | 0.16 | 0.23 | 0.17 | |
| Nlec | 0.00 | 0.00 | 0.05 | 0.01 | 0.06 | 0.03 | 0.01 | 0.01 | 0.03 | 0.02 | 0.00 | 0.04 | 0.03 | 1.28 | 0.03 | |
| Npin | 0.00 | 0.01 | 0.03 | 0.01 | 0.05 | 0.00 | 0.00 | 0.02 | 0.01 | 0.01 | 0.00 | 0.00 | 0.00 | 0.00 | 0.15 | |
| Ncom | 0.02 | 0.00 | 0.02 | 0.03 | 0.09 | 0.07 | 0.02 | 0.00 | 0.08 | 0.00 | 0.00 | 0.00 | 0.02 | 0.00 | 0.04 |
Average population gene flow estimates for COI/COII (above diagonal) and EF1α (below diagonal). Each of 16 species is listed along the top row and first column, and gene flow estimates (2Nm) for each pair are given in the cells where they intersect. Underlined values indicated maximum 2Nm values observed across all loci for a given comparison. Asterisks indicate values where there was a tie for the highest 2Nm. Dashes indicate species comparisons for which one or more migration rate parameters could not be estimated after multiple independent analyses. Species names are abbreviated as follows: N. species 1 (Nsp1); N. abbotii (Nabb); N. nigroscutum(Nnig);N. rugifrons(Nrug);N. virginiana(Nvir);N. warreni(Nwar);N. dubiosus(Ndub);N. pratti(Npra);N. taedae linearis(Ntdl);N. excitans(Nexc);N. pinusrigidae(Npri);N. swainei(Nswa);N. hetricki(Nhet);N. lecontei(Nlec);N. pinetum(Npin); andN. compar (Ncom).
| Nsp1 | Nabb | Nnig | Nrug | Nvir | Nwar | Ndub | Npra | Ntdl | Nexc | Npri | Nswa | Nhet | Nlec | Npin | Ncom | |
| Nsp1 | 0.50 | 0.04 | 0.28 | 1.04 | 0.33 | 0.35 | 0.29 | 0.27 | 0.17 | 0.23 | 0.48 | 0.38 | 0.24 | 0.39 | 0.22 | |
| Nabb | 0.00 | 0.77 | 41.60 | 0.29 | 0.58 | 0.04 | 0.70 | 0.19 | 0.21 | 1.34 | 2.39 | 0.27 | 0.22 | 0.05 | ||
| Nnig | 0.04 | – | 12.55 | 0.53 | – | 7.69 | 0.25 | 0.29 | 0.03 | – | – | 0.18 | 0.26 | 0.33 | 0.17 | |
| Nrug | 0.00 | 0.00 | 0.14 | 0.46 | 0.13 | 2.34 | 0.23 | 0.15 | 0.09 | 0.08 | 6.58 | 0.15 | 0.22 | 0.19 | 0.19 | |
| Nvir | 0.00 | 9.30 | 0.21 | 0.09 | – | 0.19 | 0.22 | 0.22 | 0.06 | 0.09 | 0.55 | 0.43 | 0.25 | 0.28 | 0.21 | |
| Nwar | 0.04 | 0.00 | 0.02 | 0.00 | 0.54 | 0.71 | 0.16 | 0.27 | 0.18 | 0.28 | 0.25* | 0.13 | ||||
| Ndub | 0.02 | 0.00 | 0.00 | 0.00 | 0.03 | 0.00 | 0.16 | 0.09* | 0.02 | 0.02 | 1.08 | 0.09 | 0.16 | 0.16 | 0.12 | |
| Npra | 0.00 | 0.00 | 0.14 | 0.00 | 0.05 | 0.05 | 0.01 | 39.53 | 0.28 | 0.29 | 0.23 | 0.27 | 0.22 | 0.29 | 0.02 | |
| Ntdl | 0.01 | 0.00 | 0.05 | 0.00 | 0.10 | 0.00 | 0.00 | 0.00 | 0.18 | 0.27 | 0.21 | 0.21 | 0.22 | 0.33 | 0.22 | |
| Nexc | 0.00 | 0.00 | 0.02 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.04 | 0.13 | 0.10 | 0.08 | |||
| Npri | 0.02 | 0.00 | – | 0.00 | 0.00 | – | 0.00 | 0.01 | 0.00 | – | 0.07 | 0.05 | 0.12 | 0.11 | 0.07 | |
| Nswa | 0.03 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.01 | 0.00 | 0.00 | 0.11 | 0.21 | 0.33 | 0.22 | |||
| Nhet | 0.01 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.03 | 0.00 | 0.01 | 0.00 | 0.00 | 0.16 | 0.23 | 0.17 | |
| Nlec | 0.00 | 0.00 | 0.05 | 0.01 | 0.06 | 0.03 | 0.01 | 0.01 | 0.03 | 0.02 | 0.00 | 0.04 | 0.03 | 1.28 | 0.03 | |
| Npin | 0.00 | 0.01 | 0.03 | 0.01 | 0.05 | 0.00 | 0.00 | 0.02 | 0.01 | 0.01 | 0.00 | 0.00 | 0.00 | 0.00 | 0.15 | |
| Ncom | 0.02 | 0.00 | 0.02 | 0.03 | 0.09 | 0.07 | 0.02 | 0.00 | 0.08 | 0.00 | 0.00 | 0.00 | 0.02 | 0.00 | 0.04 |
| Nsp1 | Nabb | Nnig | Nrug | Nvir | Nwar | Ndub | Npra | Ntdl | Nexc | Npri | Nswa | Nhet | Nlec | Npin | Ncom | |
| Nsp1 | 0.50 | 0.04 | 0.28 | 1.04 | 0.33 | 0.35 | 0.29 | 0.27 | 0.17 | 0.23 | 0.48 | 0.38 | 0.24 | 0.39 | 0.22 | |
| Nabb | 0.00 | 0.77 | 41.60 | 0.29 | 0.58 | 0.04 | 0.70 | 0.19 | 0.21 | 1.34 | 2.39 | 0.27 | 0.22 | 0.05 | ||
| Nnig | 0.04 | – | 12.55 | 0.53 | – | 7.69 | 0.25 | 0.29 | 0.03 | – | – | 0.18 | 0.26 | 0.33 | 0.17 | |
| Nrug | 0.00 | 0.00 | 0.14 | 0.46 | 0.13 | 2.34 | 0.23 | 0.15 | 0.09 | 0.08 | 6.58 | 0.15 | 0.22 | 0.19 | 0.19 | |
| Nvir | 0.00 | 9.30 | 0.21 | 0.09 | – | 0.19 | 0.22 | 0.22 | 0.06 | 0.09 | 0.55 | 0.43 | 0.25 | 0.28 | 0.21 | |
| Nwar | 0.04 | 0.00 | 0.02 | 0.00 | 0.54 | 0.71 | 0.16 | 0.27 | 0.18 | 0.28 | 0.25* | 0.13 | ||||
| Ndub | 0.02 | 0.00 | 0.00 | 0.00 | 0.03 | 0.00 | 0.16 | 0.09* | 0.02 | 0.02 | 1.08 | 0.09 | 0.16 | 0.16 | 0.12 | |
| Npra | 0.00 | 0.00 | 0.14 | 0.00 | 0.05 | 0.05 | 0.01 | 39.53 | 0.28 | 0.29 | 0.23 | 0.27 | 0.22 | 0.29 | 0.02 | |
| Ntdl | 0.01 | 0.00 | 0.05 | 0.00 | 0.10 | 0.00 | 0.00 | 0.00 | 0.18 | 0.27 | 0.21 | 0.21 | 0.22 | 0.33 | 0.22 | |
| Nexc | 0.00 | 0.00 | 0.02 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.04 | 0.13 | 0.10 | 0.08 | |||
| Npri | 0.02 | 0.00 | – | 0.00 | 0.00 | – | 0.00 | 0.01 | 0.00 | – | 0.07 | 0.05 | 0.12 | 0.11 | 0.07 | |
| Nswa | 0.03 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.01 | 0.00 | 0.00 | 0.11 | 0.21 | 0.33 | 0.22 | |||
| Nhet | 0.01 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.03 | 0.00 | 0.01 | 0.00 | 0.00 | 0.16 | 0.23 | 0.17 | |
| Nlec | 0.00 | 0.00 | 0.05 | 0.01 | 0.06 | 0.03 | 0.01 | 0.01 | 0.03 | 0.02 | 0.00 | 0.04 | 0.03 | 1.28 | 0.03 | |
| Npin | 0.00 | 0.01 | 0.03 | 0.01 | 0.05 | 0.00 | 0.00 | 0.02 | 0.01 | 0.01 | 0.00 | 0.00 | 0.00 | 0.00 | 0.15 | |
| Ncom | 0.02 | 0.00 | 0.02 | 0.03 | 0.09 | 0.07 | 0.02 | 0.00 | 0.08 | 0.00 | 0.00 | 0.00 | 0.02 | 0.00 | 0.04 |
Population gene flow estimates (2Nm) for CAD (above diagonal) and ANL43 (below diagonal). Numbers, symbols, and abbreviations are as described for Table 5.
| Nsp1 | Nabb | Nnig | Nrug | Nvir | Nwar | Ndub | Npra | Ntdl | Nexc | Npri | Nswa | Nhet | Nlec | Npin | Ncom | |
| Nsp1 | 0.22 | 0.00 | 0.10 | 0.08 | 0.20 | 0.00 | 0.12 | 0.20 | 0.00 | 0.02 | 0.03 | 0.01 | 0.00 | 0.01 | 0.00 | |
| Nabb | 0.00 | 0.18 | 12.73 | 4.27 | 0.00 | 0.11 | 6.68 | 0.03 | 0.10 | 0.05 | 0.00 | 0.00 | 0.06 | 0.00 | ||
| Nnig | 0.08 | – | 3.80 | 0.25 | – | 0.00 | 0.09 | 0.09 | 0.07 | – | – | 0.00 | 0.10 | 0.00 | 0.33 | |
| Nrug | 0.00 | 0.10 | 1.02 | 0.55 | 1.60 | 0.13 | 0.18 | 0.21 | 0.17 | 0.16 | 0.07 | 0.27 | 0.10 | 0.13 | 0.60 | |
| Nvir | 0.00 | 0.00 | 0.05 | 0.00 | – | 0.00 | 0.12 | 0.15 | 0.11* | 0.13 | 0.08 | 0.05 | 0.09 | 0.09 | 0.67 | |
| Nwar | 0.02 | 0.09 | 0.62 | 0.13 | 0.34 | 0.50 | 0.18 | 0.16 | 0.14 | 0.14 | 0.25* | 0.36 | ||||
| Ndub | 0.07 | 0.42 | 2.30 | 0.17 | 0.03 | 1.87 | 0.00 | 0.01 | 0.05 | 0.06 | 0.02 | 0.01 | 0.03 | 0.03 | 0.05 | |
| Npra | 0.00 | 0.04 | 0.19 | 0.06 | 0.00 | 0.14 | 0.57 | 5.15 | 0.04 | 0.06 | 0.03 | 0.02 | 0.03 | 0.02 | 0.01 | |
| Ntdl | 0.01 | 0.12 | 0.08 | 0.04 | 0.00 | 0.04 | 0.09* | 0.00 | 0.05 | 0.08 | 0.03 | 0.01 | 0.05 | 0.03 | 0.29 | |
| Nexc | 0.09 | 0.13 | 0.43 | 0.08 | 0.11* | 0.26 | 0.10 | 0.18 | 0.13 | 0.10 | 0.10 | 0.06 | 0.10 | |||
| Npri | 0.09 | 0.24 | – | 0.11 | 0.16 | – | 0.14 | 0.16 | 0.15 | – | 2.03 | 0.24 | 0.10 | 0.10 | 0.09 | |
| Nswa | 0.03 | 0.02 | 0.00 | 0.00 | 0.00 | 0.02 | 0.05 | 0.00 | 0.05 | 0.04 | 0.07 | 0.06 | 0.13 | |||
| Nhet | 0.03 | 0.06 | 0.07 | 0.04 | 0.02 | 0.12 | 0.06 | 0.06 | 0.04 | 0.07 | 0.08 | 0.02 | 0.25 | 0.06 | 0.09 | |
| Nlec | 0.00 | 0.00 | 0.05 | 0.02 | 0.04 | 0.03 | 0.05 | 0.08 | 0.03 | 0.07 | 0.09 | 0.02 | 0.04 | 0.03 | 0.01 | |
| Npin | 0.03 | 0.01 | 0.06 | 0.05 | 0.00 | 0.07 | 0.05 | 0.09 | 0.03 | 0.10 | 0.08 | 0.00 | 0.04 | 0.00 | 0.10 | |
| Ncom | 0.00 | 0.00 | 0.00 | 0.03 | 0.04 | 0.00 | 0.05 | 0.00 | 0.01 | 0.07 | 0.08 | 0.00 | 0.03 | 0.01 | 0.01 |
| Nsp1 | Nabb | Nnig | Nrug | Nvir | Nwar | Ndub | Npra | Ntdl | Nexc | Npri | Nswa | Nhet | Nlec | Npin | Ncom | |
| Nsp1 | 0.22 | 0.00 | 0.10 | 0.08 | 0.20 | 0.00 | 0.12 | 0.20 | 0.00 | 0.02 | 0.03 | 0.01 | 0.00 | 0.01 | 0.00 | |
| Nabb | 0.00 | 0.18 | 12.73 | 4.27 | 0.00 | 0.11 | 6.68 | 0.03 | 0.10 | 0.05 | 0.00 | 0.00 | 0.06 | 0.00 | ||
| Nnig | 0.08 | – | 3.80 | 0.25 | – | 0.00 | 0.09 | 0.09 | 0.07 | – | – | 0.00 | 0.10 | 0.00 | 0.33 | |
| Nrug | 0.00 | 0.10 | 1.02 | 0.55 | 1.60 | 0.13 | 0.18 | 0.21 | 0.17 | 0.16 | 0.07 | 0.27 | 0.10 | 0.13 | 0.60 | |
| Nvir | 0.00 | 0.00 | 0.05 | 0.00 | – | 0.00 | 0.12 | 0.15 | 0.11* | 0.13 | 0.08 | 0.05 | 0.09 | 0.09 | 0.67 | |
| Nwar | 0.02 | 0.09 | 0.62 | 0.13 | 0.34 | 0.50 | 0.18 | 0.16 | 0.14 | 0.14 | 0.25* | 0.36 | ||||
| Ndub | 0.07 | 0.42 | 2.30 | 0.17 | 0.03 | 1.87 | 0.00 | 0.01 | 0.05 | 0.06 | 0.02 | 0.01 | 0.03 | 0.03 | 0.05 | |
| Npra | 0.00 | 0.04 | 0.19 | 0.06 | 0.00 | 0.14 | 0.57 | 5.15 | 0.04 | 0.06 | 0.03 | 0.02 | 0.03 | 0.02 | 0.01 | |
| Ntdl | 0.01 | 0.12 | 0.08 | 0.04 | 0.00 | 0.04 | 0.09* | 0.00 | 0.05 | 0.08 | 0.03 | 0.01 | 0.05 | 0.03 | 0.29 | |
| Nexc | 0.09 | 0.13 | 0.43 | 0.08 | 0.11* | 0.26 | 0.10 | 0.18 | 0.13 | 0.10 | 0.10 | 0.06 | 0.10 | |||
| Npri | 0.09 | 0.24 | – | 0.11 | 0.16 | – | 0.14 | 0.16 | 0.15 | – | 2.03 | 0.24 | 0.10 | 0.10 | 0.09 | |
| Nswa | 0.03 | 0.02 | 0.00 | 0.00 | 0.00 | 0.02 | 0.05 | 0.00 | 0.05 | 0.04 | 0.07 | 0.06 | 0.13 | |||
| Nhet | 0.03 | 0.06 | 0.07 | 0.04 | 0.02 | 0.12 | 0.06 | 0.06 | 0.04 | 0.07 | 0.08 | 0.02 | 0.25 | 0.06 | 0.09 | |
| Nlec | 0.00 | 0.00 | 0.05 | 0.02 | 0.04 | 0.03 | 0.05 | 0.08 | 0.03 | 0.07 | 0.09 | 0.02 | 0.04 | 0.03 | 0.01 | |
| Npin | 0.03 | 0.01 | 0.06 | 0.05 | 0.00 | 0.07 | 0.05 | 0.09 | 0.03 | 0.10 | 0.08 | 0.00 | 0.04 | 0.00 | 0.10 | |
| Ncom | 0.00 | 0.00 | 0.00 | 0.03 | 0.04 | 0.00 | 0.05 | 0.00 | 0.01 | 0.07 | 0.08 | 0.00 | 0.03 | 0.01 | 0.01 |
Population gene flow estimates (2Nm) for CAD (above diagonal) and ANL43 (below diagonal). Numbers, symbols, and abbreviations are as described for Table 5.
