-
PDF
- Split View
-
Views
-
Cite
Cite
Shinichi Takaichi, Mari Mochimaru, Takashi Maoka, Presence of Free Myxol and 4-Hydroxymyxol and Absence of Myxol Glycosides in Anabaena variabilis ATCC 29413, and Proposal of a Biosynthetic Pathway of Carotenoids, Plant and Cell Physiology, Volume 47, Issue 2, February 2006, Pages 211–216, https://doi.org/10.1093/pcp/pci236
Close - Share Icon Share
Abstract
We identified the molecular structures of all carotenoids in Anabaena variabilis ATCC 29413 (= IAM M-204). The major carotenoids were β-carotene, echinenone and canthaxanthin. Myxol glycosides were absent, while free forms of myxol and 4-hydroxymyxol were present. The 4-hydroxyl group of the latter was a mixture of (4R) and (4S) configurations, which is a rare mixture in carotenoids. Thus, this strain was the first cyanobacterium found to have free myxol and not myxol glycosides, and seemed to lack the gene for or activity of glycosyl transferase. In another strain of A. variabilis IAM M-3 (= PCC 7118), we recently identified (3R,2′S)-myxol 2′-fucoside and (3S,2′S)-4-ketomyxol 2′-fucoside, and hence the strain ATCC 29413 might be useful for investigating the characteristics of myxol glycosides in cyanobacteria. Based on the identification of the carotenoids and the completion of the entire nucleotide sequence of the genome in A. variabilis ATCC 29413, we proposed a biosynthetic pathway of the carotenoids and the corresponding genes and enzymes. The homologous genes were searched by sequence homology only from the functionally confirmed genes.
Introduction
Anabaena variabilis ATCC 29413 (= IAM M-204) is a filamentous and heterocystous cyanobacterium, that has been studied for >40 years, and the sequencing of its genome is nearly complete. One reason for the intense interest in this bacterium is its multiple types of differentiated cells, which include heterocysts, akinetes and hormogonia, and which are useful for the study of the physiological aspects and differentiation processes of various cell types. Since the 1990s, the nitrogen and hydrogen metabolism of this strain has been widely investigated. The presence of three types of nitrogenase in heterocysts and vegetative cells has been reported, and their relationship and control of nitrogenase systems has been investigated (Thiel and Pratte 2001). This strain has recently been used to study the development of biological hydrogen production systems as the source of renewable energy. Recently developed gene manipulation techniques have made it possible to analyze and disrupt the uptake of hydrogenase genes, and to increase the amount of hydrogen produced by nitrogenase (Happe et al. 2000).
We recently identified the molecular structures of the carotenoids in some Anabaena and Nostoc species (Takaichi et al. 2005), and found that the myxoxanthophyll and ketomyxoxanthophyll in Anabaena (also known as Nostoc) sp. PCC 7120, Anabaena variabilis IAM M-3 (= PCC 7118, ATCC 27892) and Nostoc punctiforme PCC 73102 were (3R,2′S)-myxol 2′-fucoside and (3S,2′S)-4-ketomyxol 2′-fucoside, respectively. Further, the glycoside moiety of the pigments was found to be fucose, and not rhamnose. The major carotenoids were found to be β-carotene and echinenone, and the minors were β-cryptoxanthin, zeaxanthin, canthaxanthin and 3′-hydroxyechinenone. We also previously identified the carotenoids in Synechocystis sp. PCC 6803 (Takaichi et al. 2001): the myxoxanthophyll was found to be (3R,2′S)-myxol 2′-dimethyl-fucoside, and the glycoside moiety of the pigment was found to be 2,4-di-O-methyl-α-l-fucose. The other carotenoids were β-carotene, (3R,3′R)-zeaxanthin, echinenone, (3′R)-3′-hydroxyechinenone and deoxymyxol 2′-dimethyl-fucoside. We also previously proposed the biosynthetic pathways of the carotenoids and their enzymes and genes in these bacteria (Takaichi et al. 2001, Takaichi et al. 2005).
In the present study, we identified the carotenoids in A. variabilis ATCC 29413. Myxol glycosides were absent, while free myxol and 4-hydroxymyxol were present. The 4-hydroxyl group of the latter was a mixture of (4R) and (4S) configurations. This strain was the first cyanobacterium found to contain free myxol and not myxol glycosides. The major carotenoids were β-carotene, echinenone and canthaxanthin. Further, the present study proposes a biosynthetic pathway of these carotenoids and the corresponding enzymes and genes.