| Nsp1 | Nabb | Nnig | Nrug | Nvir | Nwar | Ndub | Npra | Ntdl | Nexc | Npri | Nswa | Nhet | Nlec | Npin | Ncom | |
| Nsp1 | 0.22 | 0.00 | 0.10 | 0.08 | 0.20 | 0.00 | 0.12 | 0.20 | 0.00 | 0.02 | 0.03 | 0.01 | 0.00 | 0.01 | 0.00 | |
| Nabb | 0.00 | 0.18 | 12.73 | 4.27 | 0.00 | 0.11 | 6.68 | 0.03 | 0.10 | 0.05 | 0.00 | 0.00 | 0.06 | 0.00 | ||
| Nnig | 0.08 | – | 3.80 | 0.25 | – | 0.00 | 0.09 | 0.09 | 0.07 | – | – | 0.00 | 0.10 | 0.00 | 0.33 | |
| Nrug | 0.00 | 0.10 | 1.02 | 0.55 | 1.60 | 0.13 | 0.18 | 0.21 | 0.17 | 0.16 | 0.07 | 0.27 | 0.10 | 0.13 | 0.60 | |
| Nvir | 0.00 | 0.00 | 0.05 | 0.00 | – | 0.00 | 0.12 | 0.15 | 0.11* | 0.13 | 0.08 | 0.05 | 0.09 | 0.09 | 0.67 | |
| Nwar | 0.02 | 0.09 | 0.62 | 0.13 | 0.34 | 0.50 | 0.18 | 0.16 | 0.14 | 0.14 | 0.25* | 0.36 | ||||
| Ndub | 0.07 | 0.42 | 2.30 | 0.17 | 0.03 | 1.87 | 0.00 | 0.01 | 0.05 | 0.06 | 0.02 | 0.01 | 0.03 | 0.03 | 0.05 | |
| Npra | 0.00 | 0.04 | 0.19 | 0.06 | 0.00 | 0.14 | 0.57 | 5.15 | 0.04 | 0.06 | 0.03 | 0.02 | 0.03 | 0.02 | 0.01 | |
| Ntdl | 0.01 | 0.12 | 0.08 | 0.04 | 0.00 | 0.04 | 0.09* | 0.00 | 0.05 | 0.08 | 0.03 | 0.01 | 0.05 | 0.03 | 0.29 | |
| Nexc | 0.09 | 0.13 | 0.43 | 0.08 | 0.11* | 0.26 | 0.10 | 0.18 | 0.13 | 0.10 | 0.10 | 0.06 | 0.10 | |||
| Npri | 0.09 | 0.24 | – | 0.11 | 0.16 | – | 0.14 | 0.16 | 0.15 | – | 2.03 | 0.24 | 0.10 | 0.10 | 0.09 | |
| Nswa | 0.03 | 0.02 | 0.00 | 0.00 | 0.00 | 0.02 | 0.05 | 0.00 | 0.05 | 0.04 | 0.07 | 0.06 | 0.13 | |||
| Nhet | 0.03 | 0.06 | 0.07 | 0.04 | 0.02 | 0.12 | 0.06 | 0.06 | 0.04 | 0.07 | 0.08 | 0.02 | 0.25 | 0.06 | 0.09 | |
| Nlec | 0.00 | 0.00 | 0.05 | 0.02 | 0.04 | 0.03 | 0.05 | 0.08 | 0.03 | 0.07 | 0.09 | 0.02 | 0.04 | 0.03 | 0.01 | |
| Npin | 0.03 | 0.01 | 0.06 | 0.05 | 0.00 | 0.07 | 0.05 | 0.09 | 0.03 | 0.10 | 0.08 | 0.00 | 0.04 | 0.00 | 0.10 | |
| Ncom | 0.00 | 0.00 | 0.00 | 0.03 | 0.04 | 0.00 | 0.05 | 0.00 | 0.01 | 0.07 | 0.08 | 0.00 | 0.03 | 0.01 | 0.01 |
| Nsp1 | Nabb | Nnig | Nrug | Nvir | Nwar | Ndub | Npra | Ntdl | Nexc | Npri | Nswa | Nhet | Nlec | Npin | Ncom | |
| Nsp1 | 0.22 | 0.00 | 0.10 | 0.08 | 0.20 | 0.00 | 0.12 | 0.20 | 0.00 | 0.02 | 0.03 | 0.01 | 0.00 | 0.01 | 0.00 | |
| Nabb | 0.00 | 0.18 | 12.73 | 4.27 | 0.00 | 0.11 | 6.68 | 0.03 | 0.10 | 0.05 | 0.00 | 0.00 | 0.06 | 0.00 | ||
| Nnig | 0.08 | – | 3.80 | 0.25 | – | 0.00 | 0.09 | 0.09 | 0.07 | – | – | 0.00 | 0.10 | 0.00 | 0.33 | |
| Nrug | 0.00 | 0.10 | 1.02 | 0.55 | 1.60 | 0.13 | 0.18 | 0.21 | 0.17 | 0.16 | 0.07 | 0.27 | 0.10 | 0.13 | 0.60 | |
| Nvir | 0.00 | 0.00 | 0.05 | 0.00 | – | 0.00 | 0.12 | 0.15 | 0.11* | 0.13 | 0.08 | 0.05 | 0.09 | 0.09 | 0.67 | |
| Nwar | 0.02 | 0.09 | 0.62 | 0.13 | 0.34 | 0.50 | 0.18 | 0.16 | 0.14 | 0.14 | 0.25* | 0.36 | ||||
| Ndub | 0.07 | 0.42 | 2.30 | 0.17 | 0.03 | 1.87 | 0.00 | 0.01 | 0.05 | 0.06 | 0.02 | 0.01 | 0.03 | 0.03 | 0.05 | |
| Npra | 0.00 | 0.04 | 0.19 | 0.06 | 0.00 | 0.14 | 0.57 | 5.15 | 0.04 | 0.06 | 0.03 | 0.02 | 0.03 | 0.02 | 0.01 | |
| Ntdl | 0.01 | 0.12 | 0.08 | 0.04 | 0.00 | 0.04 | 0.09* | 0.00 | 0.05 | 0.08 | 0.03 | 0.01 | 0.05 | 0.03 | 0.29 | |
| Nexc | 0.09 | 0.13 | 0.43 | 0.08 | 0.11* | 0.26 | 0.10 | 0.18 | 0.13 | 0.10 | 0.10 | 0.06 | 0.10 | |||
| Npri | 0.09 | 0.24 | – | 0.11 | 0.16 | – | 0.14 | 0.16 | 0.15 | – | 2.03 | 0.24 | 0.10 | 0.10 | 0.09 | |
| Nswa | 0.03 | 0.02 | 0.00 | 0.00 | 0.00 | 0.02 | 0.05 | 0.00 | 0.05 | 0.04 | 0.07 | 0.06 | 0.13 | |||
| Nhet | 0.03 | 0.06 | 0.07 | 0.04 | 0.02 | 0.12 | 0.06 | 0.06 | 0.04 | 0.07 | 0.08 | 0.02 | 0.25 | 0.06 | 0.09 | |
| Nlec | 0.00 | 0.00 | 0.05 | 0.02 | 0.04 | 0.03 | 0.05 | 0.08 | 0.03 | 0.07 | 0.09 | 0.02 | 0.04 | 0.03 | 0.01 | |
| Npin | 0.03 | 0.01 | 0.06 | 0.05 | 0.00 | 0.07 | 0.05 | 0.09 | 0.03 | 0.10 | 0.08 | 0.00 | 0.04 | 0.00 | 0.10 | |
| Ncom | 0.00 | 0.00 | 0.00 | 0.03 | 0.04 | 0.00 | 0.05 | 0.00 | 0.01 | 0.07 | 0.08 | 0.00 | 0.03 | 0.01 | 0.01 |
Frequency distributions of nuclear (EF1α, CAD, ANL43, and average nuclear) to mitochondrial gene flow (2Nm) ratios. Distributions include 111 out of 120 pairwise species comparisons. One comparison is not included because the observed mitochondrial 2Nm was 0; eight comparisons are not included because their results were inconclusive, see text. Observed ratios were divided into bins and all values greater than 2 were combined into a single bin. For each bar, the leftmost number represents the lower bound. Dashed lines indicate the median ratio expected under the null hypothesis that mitochondrial and nuclear gene flow rates are equal.
Wilcoxon matched-pairs signed-ranks tests confirmed that mitochondrial gene flow was very significantly higher than each of the three nuclear genes, as well as the average across all three genes (Table 7). These patterns were evident in IMset, ALLset, and UNAMBset datasets. A complete list of results for all comparisons and all IM runs is available online (Supplementary Materials Table S3).
Results of Wilcoxon signed-ranks matched-pairs tests. Comparisons are between 2Nmestimates obtained for COI/COII and each of the nuclear genes and the average across all three nuclear genes (avgNUC). Matched pairs are individual species comparisons.W+, W-,andnare the values used to calculate the test statisticZ. AllPvalues are significant atα=.05 (after Bonferroni correction for multiple comparisons). Significance results are given for each of the three sets of data files examined in IM.
| Comparison | Dataset | W+ | W− | N | Z | P value |
| COI/COII-EF1α | Imset | 6105 | 0 | 110 | 494.98 | 9.0×10−20 |
| ALLset | 4281 | 90 | 93 | 376.05 | 1.0×10−15 | |
| UNAMBset | 5676 | 102 | 107 | 466.21 | 4.8×10−18 | |
| COI/COII-CAD | Imset | 5027 | 1189 | 111 | 404.05 | 1.7×10−8 |
| ALLset | 3555 | 1596 | 101 | 297.60 | 9.1×10−4 | |
| UNAMBset | 4697 | 1298 | 109 | 380.53 | 2.8×10−7 | |
| COI/COII-ANL43 | Imset | 5544 | 561 | 110 | 448.66 | 1.1×10−13 |
| ALLset | 3759 | 801 | 95 | 325.51 | 4.1×10−8 | |
| UNAMBset | 4624 | 1154 | 107 | 378.13 | 7.0×10−8 | |
| COI/COII-avgNUC | Imset | 5542 | 674 | 111 | 446.38 | 8.1×10−13 |
| ALLset | 4627 | 1368 | 109 | 374.73 | 8.5×10−7 | |
| UNAMBset | 5575 | 1095 | 115 | 440.89 | 4.1×10−10 |
| Comparison | Dataset | W+ | W− | N | Z | P value |
| COI/COII-EF1α | Imset | 6105 | 0 | 110 | 494.98 | 9.0×10−20 |
| ALLset | 4281 | 90 | 93 | 376.05 | 1.0×10−15 | |
| UNAMBset | 5676 | 102 | 107 | 466.21 | 4.8×10−18 | |
| COI/COII-CAD | Imset | 5027 | 1189 | 111 | 404.05 | 1.7×10−8 |
| ALLset | 3555 | 1596 | 101 | 297.60 | 9.1×10−4 | |
| UNAMBset | 4697 | 1298 | 109 | 380.53 | 2.8×10−7 | |
| COI/COII-ANL43 | Imset | 5544 | 561 | 110 | 448.66 | 1.1×10−13 |
| ALLset | 3759 | 801 | 95 | 325.51 | 4.1×10−8 | |
| UNAMBset | 4624 | 1154 | 107 | 378.13 | 7.0×10−8 | |
| COI/COII-avgNUC | Imset | 5542 | 674 | 111 | 446.38 | 8.1×10−13 |
| ALLset | 4627 | 1368 | 109 | 374.73 | 8.5×10−7 | |
| UNAMBset | 5575 | 1095 | 115 | 440.89 | 4.1×10−10 |
Results of Wilcoxon signed-ranks matched-pairs tests. Comparisons are between 2Nmestimates obtained for COI/COII and each of the nuclear genes and the average across all three nuclear genes (avgNUC). Matched pairs are individual species comparisons.W+, W-,andnare the values used to calculate the test statisticZ. AllPvalues are significant atα=.05 (after Bonferroni correction for multiple comparisons). Significance results are given for each of the three sets of data files examined in IM.
| Comparison | Dataset | W+ | W− | N | Z | P value |
| COI/COII-EF1α | Imset | 6105 | 0 | 110 | 494.98 | 9.0×10−20 |
| ALLset | 4281 | 90 | 93 | 376.05 | 1.0×10−15 | |
| UNAMBset | 5676 | 102 | 107 | 466.21 | 4.8×10−18 | |
| COI/COII-CAD | Imset | 5027 | 1189 | 111 | 404.05 | 1.7×10−8 |
| ALLset | 3555 | 1596 | 101 | 297.60 | 9.1×10−4 | |
| UNAMBset | 4697 | 1298 | 109 | 380.53 | 2.8×10−7 | |
| COI/COII-ANL43 | Imset | 5544 | 561 | 110 | 448.66 | 1.1×10−13 |
| ALLset | 3759 | 801 | 95 | 325.51 | 4.1×10−8 | |
| UNAMBset | 4624 | 1154 | 107 | 378.13 | 7.0×10−8 | |
| COI/COII-avgNUC | Imset | 5542 | 674 | 111 | 446.38 | 8.1×10−13 |
| ALLset | 4627 | 1368 | 109 | 374.73 | 8.5×10−7 | |
| UNAMBset | 5575 | 1095 | 115 | 440.89 | 4.1×10−10 |
| Comparison | Dataset | W+ | W− | N | Z | P value |
| COI/COII-EF1α | Imset | 6105 | 0 | 110 | 494.98 | 9.0×10−20 |
| ALLset | 4281 | 90 | 93 | 376.05 | 1.0×10−15 | |
| UNAMBset | 5676 | 102 | 107 | 466.21 | 4.8×10−18 | |
| COI/COII-CAD | Imset | 5027 | 1189 | 111 | 404.05 | 1.7×10−8 |
| ALLset | 3555 | 1596 | 101 | 297.60 | 9.1×10−4 | |
| UNAMBset | 4697 | 1298 | 109 | 380.53 | 2.8×10−7 | |
| COI/COII-ANL43 | Imset | 5544 | 561 | 110 | 448.66 | 1.1×10−13 |
| ALLset | 3759 | 801 | 95 | 325.51 | 4.1×10−8 | |
| UNAMBset | 4624 | 1154 | 107 | 378.13 | 7.0×10−8 | |
| COI/COII-avgNUC | Imset | 5542 | 674 | 111 | 446.38 | 8.1×10−13 |
| ALLset | 4627 | 1368 | 109 | 374.73 | 8.5×10−7 | |
| UNAMBset | 5575 | 1095 | 115 | 440.89 | 4.1×10−10 |
Discussion
There is extensive disagreement between mitochondrial and nuclear genes with respect to inter- and intraspecific relationships in the lecontei group of the sawfly genus Neodiprion. These patterns were readily apparent in a phylogenetic analysis that employed dense taxonomic sampling, multiple markers, and multiple methods of analysis. Bayesian tests of species monophyly and SH tests of data partition congruence were consistent with a hypothesis of rampant hybridization and mitochondrial introgression. We explicitly tested this hypothesis by comparing interspecific mitochondrial and nuclear gene flow. In support of our hypothesis, estimates of mitochondrial gene flow were consistently higher than nuclear gene flow.
VIOLATION OF IM ASSUMPTIONS
We have interpreted our observation of consistently higher mitochondrial gene flow as evidence that hybridization and mitochondrial introgression have been prevalent throughout Neodiprion's evolutionary history, but this observation could be accounted for by processes other than introgression if systematic differences between mitochondrial and nuclear loci bias interspecific coalescence times toward the present for mitochondrial loci and/or toward the past for nuclear loci. Three IM assumptions that are likely to be violated by our dataset and that may potentially systematically bias coalescence times are selective neutrality, equal sex ratios, and no among site rate variation.
First, IM assumes that loci examined are selectively neutral (Hey and Nielsen 2004). Due to an absence of recombination in mitochondria, selective sweeps may be more common in mitochondrial genes than nuclear genes (Hudson and Turelli 2003; Ballard and Whitlock 2004). However, selective sweeps that have occurred in extant and ancestral populations are expected to stretch inferred gene tree depths in opposite directions (Berry et al. 1991; Langley et al. 1993; Guttman and Dykhuizen 1994; Hilton et al. 1994; Schlenke and Begun 2004). It is therefore unclear in which, if any, direction mitochondrial gene tree depths would be biased when IM's assumption of selective neutrality is violated by recurrent selective sweeps.
Second, we included inheritance scalars (h) of 0.25 and 0.75 for haplodiploid mitochondrial and nuclear loci, respectively. This 3:1 ratio assumes equal effective population sizes in both sexes and almost certainly does not hold for Neodiprion, which generally has female-biased sex ratios (Craig and Mopper 1993). However, under the most extreme female-biased sex ratio, the ratio of nuclear to mitochondrial gene copies will approach 2:1. When we multiplied all of our mitochondrial 2Nm estimates by 2/3 (to replace a 3:1 effective population size correction with a 2:1 correction), mitochondrial gene flow remained significantly higher than nuclear gene flow (Wilcoxon signed-ranks matched-pairs test; mitochondrial 2Nm vs. average nuclear 2Nm; Z= 417; P < 1 × 10−9).
Third, an HKY model of sequence evolution with no mutation rate variation was assumed for each locus. However, within-locus rate heterogeneity is expected to cause an overestimation in time to most recent common ancestor (Markovtsova et al. 2000). Therefore, if rate heterogeneity is lowest at the mitochondrial locus, divergence time estimates for nuclear loci, and therefore mitochondrial gene flow estimates, will have been systematically biased upward. We assessed among site rate variation (“ρ” from Gu et al. 1995) for each locus using estimates of gamma shape parameters and proportions of invariant sites obtained in PHYML (ρ for COI/COII, EF1α, CAD, and ANL43 was 0.88, 0.62, 0.81, and 0.79, respectively) and MrBayes (ρ estimates were similar across loci and ranged from 0.93–0.96); neither set of estimates indicates that COI/COII has substantially less rate heterogeneity than the nuclear loci.
Finally, because the program IM can only accommodate pairs of taxa, we estimated locus-specific gene flow rates for every possible pairwise species comparison. This approach is a clear violation of the IM assumption that the entities examined are sister taxa and have not exchanged genes with a third taxon. However, this violation impacts mitochondrial and nuclear loci equally and should not introduce systematic bias. Still, we must interpret our results with caution because gene flow may be erroneously inferred between nonhybridizing species due to gene flow with a third species and/or gene flow between ancestral taxa (Won and Hey 2005; Hey and Nielsen 2006). Phylogenetic evidence must be used in conjunction with estimates of interspecific gene flow to identify likely cases of genetic exchange (see below). In summary, violations of IM assumptions do not adequately explain the consistent differences we have observed between mitochondrial and nuclear gene flow rates. Instead, repeated mitochondrial introgression appears to be the most likely explanation for observed gene flow patterns.
MITOCHONDRIAL INTROGRESSION THROUGHOUT NEODIPRION HISTORY
Recent mitochondrial introgression
In the absence of introgression, mitochondrial gene trees are expected to achieve reciprocal monophyly more quickly than nuclear gene trees (Palumbi et al. 2001; Ballard and Whitlock 2004); but if mitochondrial introgression has been prevalent and recent, these gene trees may contain as many or more nonmonophyletic species as the nuclear gene trees. In support of this prediction, we found that monophyly was rejected for 10 out of 16 species by the mitochondrial dataset. The only nuclear dataset that rejected monophyly for a comparable number of species was CAD (11 out of 16). CAD's failure to recover species monophyly may be explained by a relative lack of variation (Table 2). In contrast, COI/COII had the most (numerically and percentagewise) variable and PI characters (Table 2); therefore, lack of resolution does not adequately explain COI/COII's tendency to reject species monophyly. Instead, estimates of interspecific mitochondrial gene flow suggest that this tendency may result from recent mitochondrial introgression.
Observations of polyphyletic species coupled with high mitochondrial gene flow estimates (i.e., 2Nm > 1, the amount of gene flow between populations that is expected to prevent divergence, Wright 1931) are consistent with several recent introgression events: introgression of N. lecontei mitochondria into N. pinetum (“A” in Fig. 1; 2Nm= 1.28); introgression of N. pratti mitochondria into N. taedae linearis (“B” in Fig. 1; 2Nm= 39.53); and a massive introgression episode involving five species in northeastern North America: N. dubiosus, N. rugifrons, N. swainei, N. nigroscutum, and N. abbotii (clades “C” and “D” in Fig. 1; 2Nm values range from 0.58 to 12.55). This last set of introgressing taxa is particularly intriguing and may be comprised of multiple geographically and temporally distinct introgression episodes. More intensive population-level sampling and analysis will be required to reconstruct an exact sequence of introgression events for these five species, but it is clear that mitochondrial introgression has been pervasive throughout their recent evolutionary history.
Ancient mitochondrial introgression
Mitochondrial introgression between nonsister taxa is expected to result in topological differences between mitochondrial and nuclear gene trees. In support of this prediction, SH tests revealed that partitions containing mitochondrial data (COI/COII and ALL) were reciprocally incongruent with all nuclear partitions (EF1α, CAD, ANL43, and NUC). In contrast, the failure of each nuclear locus to reject the combined nuclear topology (and vice versa) suggests that they are largely congruent (Hipp et al. 2004; Struck et al. 2006). The case for mitochondrial introgression is bolstered by the observation that mitochondrial gene flow was significantly higher than nuclear gene flow across all pairwise species comparisons (Table 7). This difference remains significant even after the removal of all comparisons involving species for which monophyly was rejected by the mitochondrial dataset (Wilcoxon signed-ranks matched-pairs test; mitochondrial vs. average nuclear gene flow; Z= 17.7; P= 0.013); therefore, recent introgression cannot fully explain discrepancies between mitochondrial and nuclear gene flow estimates.
Pinpointing ancient introgression events is difficult because mitochondrial lineages will have become reciprocally monophyletic in formerly hybridizing taxa and direct evidence for donor taxa is erased. However, a case for ancient introgression between nonsister taxa can be made when strongly supported topological conflicts between mitochondrial and nuclear gene trees are accompanied by appreciable estimates of mitochondrial gene flow. For example, N. hetricki monophyly was not rejected by any of the datasets (Table 3), yet this species falls out in distinctly different, strongly supported (i.e., ≥ 95% under all criteria) clades in mitochondrial and nuclear gene trees (Figs. 1 and 2). Gene flow estimates agree with the interpretation that the nuclear relationships reflect true branching history and the mitochondrial relationships reflect ancient introgression—across all comparisons involving N. hetricki, mitochondrial gene flow was consistently higher than average nuclear gene flow (Z= 22.25; P < 0.001). Average nuclear gene flow between N. hetricki and each of the three species it grouped with in the nuclear phylogeny was 0.11; average mitochondrial gene flow for species N. hetricki grouped with in the mitochondrial phylogeny was 0.34. Notably, the mitochondrial 2Nm estimate for the N. hetricki/N. abbotii comparison was substantial (2.40), suggesting that hybridization and mitochondrial introgression may have occurred somewhere in the histories of these two species.
Other hybridization outcomes
Given the apparently frequent opportunities Neodiprion species have had for gene exchange, one might expect that nuclear genes would have occasionally crossed species boundaries as well. Indeed, although mitochondrial gene flow was generally higher than nuclear gene flow, there were some comparisons in which considerable nuclear gene flow was apparent. For example, estimated gene flow between N. abbotii and N. virginiana was high (2Nm > 1) for COI/COII, EF1α, and CAD (Tables 5 and 6). These patterns could be explained by frequent hybridization and introgression, which could either lead to the eventual collapse of species (e.g., Taylor et al. 2006) or persist indefinitely if selection against introgression is present at other loci (e.g., Barton and Hewitt 1985; Shaw and Danley 2003). Alternatively, multiple high gene flow estimates could indicate that a species is of hybrid origin, a speciation mode that has already been suggested to account for the origin of one Neodiprion species (N. merkeli, Ross 1961; unfortunately gene flow could not be assessed for this species because only a single population was collected).
WHY DO NEODIPRION SAWFLIES HYBRIDIZE?
Based on our estimates of gene flow, hybridization in Neodiprion appears to have been frequent, but not universal. This observation raises several questions to be addressed by future work: (1) Why do some species hybridize and not others? (2) How do hybridizing species remain distinct in the face of gene flow? (3) What are the evolutionary consequences of different amounts of historical hybridization? At present, it seems that the extent to which species hybridize may be directly linked to how much they overlap in host use. As may be expected if host plants generally represent primary barriers in herbivorous insect speciation (Bush 1969, 1975a,b; Berlocher and Feder 2002), all of the instances of recent hybridization in Neodiprion involve species that at least sometimes share hosts. Indeed, the highest incidence of hybridization occurs between species that are monophagous (or nearly so) on jack pine. The continued existence of these species despite evident gene flow and ecological overlap is surprising and suggests that there must be nonhost-related barriers to reproduction and possibly within-host niche-partitioning (by host size or age class, e.g., Lyons 1964; McMillin and Wagner 1993). Also, some Neodiprion species that overlap in host use show no evidence of hybridization (e.g., N. compar also feeds on jack pine and has experienced very little gene exchange with other species, Tables 4 and 5), but it is not yet clear how these species differ from those that exchange genes. Finally, the propensity for males to respond to similar female pheromone blends may provide another avenue for hybridization between some species pairs (e.g., Kraemer et al. 1979, 1981, 1983, 1984; Kraemer and Coppel 1983; Anderbrant 1993). However, shared pheromone responses are not uncommon in nonhybridizing pairs (e.g., Olaifa et al. 1987) and female choice may ultimately prove to be more important in preventing interspecific hybridization events (Chan and Levin 2005; McPeek and Gavrilets 2006).