Results
Identification of non-polar pigments
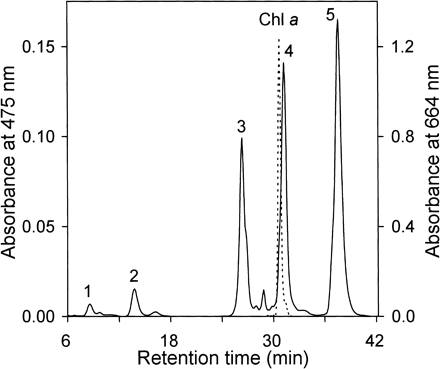
Fig. 1 shows an HPLC elution profile for the organic solvent-soluble pigments extracted from A. variabilis ATCC 29413. The pigment eluted at 31.0 min was identified as Chl a by its absorption spectrum and retention time on the HPLC system as compared with Chl a from Anabaena sp. PCC 7120 (Takaichi et al. 2005).
The absorption maxima of the peak-5 pigment in methanol were 429 (shoulder), 449 and 475 nm, and the spectral fine structure of %III/II, which is the ratio of the peak heights of the longest and the middle wavelength absorption bands from the trough between the two peaks (Takaichi and Shimada 1992), was 20. It had a relative molecular mass of 536. Hence, the peak-5 pigment was identified as β-carotene. The peak-4 pigment was identified as echinenone based on a broad absorption maximum at around 460 nm, its retention time on the HPLC system as compared with echinenone from Anabaena sp. PCC 7120, and its relative molecular mass of 550. The peak-3 pigment was identified as canthaxanthin based on the broad absorption maximum at around 475 nm, its retention time on the HPLC system as compared with canthaxanthin from Anabaena sp. PCC 7120, and its relative molecular mass of 564.
Identification of polar pigments
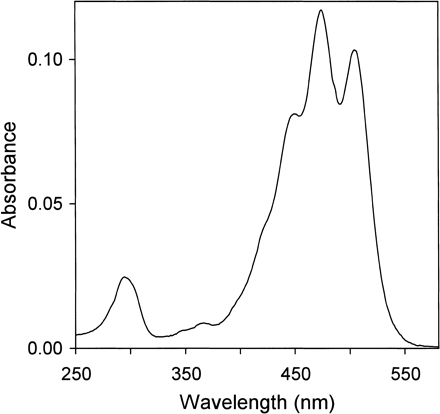
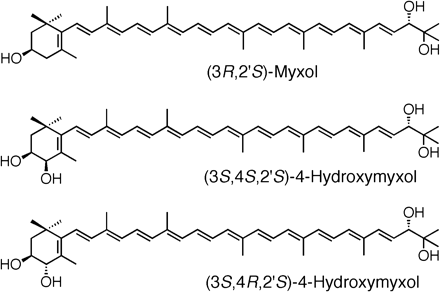
The absorption maxima of the purified peak-2 pigment were 294, 366, 450, 474 and 504 nm in methanol/water (9 : 1, v/v) (Fig. 2), and the spectral fine structure of %III/II was 58. From these results, the carotenoid was determined to be a derivative of γ-carotene with 12 conjugated double bonds (Takaichi and Shimada 1992). The relative molecular mass was 584. The formation of a diacetyl derivative and a tri-trimethylsilyl derivative indicated the presence of two primary and/or secondary hydroxyl groups and one tertiary hydroxyl group (Takaichi 1993). The 1H-nuclear magnetic resonance (NMR) spectrum corresponded to myxol (Yokoyama and Miki 1995, Takaichi et al. 2001). The circular dichroism (CD) spectrum was almost comparable with that of (3R,2′S)-myxol 2′-dimethyl-fucoside from Synechocystis sp. PCC 6803 (Takaichi et al. 2001). Thus, the structure of the peak-2 pigment was identified to be (3R,2′S)-myxol (Fig. 3). The IUPAC-IUB semi-systematic name is (3R,2′S)-3′,4′-didehydro-1′,2′-dihydro-β,ψ-carotene-3,1′,2′-triol.