WHY IS MITOCHONDRIAL INTROGRESSION PREVALENT IN NEODIPRION?
Given that Neodiprion species often exchange genes, it remains to be established why mitochondrial introgression is the most frequent result. Chan and Levin (2005) suggest that frequency-dependent prezygotic barriers to mating (e.g., normally choosy females are more willing to accept heterospecific males when conspecific males are rare) may provide a general explanation for biased cytoplasmic introgression. Under a wide range of model parameters, they found that whereas both nuclear and mitochondrial genes from a rare species readily introgressed into a common species, mitochondrial introgression consistently exceeded nuclear introgression. Also, this discrepancy was most pronounced when the proportion of immigrants (rare species) was small. Thus, one prediction of this model is that biased mitochondrial introgression will be most prevalent “when two potentially hybridizing species meet in circumstances of disparate abundance” (Chan and Levin 2005).
Intriguingly, there are several biological attributes of Neodiprion species, manifest over multiple time scales, which may lead to profound differences in the local abundance of co-occurring species. Within a single year, adults of all species are short lived (Coppel and Benjamin 1965) and have well-defined, species-specific emergence peaks in response to environmental cues (Knerer 1993). If emergence peaks between two species are only partially overlapping, stragglers of one species may find themselves in an emergence peak of a second species. Moreover, and perhaps most notably, many Neodiprion species are considered “outbreak” species, meaning that they experience dramatic changes in population densities from low densities (endemic) to extremely high densities (epidemic) and vice versa in different years (Larsson et al. 1993). If one species is in a population “boom” and another in a “bust,” the rarer female may be more willing to accept heterospecific males (i.e., she is choosy unless given little choice), and her mitochondrial haplotype may introgress into the “boom” species more readily than her nuclear genes. Similarly, nonoutbreak species may also act as mitochondrial donors during outbreaks of sympatric species. Finally, historical factors may have also led to numerical imbalance between hybridizing species. In particular, as species shifted their ranges in response to glacial advances and retreats during the Pleistocene, asymmetries may have arisen simply due to species-specific colonization patterns—rare colonists of one species may have encountered an abundant, established species (McPeek and Gavrilets 2006).
Significance and Conclusions
Our results have several implications for phylogenetic analysis of groups that, like Neodiprion, have experienced frequent hybridization and differential introgression. First, biased introgression compromises the utility of some gene trees as estimates of the species tree. Second, introgression is not just a “young species” problem. Although we have explicitly tested the mitochondrial introgression hypothesis in the lecontei clade only, extensive mito-nuclear discordance is also evident in the genus as a whole and in gene trees estimated for the family Diprionidae (C. Linnen, unpubl. data). It seems plausible that these patterns also result from mitochondrial introgression, given that evidence of introgression was observed at all levels of divergence within the lecontei clade. Third, genes that show the least gene flow throughout their history should be the most reliable markers for species delimitation and estimation of relationships. Finally, loci that are prone to introgression (such as mitochondria in Neodiprion) provide invaluable records of interspecific hybridization events.
This study also presents a novel methodology for comparative studies of genetic exchange between species and across loci. Several intriguing patterns have emerged from our analysis in Neodiprion—in particular, host use and frequency-dependent mate choice may mediate which species exchange genes and which loci cross species boundaries. These attributes may also be central to the evolution of the barriers that prevent hybridization. Moreover, the observation that species are able to remain distinct despite hybridization raises the issue of whether initial barriers to reproduction have also arisen in the face of gene flow (i.e., sympatric speciation). Comparative studies of speciation patterns in Neodiprion will address these questions and are currently underway. For now it is evident that a complex history of opposing processes has shaped the diversification of Neodiprion.
Associate Editor: M. Peterson
NOTE ADDED TO PROOF
Further morphological study of Florida N. virginiana populations suggests that they represent a new species. Additional IM analyses performed under this alternative taxonomy remain consistent with the results presented here (mitochondrial gene flow significantly higher than nuclear gene flow; p < 1 × 10–12).
ACKNOWLEDGMENTS
We thank S. Li, K. Nystrom, J. Rousselet, G. Sanchez-Martinez, and A. Sequeira for providing specimens included in this study. For collecting advice, rearing advice, logistical support, or assistance in the field, we thank the following individuals: C. Asaro, M. Breon, S. Codella, D. Conser, E. Czerwinski, A. Eglitis, W. Ingram, M. Linnen, A. Lynch, B. Mayfield, K. Raffa, D. Smith, L. Thompson, T. Tigner, M. Wagner, and many kind foresters and entomologists. A. Thornton provided lab assistance, and B. Jennings provided advice and assistance during the development of the anonymous nuclear locus. Collecting permits were provided by the Florida Department of Environmental Protection and The Nature Conservancy (Florida and Maine Chapters). We thank M. Peterson and two anonymous reviewers for helpful comments on the manuscript. We are also grateful to B. Bossert, D. Haig, N. Pierce, and J. Wakeley for discussions and/or comments on this manuscript, and to J. Hey for answering questions regarding IM. Funding for this research was provided by a Graduate Research Fellowship and a Dissertation Improvement Grant (DEB-0308815) from the National Science Foundation, a Science to Achieve Results Graduate Fellowship from the Environmental Protection Agency, the Putnam Expeditionary Fund at the Museum of Comparative Zoology, the Theodore Roosevelt Memorial Fund at the American Museum of Natural History, and the Department of Organismic and Evolutionary Biology at Harvard University.
LITERATURE CITED
Appendix
Collection data for Neodiprion specimens included in this study. For each specimen, an ID number is given to permit cross-reference with more detailed collection and rearing data, museum specimens, and future publications.
| Specimen1 | ID2 | Date | Location | Host3 |
| N. abbotii FL-1 | 001-04 | April 2004 | USA: Florida: E of Macclenny | P. palustris |
| N. abbotii FL-2 | 162-03 | November 2003 | USA: Florida: Gainesville | P. taeda |
| N. abbotii FL-3 | 184-03.2 | November 2003 | USA: Florida: Palmdale | P. elliottii |
| N. abbotii FL-4 | 188-03 | November 2003 | USA: Florida: Ocala National Forest | P. palustris |
| N. abbotii GA | 015-04 | April 2004 | USA: Georgia: Resaca | P. taeda |
| N. abbotii MD | 131-04 | July 2004 | USA: MD: N of Easton | P. taeda |
| N. abbotii ME | 014-01 | July 2001 | USA: Maine: Bar Harbor | P. resinosa |
| N. abbotii TN | 120-04.1 | July 2004 | USA: Tennessee: W of Rockwood | P. echinata |
| N. abbotii VA | 055-04 | May 2004 | USA: Virginia: W of Petersburg | P. taeda |
| N. compar FL-1 | 089-04 | July 2004 | USA: Florida: Gainesville | P. palustris |
| N. compar FL-2 | 161-03A | November 2003 | USA: Florida: Gainesville | P. palustris |
| N. compar GA | 104-04A | July 2004 | USA: Georgia: Sylvester | P. elliottii |
| N. compar MA-1 | 371-02 | September 2002 | USA: Massachusetts: Myles Standish State Forest | P. rigida |
| N. compar MA-2 | 378-02 | September 2002 | USA: Massachusetts: Myles Standish State Forest | P. rigida |
| N. compar ME | 163-04 | July 2004 | USA: Maine: W of Lake Parlin | P. banksiana |
| N. compar NC | 122-04A | July 2004 | USA: North Carolina: Kings Mountain | P. taeda |
| N. compar ON-1 | 148-02 | August 2002 | Canada: Ontario: Petawawa | P. banksiana |
| N. compar ON-2 | 241-02 | August 2002 | Canada: Ontario: SE of Kenora | P. banksiana |
| N. compar ON-3 | 262-02 | August 2002 | Canada: Ontario: Terrace Bay | P. banksiana |
| N. compar TN | 111-04A | July 2004 | USA: Tennessee: Cookeville | P. taeda |
| N. compar WI | 215-04 | August –2004 | USA: Wisconsin: Cedar | P. banksiana |
| N. dubiosus ME | 162-04 | July 2004 | USA: Maine: W of Lake Parlin | P. banksiana |
| N. dubiosus MN | 221-04 | August 2004 | USA: Minnesota: N of Cusson | P. banksiana |
| N. dubiosus ON-1 | 207-02 | August 2002 | Canada: Ontario: NW of Onaping | P. banksiana |
| N. dubiosus ON-2 | 232-02 | August 2002 | Canada: Ontario: W of Kaministiquia | P. banksiana |
| N. dubiosus ON-3 | 272-02 | August 2002 | Canada: Ontario: Hawk Junction | P. banksiana |
| N. dubiosus ON-4 | 330-02 | August 2002 | Canada: Ontario: W of Gowganda | P. banksiana |
| N. excitans FL-1 | 080-04 | July 2004 | USA: Florida: NW of Okahumpka | P. taeda |
| N. excitans FL-2 | 092-04 | July 2004 | USA: Florida: Bristol | P. glabra |
| N. excitans FL-3 | 094-04 | July 2004 | USA: Florida: Greensboro | P. taeda |
| N. excitans FL-4 | 163-03 | November 2003 | USA: Florida: Gainesville | P. taeda |
| N. excitans FL-5 | 165-03 | November 2003 | USA: Florida: Gainesville | P. glabra |
| N. excitans FL-6 | 175-03 | November 2003 | USA: Florida: NW of Cross City | P. taeda |
| N. excitans FL-7 | 179-03 | November 2003 | USA: Florida: NW of Okahumpka | P. taeda |
| N. excitans GA | 099-04 | July 2004 | USA: Georgia: S of Morgan | P. glabra |
| N. excitans NC-1 | 033-04 | May 2004 | USA: North Carolina: NW of Richfield | P. taeda |
| N. excitans NC-2 | 034-04 | May 2004 | USA: North Carolina: NW of Richfield | P. echinata |
| N. excitans NC-3 | 123-04 | July 2004 | USA: North Carolina: Kings Mountain | P. taeda |
| N. hetricki NC-1 | 046-04 | May 2004 | USA: North Carolina: W of Roanoke Rapids | P. taeda |
| N. hetricki NC-2 | 049-04 | May 2004 | USA: North Carolina: W of Roanoke Rapids | P. taeda |
| N. hetricki TN | 010-04 | April 2004 | USA: Tennessee: NW of Murfreesboro | P. taeda |
| N. hetricki VA | 057-04 | May 2004 | USA: Virginia: W of Petersburg | P. taeda |
| N. lecontei FL-1 | 174-03A | November 2003 | USA: Florida: NW of Cross City | P. taeda |
| N. lecontei FL-2 | 178-03 | November 2003 | USA: Florida: E of Crystal Lake | P. palustris |
| N. lecontei FL-3 | 185-03 | November 2003 | USA: Florida: Palmdale | P. elliottii |
| N. lecontei GA-1 | 096-04 | July 2004 | USA: Georgia: S of Morgan | P. taeda |
| N. lecontei GA-2 | 097-04 | July 2004 | USA: Georgia: S of Morgan | P. glabra |
| N. lecontei GA-3 | 102-04 | July 2004 | USA: Georgia: E of Albany | P. elliottii |
| N. lecontei MA | 372-02 | September 2002 | USA: Massachusetts: Myles Standish State Forest | P. rigida |
| N. lecontei MD | 132-04 | July 2004 | USA: Maryland: S of Easton | P. virginiana |
| N. lecontei NH-1 | 145-04 | July 2004 | USA: New Hampshire: West Ossippee | P. rigida |
| N. lecontei NH-2(a) | 125-02 | July 2002 | USA: New Hampshire: Nottingham | P. sylvestris |
| N. lecontei ON | 343-02 | August 2002 | Canada: Ontario: Redbridge | P. banksiana |
| N. lecontei TN | 116-04 | July 2004 | USA: Tennessee: NW of Crossville | P. virginiana |
| N. lecontei VT | 018-01 | August 2001 | USA: Vermont: Mallets Bay | P. resinosa |
| N. lecontei WI | 196-04 | August 2004 | USA: Wisconsin: W of Sparta | P. banksiana |
| N. maurus ON(b) | 035-0321B | July 2003 | Canada: Ontario: Kashabowie | P. banksiana |
| N. merkeli merkeli FL | 184-03.1 | November 2003 | USA: Florida: Palmdale | P. elliottii |
| N. nigroscutum ON-1 | 219-02 | August 2002 | Canada: Ontario: Kakabeka Falls | P. banksiana |
| N. nigroscutum ON-2 | 247-02 | August 2002 | Canada: Ontario: N of Sioux Narrows | P. banksiana |
| N. nigroscutum WI | 209-04 | August 2004 | USA: Wisconsin: Peeksville | P. banksiana |
| N. pinetum MA-1 | 377-02 | September 2002 | USA: Massachusetts: Myles Standish State Forest | P. rigida |
| N. pinetum MA-2 | 004-01 | July 2001 | USA: Massachusetts: Northborough | P. strobus |
| N. pinetum ME | 151-04 | July 2004 | USA: Maine: West Kennebunk | P. strobus |
| N. pinetum TN | 113-04 | July 2004 | USA: Tennessee: Crossville | P. strobus |
| N. pinetum VA | 130-04 | July 2004 | USA: Virginia: N of Bland | P. strobus |
| N. pinusrigidae MA-1 | 010-01 | July 2001 | USA: Massachusetts: N of Bourne | P. rigida |
| N. pinusrigidae MA-2 | 373-02 | September 2002 | USA: Massachusetts: Myles Standish State Forest | P. strobus |
| N. pinusrigidae MA-3 | 375-02 | September 2002 | USA: Massachusetts: Myles Standish State Forest | P. rigida |
| N. pinusrigidae ME | 148-04 | July 2004 | USA: Maine: Cornish | P. rigida |
| N. pinusrigidae NH | 142-04 | July 2004 | USA: New Hampshire: Effingham Falls | P. rigida |
| N. pratti banksianae ON-1 | 075-02 | June 2002 | Canada: Ontario: W of Nairn | P. banksiana |
| N. pratti banksianae ON-2(c) | NPtb | Canada: Ontario | – | |
| N. pratti paradoxicus ME-1 | 155-04 | July 2004 | USA: Maine: Great Wass Island | P. banksiana |
| N. pratti paradoxicus ME-2 | 156-04 | July 2004 | USA: Maine: Great Wass Island | P. banksiana |
| N. pratti paradoxicus ON-1 | 103-02 | June 2002 | Canada: Ontario: Petawawa | P. banksiana |
| N. pratti paradoxicus ON-2(b) | 120-02 | June 2002 | Canada: Ontario: Gallingertown | P. banksiana |
| N. pratti paradoxicus ON-3(b) | 133-02 | June 2002 | Canada: Ontario: Burrits Rapids | P. sylvestris |
| N. pratti pratti NC-1 | 027-04 | May 2004 | USA: North Carolina: Casar | P. virginiana |
| N. pratti pratti NC-2 | 032-04 | May 2004 | USA: North Carolina: W of Mt Pleasant | P. echinata |
| N. pratti pratti NC-3 | 043-04 | May 2004 | USA: North Carolina: Roanoke Rapids | P. virginiana |
| N. pratti pratti NC-4 | 044-04 | May 2004 | USA: North Carolina: Roanoke Rapids | P. taeda |
| N. pratti pratti NC-5 | 048-04 | May 2004 | USA: North Carolina: W of Roanoke Rapids | P. echinata |
| N. pratti pratti TN | 004-04 | April 2004 | USA: Tennessee: E of Lebanon | P. virginiana |
| N. pratti pratti VA | 054-04 | May 2004 | USA: Virginia: W of Petersburg | P. taeda |
| N. rugifrons MN | 222-04 | August 2004 | USA: Minnesota: N of Cusson | P. banksiana |
| N. rugifrons ON-1 | 155-02 | August 2002 | Canada: Ontario: Petawawa | P. banksiana |
| N. rugifrons ON-2 | 183-02 | August 2002 | Canada: Ontario: Britt | P. banksiana |
| N. rugifrons ON-3 | 184-02 | August 2002 | Canada: Ontario: Britt | P. banksiana |
| N. rugifrons ON-4 | 208-02 | August 2002 | Canada: Ontario: NW of Onaping | P. banksiana |
| N. rugifrons ON-5 | 231-02 | August 2002 | Canada: Ontario: W of Kaministiquia | P. banksiana |
| N. rugifrons ON-6 | 274-02 | August 2002 | Canada: Ontario: Hawk Junction | P. banksiana |
| N. rugifrons ON-7 | 316-02 | August 2002 | Canada: Ontario: W of Shining Tree | P. banksiana |
| N. rugifrons ON-8(b) | NRg1 | Canada | - | |
| N. rugifrons ON-9(b) | 035-0321A | July 2003 | Canada: Ontario: Kashabowie | P. banksiana |
| N. sp. AR | 002-04B | April 2004 | USA: Arkansas: NW of Artesian | P. taeda |
| N. sp. FL-1 | 091-04 | July 2004 | USA: Florida: S of Greensboro | P. taeda |
| N. sp. FL-2 | 095-04 | July 2004 | USA: Florida: Greensboro | P. taeda |
| N. sp. GA | 016-04 | April 2004 | USA: Georgia: S of Thomson | P. taeda |
| N. sp. NC | 037-04 | May 2004 | USA: North Carolina: N of New Bern | P. taeda |
| N. sp. TN-1 | 006-04 | April 2004 | USA: Tennessee: E of Lebanon | P. taeda |
| N. sp. TN-2 | 107-04C | July 2004 | USA: Tennessee: Murfreesboro | P. taeda |
| N. sp. TN-3 | 109-04 | July 2004 | USA: Tennessee: N of Murfreesboro | P. taeda |
| N. sp. TN-4 | 117-04 | July 2004 | USA: Tennessee: NW of Crossville | P. taeda |
| N. sp. TN-5 | 120-04.2 | July 2004 | USA: Tennessee W of Rockwood | P. echinata |
| N. sp. VA | 061-04 | May 2004 | USA: Virginia: Petersburg | P. taeda |
| N. taedae linearis AR | 002-04A | April 2004 | USA: Arkansas: NW of Artesian | P. taeda |
| N. taedae linearis LA(c) | NTdl | USA: Louisiana | ||
| N. taedae linearis TN | 011-04 | April 2004 | USA: Tennessee: NW of Murfreesboro | P. taeda |
| N. virginiana FL-1 | 081-04 | July 2004 | USA: Florida: Ocala National Forest | P. clausa |
| N. virginiana FL-2 | 082-04 | July 2004 | USA: Florida: Ocala National Forest | P. clausa |
| N. virginiana FL-3 | 090-04 | July 2004 | USA: Florida: N of Bronson | P. clausa |
| N. virginiana VA-1 | 126-04.1 | July 2004 | USA: Virginia: Blackstone | P. virginiana |
| N. virginiana VA-2 | 126-04.2 | July 2004 | USA: Virginia: Blackstone | P. virginiana |
| N. warreni FL-1 | 093-04 | July 2004 | USA: Florida: Bristol | P. glabra |
| N. warreni FL-2 | 168-03 | November 2003 | USA: Florida: Gainesville | P. glabra |
| N. warreni GA-1 | 098-04 | July 2004 | USA: Georgia: S of Morgan | P. glabra |
| N. warreni GA-2 | 100-04 | July 2004 | USA: Georgia: S of Morgan | P. taeda |
| N. swainei ON-1 | 257-02 | August 2002 | Canada: Ontario: W of Rainy Lake | P. banksiana |
| N. swainei ON-2 | 301-02 | August 2002 | Canada: Ontario: W of Mattagami | P. banksiana |
| N. swainei WI-1 | 179-04 | August 2004 | USA: Wisconsin: E of Mondovi | P. banksiana |
| N. swainei WI-2 | 180-04 | August 2004 | USA: Wisconsin: E of Mondovi | P. banksiana |
| N. swainei WI-3 | 197-04 | August 2004 | USA: Wisconsin: W of Sparta | P. banksiana |
| N. swainei WI-4 | 206-04 | August 2004 | USA: Wisconsin: Eagle's River | P. banksiana |
| N. swainei WI-5 | 208-04 | August 2004 | USA: Wisconsin: S of Butternut | P. banksiana |
| N. gillettei OG | 023-03 | April 2003 | USA: Arizona: W of Springerville | P. ponderosa |
| N. nearomosus OG(d) | 068-04C | Febuary 2004 | Mexico | P. michoacana |
| N. autumnalis OG | 044-03 | June 2003 | USA: Arizona: Flagstaff | P. ponderosa |
| N. nanulus nanulus OG | 009-04 | April 2004 | USA: Tennessee: W of Baxter | P. virginiana |
| N. nanulus nanulus 1 OG | 154-04 | July 2004 | USA: Maine: Great Wass Island | P. banksiana |
| N. sertifer OG | 069-02 | June 2002 | Canada: Ontario: E of Bancroft (introduced from Europe) | P. mugho |
| N. abietis OG(e) | 369-02 | July 2002 | Canada: Newfoundland: Corner Brook | Picea glauca |
| N. nanulus nanulus 2 OG | 088-02 | June 2002 | Canada: Ontario: NW of Onaping | P. banksiana |
| N. dailingensis OG(c) | 006-05 | China | ||
| N. scutellatus OG | 143-03 | July 2003 | USA: Washington: Acme | Pseudotsuga |
| menziesii | ||||
| Specimen1 | ID2 | Date | Location | Host3 |
| N. abbotii FL-1 | 001-04 | April 2004 | USA: Florida: E of Macclenny | P. palustris |
| N. abbotii FL-2 | 162-03 | November 2003 | USA: Florida: Gainesville | P. taeda |
| N. abbotii FL-3 | 184-03.2 | November 2003 | USA: Florida: Palmdale | P. elliottii |
| N. abbotii FL-4 | 188-03 | November 2003 | USA: Florida: Ocala National Forest | P. palustris |
| N. abbotii GA | 015-04 | April 2004 | USA: Georgia: Resaca | P. taeda |
| N. abbotii MD | 131-04 | July 2004 | USA: MD: N of Easton | P. taeda |
| N. abbotii ME | 014-01 | July 2001 | USA: Maine: Bar Harbor | P. resinosa |
| N. abbotii TN | 120-04.1 | July 2004 | USA: Tennessee: W of Rockwood | P. echinata |
| N. abbotii VA | 055-04 | May 2004 | USA: Virginia: W of Petersburg | P. taeda |
| N. compar FL-1 | 089-04 | July 2004 | USA: Florida: Gainesville | P. palustris |
| N. compar FL-2 | 161-03A | November 2003 | USA: Florida: Gainesville | P. palustris |
| N. compar GA | 104-04A | July 2004 | USA: Georgia: Sylvester | P. elliottii |
| N. compar MA-1 | 371-02 | September 2002 | USA: Massachusetts: Myles Standish State Forest | P. rigida |
| N. compar MA-2 | 378-02 | September 2002 | USA: Massachusetts: Myles Standish State Forest | P. rigida |
| N. compar ME | 163-04 | July 2004 | USA: Maine: W of Lake Parlin | P. banksiana |
| N. compar NC | 122-04A | July 2004 | USA: North Carolina: Kings Mountain | P. taeda |
| N. compar ON-1 | 148-02 | August 2002 | Canada: Ontario: Petawawa | P. banksiana |
| N. compar ON-2 | 241-02 | August 2002 | Canada: Ontario: SE of Kenora | P. banksiana |
| N. compar ON-3 | 262-02 | August 2002 | Canada: Ontario: Terrace Bay | P. banksiana |
| N. compar TN | 111-04A | July 2004 | USA: Tennessee: Cookeville | P. taeda |
| N. compar WI | 215-04 | August –2004 | USA: Wisconsin: Cedar | P. banksiana |
| N. dubiosus ME | 162-04 | July 2004 | USA: Maine: W of Lake Parlin | P. banksiana |
| N. dubiosus MN | 221-04 | August 2004 | USA: Minnesota: N of Cusson | P. banksiana |
| N. dubiosus ON-1 | 207-02 | August 2002 | Canada: Ontario: NW of Onaping | P. banksiana |
| N. dubiosus ON-2 | 232-02 | August 2002 | Canada: Ontario: W of Kaministiquia | P. banksiana |