The absorption maxima of the purified peak-1 pigment were comparable with those of the peak-2 pigment. The relative molecular mass was 600. The formation of a triacetyl derivative and a tetra-trimethylsilyl derivative indicated the presence of three primary and/or secondary hydroxyl groups and one tertiary hydroxyl group. Therefore, this pigment had one more hydroxyl group than did the peak-2 pigment. The oxymetin signal at 3.94 ppm in the 1H-NMR spectrum indicated that the position of the additional hydroxyl group was at C-4 (Tsushima et al. 1989, Englert 1995). Therefore, the structure of the peak-1 pigment was assigned to be 4-hydroxymyxol. Furthermore, comparing the 1H-NMR data with (3S,4R,3′R,6′R)- and (3S,4S,3′R,6′R)-β,ε-carotene-3,4,3′-triol (Tsushima et al. 1989, Englert 1995), it was revealed that the peak-1 pigment was a mixture of (4R) and (4S) isomers. The methyl signals 1.098 and 1.110 (s, C-16,17), 1.834 (s, C-18) and 1.989 ppm (s, C-19) were assigned to the (3S,4R) configuration of 4-hydroxymyxol, and 1.087 and 1.110 (s, C-16,17), 1.919 (s, C-18) and 1.989 ppm (s, C-19) were assigned to the (3S,4S) configuration. Since the intensities of each singlet peak were almost the same, the molar ratio of the (4R) and (4S) isomers was almost the same. The CD spectrum of the mixture of two isomers was comparable with that of the peak-2 pigment. It was previously reported that the chirality of the 4-hydroxyl group in the 3,4-dihydroxy-β-end group does not influence the shape of the CD spectrum (Matsuno et al. 1984). Thus, the structure of the peak-1 pigment was identified to be (3S,4RS,2′S)-4-hydroxymyxol, (3S,4RS,2′S)-3′,4′-didehydro-1′,2′-dihydro-β,ψ-carotene-3,4,1′,2′-tetrol (Fig. 3). [Note (3R,2′S)-myxol and (3S,4RS,2′S)-4-hydroxymyxol have equivalent absolute configurations of the 3-hydroxyl group as indicated in Fig. 3.]
Composition of pigments
The composition of the pigments in the cells cultured under light for 2 weeks was 5% myxol (mol% of total carotenoids), 2% 4-hydroxymyxol, 51% β-carotene, 20% echinenone and 22% canthaxanthin. Minor carotenoids eluted just after peaks-1 and -2 on HPLC (Fig. 1) were cis-forms of the peak-1 and -2 pigments, respectively, based on their absorption spectra. A minor peak between peaks-3 and -4 was deoxymyxol based on the same absorption spectrum as that of myxol and the retention time on HPLC. Any carotenoid glycosides could not be detected on silica gel thin-layer chromatography (TLC), and zeaxanthin and β-cryptoxanthin were also absent. The molar ratio of total carotenoids to Chl a was 0.44, which was comparable with those of some Anabaena and Nostoc species (Takaichi et al. 2005).
Discussion
We identified the molecular structures of the carotenoids in A. variabilis ATCC 29413 (Fig. 3). Myxol glycosides were absent, while the free forms of myxol and 4-hydroxymyxol were present. All of the myxol and its derivatives (4-ketomyxol, 4-hydroxymyxol and deoxymyxol) in cyanobacteria are known to be glycosides (Britton et al. 2004), and free myxol has been found in only one marine bacterium strain P99-3 (MBIC 03313; previous name, Flavobacterium sp.) (Yokoyama and Miki 1995). Thus, A. variabilis ATCC 29413 is the first cyanobacterium found to have free myxol and not myxol glycosides, and it seems to lack the gene for or activity of glycosyl transferase. Thus, this strain was considered to be of potential use in investigating the characteristics of myxol glycosides in cyanobacteria, since in another strain, A. variabilis IAM M-3 (= PCC 7118, ATCC 27892), we recently identified (3R,2′S)-myxol 2′-fucoside and (3S,2′S)-4-ketomyxol 2′-fucoside (Takaichi et al. 2005).