| N. dubiosus ON-3 | 272-02 | August 2002 | Canada: Ontario: Hawk Junction | P. banksiana |
| N. dubiosus ON-4 | 330-02 | August 2002 | Canada: Ontario: W of Gowganda | P. banksiana |
| N. excitans FL-1 | 080-04 | July 2004 | USA: Florida: NW of Okahumpka | P. taeda |
| N. excitans FL-2 | 092-04 | July 2004 | USA: Florida: Bristol | P. glabra |
| N. excitans FL-3 | 094-04 | July 2004 | USA: Florida: Greensboro | P. taeda |
| N. excitans FL-4 | 163-03 | November 2003 | USA: Florida: Gainesville | P. taeda |
| N. excitans FL-5 | 165-03 | November 2003 | USA: Florida: Gainesville | P. glabra |
| N. excitans FL-6 | 175-03 | November 2003 | USA: Florida: NW of Cross City | P. taeda |
| N. excitans FL-7 | 179-03 | November 2003 | USA: Florida: NW of Okahumpka | P. taeda |
| N. excitans GA | 099-04 | July 2004 | USA: Georgia: S of Morgan | P. glabra |
| N. excitans NC-1 | 033-04 | May 2004 | USA: North Carolina: NW of Richfield | P. taeda |
| N. excitans NC-2 | 034-04 | May 2004 | USA: North Carolina: NW of Richfield | P. echinata |
| N. excitans NC-3 | 123-04 | July 2004 | USA: North Carolina: Kings Mountain | P. taeda |
| N. hetricki NC-1 | 046-04 | May 2004 | USA: North Carolina: W of Roanoke Rapids | P. taeda |
| N. hetricki NC-2 | 049-04 | May 2004 | USA: North Carolina: W of Roanoke Rapids | P. taeda |
| N. hetricki TN | 010-04 | April 2004 | USA: Tennessee: NW of Murfreesboro | P. taeda |
| N. hetricki VA | 057-04 | May 2004 | USA: Virginia: W of Petersburg | P. taeda |
| N. lecontei FL-1 | 174-03A | November 2003 | USA: Florida: NW of Cross City | P. taeda |
| N. lecontei FL-2 | 178-03 | November 2003 | USA: Florida: E of Crystal Lake | P. palustris |
| N. lecontei FL-3 | 185-03 | November 2003 | USA: Florida: Palmdale | P. elliottii |
| N. lecontei GA-1 | 096-04 | July 2004 | USA: Georgia: S of Morgan | P. taeda |
| N. lecontei GA-2 | 097-04 | July 2004 | USA: Georgia: S of Morgan | P. glabra |
| N. lecontei GA-3 | 102-04 | July 2004 | USA: Georgia: E of Albany | P. elliottii |
| N. lecontei MA | 372-02 | September 2002 | USA: Massachusetts: Myles Standish State Forest | P. rigida |
| N. lecontei MD | 132-04 | July 2004 | USA: Maryland: S of Easton | P. virginiana |
| N. lecontei NH-1 | 145-04 | July 2004 | USA: New Hampshire: West Ossippee | P. rigida |
| N. lecontei NH-2(a) | 125-02 | July 2002 | USA: New Hampshire: Nottingham | P. sylvestris |
| N. lecontei ON | 343-02 | August 2002 | Canada: Ontario: Redbridge | P. banksiana |
| N. lecontei TN | 116-04 | July 2004 | USA: Tennessee: NW of Crossville | P. virginiana |
| N. lecontei VT | 018-01 | August 2001 | USA: Vermont: Mallets Bay | P. resinosa |
| N. lecontei WI | 196-04 | August 2004 | USA: Wisconsin: W of Sparta | P. banksiana |
| N. maurus ON(b) | 035-0321B | July 2003 | Canada: Ontario: Kashabowie | P. banksiana |
| N. merkeli merkeli FL | 184-03.1 | November 2003 | USA: Florida: Palmdale | P. elliottii |
| N. nigroscutum ON-1 | 219-02 | August 2002 | Canada: Ontario: Kakabeka Falls | P. banksiana |
| N. nigroscutum ON-2 | 247-02 | August 2002 | Canada: Ontario: N of Sioux Narrows | P. banksiana |
| N. nigroscutum WI | 209-04 | August 2004 | USA: Wisconsin: Peeksville | P. banksiana |
| N. pinetum MA-1 | 377-02 | September 2002 | USA: Massachusetts: Myles Standish State Forest | P. rigida |
| N. pinetum MA-2 | 004-01 | July 2001 | USA: Massachusetts: Northborough | P. strobus |
| N. pinetum ME | 151-04 | July 2004 | USA: Maine: West Kennebunk | P. strobus |
| N. pinetum TN | 113-04 | July 2004 | USA: Tennessee: Crossville | P. strobus |
| N. pinetum VA | 130-04 | July 2004 | USA: Virginia: N of Bland | P. strobus |
| N. pinusrigidae MA-1 | 010-01 | July 2001 | USA: Massachusetts: N of Bourne | P. rigida |
| N. pinusrigidae MA-2 | 373-02 | September 2002 | USA: Massachusetts: Myles Standish State Forest | P. strobus |
| N. pinusrigidae MA-3 | 375-02 | September 2002 | USA: Massachusetts: Myles Standish State Forest | P. rigida |
| N. pinusrigidae ME | 148-04 | July 2004 | USA: Maine: Cornish | P. rigida |
| N. pinusrigidae NH | 142-04 | July 2004 | USA: New Hampshire: Effingham Falls | P. rigida |
| N. pratti banksianae ON-1 | 075-02 | June 2002 | Canada: Ontario: W of Nairn | P. banksiana |
| N. pratti banksianae ON-2(c) | NPtb | Canada: Ontario | – | |
| N. pratti paradoxicus ME-1 | 155-04 | July 2004 | USA: Maine: Great Wass Island | P. banksiana |
| N. pratti paradoxicus ME-2 | 156-04 | July 2004 | USA: Maine: Great Wass Island | P. banksiana |
| N. pratti paradoxicus ON-1 | 103-02 | June 2002 | Canada: Ontario: Petawawa | P. banksiana |
| N. pratti paradoxicus ON-2(b) | 120-02 | June 2002 | Canada: Ontario: Gallingertown | P. banksiana |
| N. pratti paradoxicus ON-3(b) | 133-02 | June 2002 | Canada: Ontario: Burrits Rapids | P. sylvestris |
| N. pratti pratti NC-1 | 027-04 | May 2004 | USA: North Carolina: Casar | P. virginiana |
| N. pratti pratti NC-2 | 032-04 | May 2004 | USA: North Carolina: W of Mt Pleasant | P. echinata |
| N. pratti pratti NC-3 | 043-04 | May 2004 | USA: North Carolina: Roanoke Rapids | P. virginiana |
| N. pratti pratti NC-4 | 044-04 | May 2004 | USA: North Carolina: Roanoke Rapids | P. taeda |
| N. pratti pratti NC-5 | 048-04 | May 2004 | USA: North Carolina: W of Roanoke Rapids | P. echinata |
| N. pratti pratti TN | 004-04 | April 2004 | USA: Tennessee: E of Lebanon | P. virginiana |
| N. pratti pratti VA | 054-04 | May 2004 | USA: Virginia: W of Petersburg | P. taeda |
| N. rugifrons MN | 222-04 | August 2004 | USA: Minnesota: N of Cusson | P. banksiana |
| N. rugifrons ON-1 | 155-02 | August 2002 | Canada: Ontario: Petawawa | P. banksiana |
| N. rugifrons ON-2 | 183-02 | August 2002 | Canada: Ontario: Britt | P. banksiana |
| N. rugifrons ON-3 | 184-02 | August 2002 | Canada: Ontario: Britt | P. banksiana |
| N. rugifrons ON-4 | 208-02 | August 2002 | Canada: Ontario: NW of Onaping | P. banksiana |
| N. rugifrons ON-5 | 231-02 | August 2002 | Canada: Ontario: W of Kaministiquia | P. banksiana |
| N. rugifrons ON-6 | 274-02 | August 2002 | Canada: Ontario: Hawk Junction | P. banksiana |
| N. rugifrons ON-7 | 316-02 | August 2002 | Canada: Ontario: W of Shining Tree | P. banksiana |
| N. rugifrons ON-8(b) | NRg1 | Canada | - | |
| N. rugifrons ON-9(b) | 035-0321A | July 2003 | Canada: Ontario: Kashabowie | P. banksiana |
| N. sp. AR | 002-04B | April 2004 | USA: Arkansas: NW of Artesian | P. taeda |
| N. sp. FL-1 | 091-04 | July 2004 | USA: Florida: S of Greensboro | P. taeda |
| N. sp. FL-2 | 095-04 | July 2004 | USA: Florida: Greensboro | P. taeda |
| N. sp. GA | 016-04 | April 2004 | USA: Georgia: S of Thomson | P. taeda |
| N. sp. NC | 037-04 | May 2004 | USA: North Carolina: N of New Bern | P. taeda |
| N. sp. TN-1 | 006-04 | April 2004 | USA: Tennessee: E of Lebanon | P. taeda |
| N. sp. TN-2 | 107-04C | July 2004 | USA: Tennessee: Murfreesboro | P. taeda |
| N. sp. TN-3 | 109-04 | July 2004 | USA: Tennessee: N of Murfreesboro | P. taeda |
| N. sp. TN-4 | 117-04 | July 2004 | USA: Tennessee: NW of Crossville | P. taeda |
| N. sp. TN-5 | 120-04.2 | July 2004 | USA: Tennessee W of Rockwood | P. echinata |
| N. sp. VA | 061-04 | May 2004 | USA: Virginia: Petersburg | P. taeda |
| N. taedae linearis AR | 002-04A | April 2004 | USA: Arkansas: NW of Artesian | P. taeda |
| N. taedae linearis LA(c) | NTdl | USA: Louisiana | ||
| N. taedae linearis TN | 011-04 | April 2004 | USA: Tennessee: NW of Murfreesboro | P. taeda |
| N. virginiana FL-1 | 081-04 | July 2004 | USA: Florida: Ocala National Forest | P. clausa |
| N. virginiana FL-2 | 082-04 | July 2004 | USA: Florida: Ocala National Forest | P. clausa |
| N. virginiana FL-3 | 090-04 | July 2004 | USA: Florida: N of Bronson | P. clausa |
| N. virginiana VA-1 | 126-04.1 | July 2004 | USA: Virginia: Blackstone | P. virginiana |
| N. virginiana VA-2 | 126-04.2 | July 2004 | USA: Virginia: Blackstone | P. virginiana |
| N. warreni FL-1 | 093-04 | July 2004 | USA: Florida: Bristol | P. glabra |
| N. warreni FL-2 | 168-03 | November 2003 | USA: Florida: Gainesville | P. glabra |
| N. warreni GA-1 | 098-04 | July 2004 | USA: Georgia: S of Morgan | P. glabra |
| N. warreni GA-2 | 100-04 | July 2004 | USA: Georgia: S of Morgan | P. taeda |
| N. swainei ON-1 | 257-02 | August 2002 | Canada: Ontario: W of Rainy Lake | P. banksiana |
| N. swainei ON-2 | 301-02 | August 2002 | Canada: Ontario: W of Mattagami | P. banksiana |
| N. swainei WI-1 | 179-04 | August 2004 | USA: Wisconsin: E of Mondovi | P. banksiana |
| N. swainei WI-2 | 180-04 | August 2004 | USA: Wisconsin: E of Mondovi | P. banksiana |
| N. swainei WI-3 | 197-04 | August 2004 | USA: Wisconsin: W of Sparta | P. banksiana |
| N. swainei WI-4 | 206-04 | August 2004 | USA: Wisconsin: Eagle's River | P. banksiana |
| N. swainei WI-5 | 208-04 | August 2004 | USA: Wisconsin: S of Butternut | P. banksiana |
| N. gillettei OG | 023-03 | April 2003 | USA: Arizona: W of Springerville | P. ponderosa |
| N. nearomosus OG(d) | 068-04C | Febuary 2004 | Mexico | P. michoacana |
| N. autumnalis OG | 044-03 | June 2003 | USA: Arizona: Flagstaff | P. ponderosa |
| N. nanulus nanulus OG | 009-04 | April 2004 | USA: Tennessee: W of Baxter | P. virginiana |
| N. nanulus nanulus 1 OG | 154-04 | July 2004 | USA: Maine: Great Wass Island | P. banksiana |
| N. sertifer OG | 069-02 | June 2002 | Canada: Ontario: E of Bancroft (introduced from Europe) | P. mugho |
| N. abietis OG(e) | 369-02 | July 2002 | Canada: Newfoundland: Corner Brook | Picea glauca |
| N. nanulus nanulus 2 OG | 088-02 | June 2002 | Canada: Ontario: NW of Onaping | P. banksiana |
| N. dailingensis OG(c) | 006-05 | China | ||
| N. scutellatus OG | 143-03 | July 2003 | USA: Washington: Acme | Pseudotsuga |
| menziesii | ||||
Specimens without superscripts were collected by C. Linnen; superscript letters correspond to the following collectors: (a) A. Sequiera (b) K. Nystrom (c) J. Rousselet (d) G. Sanchez-Martinez (e) S. Li.
Specimens with a superscript “C” were collected as cocoons; specimens with a superscript “A” were collected as adults.
A dash indicates that the host is unknown. With the exception of one specimen on Pseudotsuga and one specimen on Picea, all specimens were collected on hosts in the genus Pinus.
Collection data for Neodiprion specimens included in this study. For each specimen, an ID number is given to permit cross-reference with more detailed collection and rearing data, museum specimens, and future publications.