We also recently identified the molecular structures of the carotenoids in some Anabaena and Nostoc species (Takaichi et al. 2005). The carotenoid moieties of the carotenoid glycosides in Anabaena sp. PCC 7120, A. variabilis IAM M-3, Nostoc punctiforme PCC 73102 and Nostoc sp. KK-01 are (3R,2′S)-myxol and (3S,2′S)-4-ketomyxol. Two newly isolated Nostoc species have only myxol as the carotenoid moiety. In the present study, A. variabilis ATCC 29413 was found to contain free forms of myxol and 4-hydroxymyxol. The phylogenetic tree of the 16S rDNA sequences of these species including ATCC 29413 suggests that these species and strains are very close (Takaichi et al. 2005), although their carotenoid moieties were different, as described above. Further, 4-hydroxymyxol glycoside is known to be distributed only in some cyanobacteria (Britton et al. 2004).
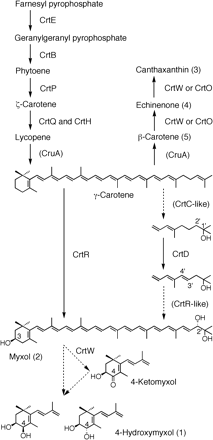
The biosynthetic pathway of 4-hydroxymyxol is not known, but there are two possibilities (Fig. 4): either myxol is oxidized to 4-ketomyxol by β-carotene ketolase (CrtW) and is reduced to 4-hydroxymyxol, or myxol is directly hydroxylated to 4-hydroxymyxol. The 4-hydroxyl group was an equimolar mixture of (4R) and (4S) configurations (Fig. 3), which is a very rare mixture in carotenoids (Britton et al. 2004). In the macular pigment of the human retina, (3R,3′R)- and (3R,3′S)-zeaxanthin are present, and two configurations of the hydroxyl group might be produced by chemical reduction of the keto group at C-3′ (Landrum and Bone 2001). The Japanese sea mussel Mytilus coruscus contains pectenol A and pectenol B, which are (3S,4R,3′S) and (3S,4S,3′S) configurations, respectively (Maoka and Matsuno 1988), and two configurations might be produced by non-stereospecific reduction of the keto group at C-4. The (R) and (S) configurations of a hydroxyl group exist at the C-31 carbon of bacteriochlorophyll c, and the (R) and (S) configurations of bacteriochlorophyllide c are known to be produced by BchlF and BchlV, respectively (Frigarrd et al. 2003). Further studies are needed to determine the formation of two configurations.
The identification of the carotenoids in the present study coupled with the near completion of the entire nucleotide sequence of the genome in A. variabilis ATCC 29413 will greatly facilitate the elucidation of the biosynthetic pathway and the corresponding enzymes and genes. Fig. 4 presents a proposed biosynthetic pathway of the carotenoids in this strain. Only CrtQ (ζ-carotene desaturase) (Linden et al. 1994), CrtW and CrtO (β-carotene ketolase) (Mochimaru et al. 2005) from Anabaena sp. PCC 7120, and two CrtWs from N. punctiforme PCC 73102 (Steiger and Sandmann 2004) have been functionally identified among Anabaena and Nostoc. We proposed the genes in A. variabilis ATCC 29413 on the basis of sequence homology (Table 1). In fact, the genes from only a limited number of species have actually been functionally identified, while many genes are simply suggested by sequence homology. In Table 1, only the functionally confirmed genes are chosen for the query sequences.
Two types of CrtQs (ζ-carotene desaturase), crtI-type and plant crtQ-type sequences, in prokaryotes are known, although they show no significant sequence homology with each other (Sandmann 2001). In prokaryotes, only three CrtQs have been functionally identified: Anabaena sp. PCC 7120 (crtI-type) (Linden et al. 1994), Synechocystis sp. PCC 6803 (plant crtQ-type) (Breitenbach et al. 1998) and Chlorobium tepidum (a green sulfur bacterium, plant crtQ-type) (Frigarrd et al. 2003). A. variabilis ATCC 29413 contains a plant crtQ-type gene based on the sequence homology (Table 1), although Anabaena sp. PCC 7120 contains a crtI-type gene.
Two β-carotene ketolases, CrtW and CrtO, are known in bacteria, although they show no significant sequence homology with each other (Mochimaru et al. 2005). At present, only six β-carotene ketolases are functionally confirmed in cyanobacteria. The reaction from myxol 2′-fucoside to ketomyxol 2′-fucoside is catalyzed by CrtW in two species, Anabaena sp. PCC 7120 and N. punctiforme PCC 73102, while that from β-carotene to echinenone is catalyzed by CrtW in two species, N. punctiforme PCC 73102 and Gloeobacter violaceus PCC 7421, and by CrtO in two species, Anabaena sp. PCC 7120 and Synechocystis sp. PCC 6803 (Fernández-González et al. 1997, Steiger and Sandmann 2004, Mochimaru et al. 2005, Tsuchiya et al. 2005). A. variabilis ATCC 29413 contains both crtW-type and crtO-type genes (Table 1).