| Specimen1 | ID2 | Date | Location | Host3 |
| N. abbotii FL-1 | 001-04 | April 2004 | USA: Florida: E of Macclenny | P. palustris |
| N. abbotii FL-2 | 162-03 | November 2003 | USA: Florida: Gainesville | P. taeda |
| N. abbotii FL-3 | 184-03.2 | November 2003 | USA: Florida: Palmdale | P. elliottii |
| N. abbotii FL-4 | 188-03 | November 2003 | USA: Florida: Ocala National Forest | P. palustris |
| N. abbotii GA | 015-04 | April 2004 | USA: Georgia: Resaca | P. taeda |
| N. abbotii MD | 131-04 | July 2004 | USA: MD: N of Easton | P. taeda |
| N. abbotii ME | 014-01 | July 2001 | USA: Maine: Bar Harbor | P. resinosa |
| N. abbotii TN | 120-04.1 | July 2004 | USA: Tennessee: W of Rockwood | P. echinata |
| N. abbotii VA | 055-04 | May 2004 | USA: Virginia: W of Petersburg | P. taeda |
| N. compar FL-1 | 089-04 | July 2004 | USA: Florida: Gainesville | P. palustris |
| N. compar FL-2 | 161-03A | November 2003 | USA: Florida: Gainesville | P. palustris |
| N. compar GA | 104-04A | July 2004 | USA: Georgia: Sylvester | P. elliottii |
| N. compar MA-1 | 371-02 | September 2002 | USA: Massachusetts: Myles Standish State Forest | P. rigida |
| N. compar MA-2 | 378-02 | September 2002 | USA: Massachusetts: Myles Standish State Forest | P. rigida |
| N. compar ME | 163-04 | July 2004 | USA: Maine: W of Lake Parlin | P. banksiana |
| N. compar NC | 122-04A | July 2004 | USA: North Carolina: Kings Mountain | P. taeda |
| N. compar ON-1 | 148-02 | August 2002 | Canada: Ontario: Petawawa | P. banksiana |
| N. compar ON-2 | 241-02 | August 2002 | Canada: Ontario: SE of Kenora | P. banksiana |
| N. compar ON-3 | 262-02 | August 2002 | Canada: Ontario: Terrace Bay | P. banksiana |
| N. compar TN | 111-04A | July 2004 | USA: Tennessee: Cookeville | P. taeda |
| N. compar WI | 215-04 | August –2004 | USA: Wisconsin: Cedar | P. banksiana |
| N. dubiosus ME | 162-04 | July 2004 | USA: Maine: W of Lake Parlin | P. banksiana |
| N. dubiosus MN | 221-04 | August 2004 | USA: Minnesota: N of Cusson | P. banksiana |
| N. dubiosus ON-1 | 207-02 | August 2002 | Canada: Ontario: NW of Onaping | P. banksiana |
| N. dubiosus ON-2 | 232-02 | August 2002 | Canada: Ontario: W of Kaministiquia | P. banksiana |
| N. dubiosus ON-3 | 272-02 | August 2002 | Canada: Ontario: Hawk Junction | P. banksiana |
| N. dubiosus ON-4 | 330-02 | August 2002 | Canada: Ontario: W of Gowganda | P. banksiana |
| N. excitans FL-1 | 080-04 | July 2004 | USA: Florida: NW of Okahumpka | P. taeda |
| N. excitans FL-2 | 092-04 | July 2004 | USA: Florida: Bristol | P. glabra |
| N. excitans FL-3 | 094-04 | July 2004 | USA: Florida: Greensboro | P. taeda |
| N. excitans FL-4 | 163-03 | November 2003 | USA: Florida: Gainesville | P. taeda |
| N. excitans FL-5 | 165-03 | November 2003 | USA: Florida: Gainesville | P. glabra |
| N. excitans FL-6 | 175-03 | November 2003 | USA: Florida: NW of Cross City | P. taeda |
| N. excitans FL-7 | 179-03 | November 2003 | USA: Florida: NW of Okahumpka | P. taeda |
| N. excitans GA | 099-04 | July 2004 | USA: Georgia: S of Morgan | P. glabra |
| N. excitans NC-1 | 033-04 | May 2004 | USA: North Carolina: NW of Richfield | P. taeda |
| N. excitans NC-2 | 034-04 | May 2004 | USA: North Carolina: NW of Richfield | P. echinata |
| N. excitans NC-3 | 123-04 | July 2004 | USA: North Carolina: Kings Mountain | P. taeda |
| N. hetricki NC-1 | 046-04 | May 2004 | USA: North Carolina: W of Roanoke Rapids | P. taeda |
| N. hetricki NC-2 | 049-04 | May 2004 | USA: North Carolina: W of Roanoke Rapids | P. taeda |
| N. hetricki TN | 010-04 | April 2004 | USA: Tennessee: NW of Murfreesboro | P. taeda |
| N. hetricki VA | 057-04 | May 2004 | USA: Virginia: W of Petersburg | P. taeda |
| N. lecontei FL-1 | 174-03A | November 2003 | USA: Florida: NW of Cross City | P. taeda |
| N. lecontei FL-2 | 178-03 | November 2003 | USA: Florida: E of Crystal Lake | P. palustris |
| N. lecontei FL-3 | 185-03 | November 2003 | USA: Florida: Palmdale | P. elliottii |
| N. lecontei GA-1 | 096-04 | July 2004 | USA: Georgia: S of Morgan | P. taeda |
| N. lecontei GA-2 | 097-04 | July 2004 | USA: Georgia: S of Morgan | P. glabra |
| N. lecontei GA-3 | 102-04 | July 2004 | USA: Georgia: E of Albany | P. elliottii |
| N. lecontei MA | 372-02 | September 2002 | USA: Massachusetts: Myles Standish State Forest | P. rigida |
| N. lecontei MD | 132-04 | July 2004 | USA: Maryland: S of Easton | P. virginiana |
| N. lecontei NH-1 | 145-04 | July 2004 | USA: New Hampshire: West Ossippee | P. rigida |
| N. lecontei NH-2(a) | 125-02 | July 2002 | USA: New Hampshire: Nottingham | P. sylvestris |
| N. lecontei ON | 343-02 | August 2002 | Canada: Ontario: Redbridge | P. banksiana |
| N. lecontei TN | 116-04 | July 2004 | USA: Tennessee: NW of Crossville | P. virginiana |
| N. lecontei VT | 018-01 | August 2001 | USA: Vermont: Mallets Bay | P. resinosa |
| N. lecontei WI | 196-04 | August 2004 | USA: Wisconsin: W of Sparta | P. banksiana |
| N. maurus ON(b) | 035-0321B | July 2003 | Canada: Ontario: Kashabowie | P. banksiana |
| N. merkeli merkeli FL | 184-03.1 | November 2003 | USA: Florida: Palmdale | P. elliottii |
| N. nigroscutum ON-1 | 219-02 | August 2002 | Canada: Ontario: Kakabeka Falls | P. banksiana |
| N. nigroscutum ON-2 | 247-02 | August 2002 | Canada: Ontario: N of Sioux Narrows | P. banksiana |
| N. nigroscutum WI | 209-04 | August 2004 | USA: Wisconsin: Peeksville | P. banksiana |
| N. pinetum MA-1 | 377-02 | September 2002 | USA: Massachusetts: Myles Standish State Forest | P. rigida |
| N. pinetum MA-2 | 004-01 | July 2001 | USA: Massachusetts: Northborough | P. strobus |
| N. pinetum ME | 151-04 | July 2004 | USA: Maine: West Kennebunk | P. strobus |
| N. pinetum TN | 113-04 | July 2004 | USA: Tennessee: Crossville | P. strobus |
| N. pinetum VA | 130-04 | July 2004 | USA: Virginia: N of Bland | P. strobus |
| N. pinusrigidae MA-1 | 010-01 | July 2001 | USA: Massachusetts: N of Bourne | P. rigida |
| N. pinusrigidae MA-2 | 373-02 | September 2002 | USA: Massachusetts: Myles Standish State Forest | P. strobus |
| N. pinusrigidae MA-3 | 375-02 | September 2002 | USA: Massachusetts: Myles Standish State Forest | P. rigida |
| N. pinusrigidae ME | 148-04 | July 2004 | USA: Maine: Cornish | P. rigida |
| N. pinusrigidae NH | 142-04 | July 2004 | USA: New Hampshire: Effingham Falls | P. rigida |
| N. pratti banksianae ON-1 | 075-02 | June 2002 | Canada: Ontario: W of Nairn | P. banksiana |
| N. pratti banksianae ON-2(c) | NPtb | Canada: Ontario | – | |
| N. pratti paradoxicus ME-1 | 155-04 | July 2004 | USA: Maine: Great Wass Island | P. banksiana |
| N. pratti paradoxicus ME-2 | 156-04 | July 2004 | USA: Maine: Great Wass Island | P. banksiana |
| N. pratti paradoxicus ON-1 | 103-02 | June 2002 | Canada: Ontario: Petawawa | P. banksiana |
| N. pratti paradoxicus ON-2(b) | 120-02 | June 2002 | Canada: Ontario: Gallingertown | P. banksiana |
| N. pratti paradoxicus ON-3(b) | 133-02 | June 2002 | Canada: Ontario: Burrits Rapids | P. sylvestris |
| N. pratti pratti NC-1 | 027-04 | May 2004 | USA: North Carolina: Casar | P. virginiana |
| N. pratti pratti NC-2 | 032-04 | May 2004 | USA: North Carolina: W of Mt Pleasant | P. echinata |
| N. pratti pratti NC-3 | 043-04 | May 2004 | USA: North Carolina: Roanoke Rapids | P. virginiana |
| N. pratti pratti NC-4 | 044-04 | May 2004 | USA: North Carolina: Roanoke Rapids | P. taeda |
| N. pratti pratti NC-5 | 048-04 | May 2004 | USA: North Carolina: W of Roanoke Rapids | P. echinata |
| N. pratti pratti TN | 004-04 | April 2004 | USA: Tennessee: E of Lebanon | P. virginiana |
| N. pratti pratti VA | 054-04 | May 2004 | USA: Virginia: W of Petersburg | P. taeda |
| N. rugifrons MN | 222-04 | August 2004 | USA: Minnesota: N of Cusson | P. banksiana |
| N. rugifrons ON-1 | 155-02 | August 2002 | Canada: Ontario: Petawawa | P. banksiana |
| N. rugifrons ON-2 | 183-02 | August 2002 | Canada: Ontario: Britt | P. banksiana |
| N. rugifrons ON-3 | 184-02 | August 2002 | Canada: Ontario: Britt | P. banksiana |
| N. rugifrons ON-4 | 208-02 | August 2002 | Canada: Ontario: NW of Onaping | P. banksiana |
| N. rugifrons ON-5 | 231-02 | August 2002 | Canada: Ontario: W of Kaministiquia | P. banksiana |
| N. rugifrons ON-6 | 274-02 | August 2002 | Canada: Ontario: Hawk Junction | P. banksiana |
| N. rugifrons ON-7 | 316-02 | August 2002 | Canada: Ontario: W of Shining Tree | P. banksiana |
| N. rugifrons ON-8(b) | NRg1 | Canada | - | |
| N. rugifrons ON-9(b) | 035-0321A | July 2003 | Canada: Ontario: Kashabowie | P. banksiana |
| N. sp. AR | 002-04B | April 2004 | USA: Arkansas: NW of Artesian | P. taeda |
| N. sp. FL-1 | 091-04 | July 2004 | USA: Florida: S of Greensboro | P. taeda |
| N. sp. FL-2 | 095-04 | July 2004 | USA: Florida: Greensboro | P. taeda |
| N. sp. GA | 016-04 | April 2004 | USA: Georgia: S of Thomson | P. taeda |
| N. sp. NC | 037-04 | May 2004 | USA: North Carolina: N of New Bern | P. taeda |
| N. sp. TN-1 | 006-04 | April 2004 | USA: Tennessee: E of Lebanon | P. taeda |
| N. sp. TN-2 | 107-04C | July 2004 | USA: Tennessee: Murfreesboro | P. taeda |
| N. sp. TN-3 | 109-04 | July 2004 | USA: Tennessee: N of Murfreesboro | P. taeda |
| N. sp. TN-4 | 117-04 | July 2004 | USA: Tennessee: NW of Crossville | P. taeda |
| N. sp. TN-5 | 120-04.2 | July 2004 | USA: Tennessee W of Rockwood | P. echinata |
| N. sp. VA | 061-04 | May 2004 | USA: Virginia: Petersburg | P. taeda |
| N. taedae linearis AR | 002-04A | April 2004 | USA: Arkansas: NW of Artesian | P. taeda |
| N. taedae linearis LA(c) | NTdl | USA: Louisiana | ||
| N. taedae linearis TN | 011-04 | April 2004 | USA: Tennessee: NW of Murfreesboro | P. taeda |
| N. virginiana FL-1 | 081-04 | July 2004 | USA: Florida: Ocala National Forest | P. clausa |
| N. virginiana FL-2 | 082-04 | July 2004 | USA: Florida: Ocala National Forest | P. clausa |
| N. virginiana FL-3 | 090-04 | July 2004 | USA: Florida: N of Bronson | P. clausa |
| N. virginiana VA-1 | 126-04.1 | July 2004 | USA: Virginia: Blackstone | P. virginiana |
| N. virginiana VA-2 | 126-04.2 | July 2004 | USA: Virginia: Blackstone | P. virginiana |
| N. warreni FL-1 | 093-04 | July 2004 | USA: Florida: Bristol | P. glabra |
| N. warreni FL-2 | 168-03 | November 2003 | USA: Florida: Gainesville | P. glabra |
| N. warreni GA-1 | 098-04 | July 2004 | USA: Georgia: S of Morgan | P. glabra |
| N. warreni GA-2 | 100-04 | July 2004 | USA: Georgia: S of Morgan | P. taeda |
| N. swainei ON-1 | 257-02 | August 2002 | Canada: Ontario: W of Rainy Lake | P. banksiana |
| N. swainei ON-2 | 301-02 | August 2002 | Canada: Ontario: W of Mattagami | P. banksiana |
| N. swainei WI-1 | 179-04 | August 2004 | USA: Wisconsin: E of Mondovi | P. banksiana |
| N. swainei WI-2 | 180-04 | August 2004 | USA: Wisconsin: E of Mondovi | P. banksiana |
| N. swainei WI-3 | 197-04 | August 2004 | USA: Wisconsin: W of Sparta | P. banksiana |
| N. swainei WI-4 | 206-04 | August 2004 | USA: Wisconsin: Eagle's River | P. banksiana |
| N. swainei WI-5 | 208-04 | August 2004 | USA: Wisconsin: S of Butternut | P. banksiana |
| N. gillettei OG | 023-03 | April 2003 | USA: Arizona: W of Springerville | P. ponderosa |
| N. nearomosus OG(d) | 068-04C | Febuary 2004 | Mexico | P. michoacana |
| N. autumnalis OG | 044-03 | June 2003 | USA: Arizona: Flagstaff | P. ponderosa |
| N. nanulus nanulus OG | 009-04 | April 2004 | USA: Tennessee: W of Baxter | P. virginiana |
| N. nanulus nanulus 1 OG | 154-04 | July 2004 | USA: Maine: Great Wass Island | P. banksiana |
| N. sertifer OG | 069-02 | June 2002 | Canada: Ontario: E of Bancroft (introduced from Europe) | P. mugho |
| N. abietis OG(e) | 369-02 | July 2002 | Canada: Newfoundland: Corner Brook | Picea glauca |
| N. nanulus nanulus 2 OG | 088-02 | June 2002 | Canada: Ontario: NW of Onaping | P. banksiana |
| N. dailingensis OG(c) | 006-05 | China | ||
| N. scutellatus OG | 143-03 | July 2003 | USA: Washington: Acme | Pseudotsuga |
| menziesii | ||||
| Specimen1 | ID2 | Date | Location | Host3 |
| N. abbotii FL-1 | 001-04 | April 2004 | USA: Florida: E of Macclenny | P. palustris |
| N. abbotii FL-2 | 162-03 | November 2003 | USA: Florida: Gainesville | P. taeda |
| N. abbotii FL-3 | 184-03.2 | November 2003 | USA: Florida: Palmdale | P. elliottii |
| N. abbotii FL-4 | 188-03 | November 2003 | USA: Florida: Ocala National Forest | P. palustris |
| N. abbotii GA | 015-04 | April 2004 | USA: Georgia: Resaca | P. taeda |
| N. abbotii MD | 131-04 | July 2004 | USA: MD: N of Easton | P. taeda |
| N. abbotii ME | 014-01 | July 2001 | USA: Maine: Bar Harbor | P. resinosa |
| N. abbotii TN | 120-04.1 | July 2004 | USA: Tennessee: W of Rockwood | P. echinata |
| N. abbotii VA | 055-04 | May 2004 | USA: Virginia: W of Petersburg | P. taeda |
| N. compar FL-1 | 089-04 | July 2004 | USA: Florida: Gainesville | P. palustris |
| N. compar FL-2 | 161-03A | November 2003 | USA: Florida: Gainesville | P. palustris |
| N. compar GA | 104-04A | July 2004 | USA: Georgia: Sylvester | P. elliottii |
| N. compar MA-1 | 371-02 | September 2002 | USA: Massachusetts: Myles Standish State Forest | P. rigida |
| N. compar MA-2 | 378-02 | September 2002 | USA: Massachusetts: Myles Standish State Forest | P. rigida |
| N. compar ME | 163-04 | July 2004 | USA: Maine: W of Lake Parlin | P. banksiana |
| N. compar NC | 122-04A | July 2004 | USA: North Carolina: Kings Mountain | P. taeda |
| N. compar ON-1 | 148-02 | August 2002 | Canada: Ontario: Petawawa | P. banksiana |
| N. compar ON-2 | 241-02 | August 2002 | Canada: Ontario: SE of Kenora | P. banksiana |
| N. compar ON-3 | 262-02 | August 2002 | Canada: Ontario: Terrace Bay | P. banksiana |
| N. compar TN | 111-04A | July 2004 | USA: Tennessee: Cookeville | P. taeda |
| N. compar WI | 215-04 | August –2004 | USA: Wisconsin: Cedar | P. banksiana |
| N. dubiosus ME | 162-04 | July 2004 | USA: Maine: W of Lake Parlin | P. banksiana |
| N. dubiosus MN | 221-04 | August 2004 | USA: Minnesota: N of Cusson | P. banksiana |
| N. dubiosus ON-1 | 207-02 | August 2002 | Canada: Ontario: NW of Onaping | P. banksiana |
| N. dubiosus ON-2 | 232-02 | August 2002 | Canada: Ontario: W of Kaministiquia | P. banksiana |
| N. dubiosus ON-3 | 272-02 | August 2002 | Canada: Ontario: Hawk Junction | P. banksiana |
| N. dubiosus ON-4 | 330-02 | August 2002 | Canada: Ontario: W of Gowganda | P. banksiana |
| N. excitans FL-1 | 080-04 | July 2004 | USA: Florida: NW of Okahumpka | P. taeda |
| N. excitans FL-2 | 092-04 | July 2004 | USA: Florida: Bristol | P. glabra |
| N. excitans FL-3 | 094-04 | July 2004 | USA: Florida: Greensboro | P. taeda |
| N. excitans FL-4 | 163-03 | November 2003 | USA: Florida: Gainesville | P. taeda |
| N. excitans FL-5 | 165-03 | November 2003 | USA: Florida: Gainesville | P. glabra |
| N. excitans FL-6 | 175-03 | November 2003 | USA: Florida: NW of Cross City | P. taeda |
| N. excitans FL-7 | 179-03 | November 2003 | USA: Florida: NW of Okahumpka | P. taeda |
| N. excitans GA | 099-04 | July 2004 | USA: Georgia: S of Morgan | P. glabra |
| N. excitans NC-1 | 033-04 | May 2004 | USA: North Carolina: NW of Richfield | P. taeda |
| N. excitans NC-2 | 034-04 | May 2004 | USA: North Carolina: NW of Richfield | P. echinata |
| N. excitans NC-3 | 123-04 | July 2004 | USA: North Carolina: Kings Mountain | P. taeda |
| N. hetricki NC-1 | 046-04 | May 2004 | USA: North Carolina: W of Roanoke Rapids | P. taeda |
| N. hetricki NC-2 | 049-04 | May 2004 | USA: North Carolina: W of Roanoke Rapids | P. taeda |
| N. hetricki TN | 010-04 | April 2004 | USA: Tennessee: NW of Murfreesboro | P. taeda |
| N. hetricki VA | 057-04 | May 2004 | USA: Virginia: W of Petersburg | P. taeda |
| N. lecontei FL-1 | 174-03A | November 2003 | USA: Florida: NW of Cross City | P. taeda |
| N. lecontei FL-2 | 178-03 | November 2003 | USA: Florida: E of Crystal Lake | P. palustris |
| N. lecontei FL-3 | 185-03 | November 2003 | USA: Florida: Palmdale | P. elliottii |
| N. lecontei GA-1 | 096-04 | July 2004 | USA: Georgia: S of Morgan | P. taeda |
| N. lecontei GA-2 | 097-04 | July 2004 | USA: Georgia: S of Morgan | P. glabra |
| N. lecontei GA-3 | 102-04 | July 2004 | USA: Georgia: E of Albany | P. elliottii |
| N. lecontei MA | 372-02 | September 2002 | USA: Massachusetts: Myles Standish State Forest | P. rigida |
| N. lecontei MD | 132-04 | July 2004 | USA: Maryland: S of Easton | P. virginiana |
| N. lecontei NH-1 | 145-04 | July 2004 | USA: New Hampshire: West Ossippee | P. rigida |
| N. lecontei NH-2(a) | 125-02 | July 2002 | USA: New Hampshire: Nottingham | P. sylvestris |
| N. lecontei ON | 343-02 | August 2002 | Canada: Ontario: Redbridge | P. banksiana |
| N. lecontei TN | 116-04 | July 2004 | USA: Tennessee: NW of Crossville | P. virginiana |
| N. lecontei VT | 018-01 | August 2001 | USA: Vermont: Mallets Bay | P. resinosa |
| N. lecontei WI | 196-04 | August 2004 | USA: Wisconsin: W of Sparta | P. banksiana |
| N. maurus ON(b) | 035-0321B | July 2003 | Canada: Ontario: Kashabowie | P. banksiana |
| N. merkeli merkeli FL | 184-03.1 | November 2003 | USA: Florida: Palmdale | P. elliottii |
| N. nigroscutum ON-1 | 219-02 | August 2002 | Canada: Ontario: Kakabeka Falls | P. banksiana |
| N. nigroscutum ON-2 | 247-02 | August 2002 | Canada: Ontario: N of Sioux Narrows | P. banksiana |
| N. nigroscutum WI | 209-04 | August 2004 | USA: Wisconsin: Peeksville | P. banksiana |
| N. pinetum MA-1 | 377-02 | September 2002 | USA: Massachusetts: Myles Standish State Forest | P. rigida |
| N. pinetum MA-2 | 004-01 | July 2001 | USA: Massachusetts: Northborough | P. strobus |
| N. pinetum ME | 151-04 | July 2004 | USA: Maine: West Kennebunk | P. strobus |
| N. pinetum TN | 113-04 | July 2004 | USA: Tennessee: Crossville | P. strobus |
| N. pinetum VA | 130-04 | July 2004 | USA: Virginia: N of Bland | P. strobus |
| N. pinusrigidae MA-1 | 010-01 | July 2001 | USA: Massachusetts: N of Bourne | P. rigida |
| N. pinusrigidae MA-2 | 373-02 | September 2002 | USA: Massachusetts: Myles Standish State Forest | P. strobus |
| N. pinusrigidae MA-3 | 375-02 | September 2002 | USA: Massachusetts: Myles Standish State Forest | P. rigida |
| N. pinusrigidae ME | 148-04 | July 2004 | USA: Maine: Cornish | P. rigida |
| N. pinusrigidae NH | 142-04 | July 2004 | USA: New Hampshire: Effingham Falls | P. rigida |
| N. pratti banksianae ON-1 | 075-02 | June 2002 | Canada: Ontario: W of Nairn | P. banksiana |
| N. pratti banksianae ON-2(c) | NPtb | Canada: Ontario | – | |
| N. pratti paradoxicus ME-1 | 155-04 | July 2004 | USA: Maine: Great Wass Island | P. banksiana |
| N. pratti paradoxicus ME-2 | 156-04 | July 2004 | USA: Maine: Great Wass Island | P. banksiana |
| N. pratti paradoxicus ON-1 | 103-02 | June 2002 | Canada: Ontario: Petawawa | P. banksiana |
| N. pratti paradoxicus ON-2(b) | 120-02 | June 2002 | Canada: Ontario: Gallingertown | P. banksiana |
| N. pratti paradoxicus ON-3(b) | 133-02 | June 2002 | Canada: Ontario: Burrits Rapids | P. sylvestris |
| N. pratti pratti NC-1 | 027-04 | May 2004 | USA: North Carolina: Casar | P. virginiana |
| N. pratti pratti NC-2 | 032-04 | May 2004 | USA: North Carolina: W of Mt Pleasant | P. echinata |
| N. pratti pratti NC-3 | 043-04 | May 2004 | USA: North Carolina: Roanoke Rapids | P. virginiana |
| N. pratti pratti NC-4 | 044-04 | May 2004 | USA: North Carolina: Roanoke Rapids | P. taeda |
| N. pratti pratti NC-5 | 048-04 | May 2004 | USA: North Carolina: W of Roanoke Rapids | P. echinata |
| N. pratti pratti TN | 004-04 | April 2004 | USA: Tennessee: E of Lebanon | P. virginiana |
| N. pratti pratti VA | 054-04 | May 2004 | USA: Virginia: W of Petersburg | P. taeda |
| N. rugifrons MN | 222-04 | August 2004 | USA: Minnesota: N of Cusson | P. banksiana |
| N. rugifrons ON-1 | 155-02 | August 2002 | Canada: Ontario: Petawawa | P. banksiana |
| N. rugifrons ON-2 | 183-02 | August 2002 | Canada: Ontario: Britt | P. banksiana |
| N. rugifrons ON-3 | 184-02 | August 2002 | Canada: Ontario: Britt | P. banksiana |
| N. rugifrons ON-4 | 208-02 | August 2002 | Canada: Ontario: NW of Onaping | P. banksiana |
| N. rugifrons ON-5 | 231-02 | August 2002 | Canada: Ontario: W of Kaministiquia | P. banksiana |
| N. rugifrons ON-6 | 274-02 | August 2002 | Canada: Ontario: Hawk Junction | P. banksiana |
| N. rugifrons ON-7 | 316-02 | August 2002 | Canada: Ontario: W of Shining Tree | P. banksiana |
| N. rugifrons ON-8(b) | NRg1 | Canada | - | |
| N. rugifrons ON-9(b) | 035-0321A | July 2003 | Canada: Ontario: Kashabowie | P. banksiana |
| N. sp. AR | 002-04B | April 2004 | USA: Arkansas: NW of Artesian | P. taeda |
| N. sp. FL-1 | 091-04 | July 2004 | USA: Florida: S of Greensboro | P. taeda |
| N. sp. FL-2 | 095-04 | July 2004 | USA: Florida: Greensboro | P. taeda |
| N. sp. GA | 016-04 | April 2004 | USA: Georgia: S of Thomson | P. taeda |
| N. sp. NC | 037-04 | May 2004 | USA: North Carolina: N of New Bern | P. taeda |
| N. sp. TN-1 | 006-04 | April 2004 | USA: Tennessee: E of Lebanon | P. taeda |
| N. sp. TN-2 | 107-04C | July 2004 | USA: Tennessee: Murfreesboro | P. taeda |
| N. sp. TN-3 | 109-04 | July 2004 | USA: Tennessee: N of Murfreesboro | P. taeda |
| N. sp. TN-4 | 117-04 | July 2004 | USA: Tennessee: NW of Crossville | P. taeda |
| N. sp. TN-5 | 120-04.2 | July 2004 | USA: Tennessee W of Rockwood | P. echinata |
| N. sp. VA | 061-04 | May 2004 | USA: Virginia: Petersburg | P. taeda |
| N. taedae linearis AR | 002-04A | April 2004 | USA: Arkansas: NW of Artesian | P. taeda |
| N. taedae linearis LA(c) | NTdl | USA: Louisiana | ||
| N. taedae linearis TN | 011-04 | April 2004 | USA: Tennessee: NW of Murfreesboro | P. taeda |
| N. virginiana FL-1 | 081-04 | July 2004 | USA: Florida: Ocala National Forest | P. clausa |
| N. virginiana FL-2 | 082-04 | July 2004 | USA: Florida: Ocala National Forest | P. clausa |
| N. virginiana FL-3 | 090-04 | July 2004 | USA: Florida: N of Bronson | P. clausa |
| N. virginiana VA-1 | 126-04.1 | July 2004 | USA: Virginia: Blackstone | P. virginiana |
| N. virginiana VA-2 | 126-04.2 | July 2004 | USA: Virginia: Blackstone | P. virginiana |
| N. warreni FL-1 | 093-04 | July 2004 | USA: Florida: Bristol | P. glabra |
| N. warreni FL-2 | 168-03 | November 2003 | USA: Florida: Gainesville | P. glabra |
| N. warreni GA-1 | 098-04 | July 2004 | USA: Georgia: S of Morgan | P. glabra |
| N. warreni GA-2 | 100-04 | July 2004 | USA: Georgia: S of Morgan | P. taeda |
| N. swainei ON-1 | 257-02 | August 2002 | Canada: Ontario: W of Rainy Lake | P. banksiana |
| N. swainei ON-2 | 301-02 | August 2002 | Canada: Ontario: W of Mattagami | P. banksiana |
| N. swainei WI-1 | 179-04 | August 2004 | USA: Wisconsin: E of Mondovi | P. banksiana |
| N. swainei WI-2 | 180-04 | August 2004 | USA: Wisconsin: E of Mondovi | P. banksiana |
| N. swainei WI-3 | 197-04 | August 2004 | USA: Wisconsin: W of Sparta | P. banksiana |
| N. swainei WI-4 | 206-04 | August 2004 | USA: Wisconsin: Eagle's River | P. banksiana |
| N. swainei WI-5 | 208-04 | August 2004 | USA: Wisconsin: S of Butternut | P. banksiana |
| N. gillettei OG | 023-03 | April 2003 | USA: Arizona: W of Springerville | P. ponderosa |
| N. nearomosus OG(d) | 068-04C | Febuary 2004 | Mexico | P. michoacana |
| N. autumnalis OG | 044-03 | June 2003 | USA: Arizona: Flagstaff | P. ponderosa |
| N. nanulus nanulus OG | 009-04 | April 2004 | USA: Tennessee: W of Baxter | P. virginiana |
| N. nanulus nanulus 1 OG | 154-04 | July 2004 | USA: Maine: Great Wass Island | P. banksiana |
| N. sertifer OG | 069-02 | June 2002 | Canada: Ontario: E of Bancroft (introduced from Europe) | P. mugho |
| N. abietis OG(e) | 369-02 | July 2002 | Canada: Newfoundland: Corner Brook | Picea glauca |
| N. nanulus nanulus 2 OG | 088-02 | June 2002 | Canada: Ontario: NW of Onaping | P. banksiana |
| N. dailingensis OG(c) | 006-05 | China | ||
| N. scutellatus OG | 143-03 | July 2003 | USA: Washington: Acme | Pseudotsuga |
| menziesii | ||||
Specimens without superscripts were collected by C. Linnen; superscript letters correspond to the following collectors: (a) A. Sequiera (b) K. Nystrom (c) J. Rousselet (d) G. Sanchez-Martinez (e) S. Li.
Specimens with a superscript “C” were collected as cocoons; specimens with a superscript “A” were collected as adults.
A dash indicates that the host is unknown. With the exception of one specimen on Pseudotsuga and one specimen on Picea, all specimens were collected on hosts in the genus Pinus.