CrtR (β-carotene hydroxylase) from Anabaena sp. PCC 7120 and Synechocystis sp. PCC 6803 catalyzes conversion of β-carotene to zeaxanthin via β-cryptoxanthin, echinenone to 3′-hydroxyechinenone, and deoxymyxol to myxol (Masamoto et al. 1998, Lagarde and Vermaas 1999, Takaichi et al. 2005). On the other hand, CrtR from strain ATCC 29413 catalyzes only conversion of deoxymyxol to myxol but not β-carotene to zeaxanthin in the cells, since zeaxanthin and β-cryptoxanthin are absent. Functional analyses are still needed for these genes.
Materials and Methods
Biological materials and cultivation
Anabaena variabilis ATCC 29413 (= IAM M-204) was grown in BG-11 medium with continuous shaking (110 rpm) at 26–28°C under continuous illumination by white fluorescent light (30–40 µE m–2 s–1) for 2 weeks. The grown cells were collected by centrifugation (Takaichi et al. 2005).
Purification and identification of pigments
Acetone/methanol-soluble pigments (7 : 2, v/v) were extracted from the cells, followed by evaporation of the solvent. Chl a and the known carotenoids were identified from their absorption spectra and specific retention times on HPLC equipped with a µBondapak C18 column (8×100 mm, RCM type; Waters, Milford, MA, USA) eluted with methanol/water (9 : 1, v/v) for 20 min and then 100% methanol (2.0 ml/min) (Takaichi et al. 2001). Anabaena sp. PCC 7120 and Synechocystis sp. PCC 6803 were also used for comparison of the pigments (Takaichi et al. 2001, Takaichi et al. 2005).
The polar carotenoids were isolated and purified as follows. We extracted the pigments with acetone/methanol (7 : 2, v/v) using an ultrasonicator, followed by evaporation of the solvent. The pigments were loaded on a column of DEAE-Toyopearl 650 M (Tosoh, Tokyo, Japan), and the carotenoids were eluted with n-hexane/acetone (1 : 1, v/v), but the Chl a and polar lipids remained on the column. The carotenoids were then loaded on a column of silica gel 60 (Merck, Darmstadt, Germany). β-Carotene was first eluted with n-hexane, and non-polar carotenoids were eluted with n-hexane/acetone (8 : 2, v/v). Finally, the polar carotenoids were eluted with n-hexane/acetone (1 : 1, v/v) and acetone. The polar carotenoids were purified by silica gel TLC (Merck) developed with dichloromethane/ethyl acetate/acetone (5 : 10 : 2, by vol.), and were finally collected from the HPLC apparatus described above (Takaichi et al. 2001).
Spectroscopic analysis
We measured the absorption spectra of the pigments using an MCPD-3600 photodiode array detector (Otsuka Electronics, Osaka, Japan) attached to the HPLC apparatus described above (Takaichi and Shimada 1992). For quantitative analysis, the molar extinction coefficients at the maximum wavelengths of each carotenoid were assumed to be the same. The CD spectra of the carotenoids were measured using a J-820 spectropolarimeter (JASCO, Tokyo, Japan) in diethyl ether/2-pentane/ethanol (5 : 5 : 2, by vol.) at room temperature. The relative molecular masses of the carotenoids and their acetyl and trimethylsilyl derivatives were measured using an FD-MS; M-2500 double-focusing gas chromatograph–mass spectrometer (Hitachi, Tokyo, Japan) equipped with a field-desorption apparatus (Takaichi 1993). The 1H-NMR (500 MHz) spectra of the carotenoids in CDCl3 at 24°C were measured using the UNITY INOVA-500 system (Varian, Palo Alto, CA, USA).
Sequence analysis
The amino acid sequences of enzymes in A. variabilis ATCC 29413 can be found in the database Ver. 26jun05 produced by JGI (http://genome.ornl.gov/microbial/avar/). Database searches were carried out with BLASTP 2.2.5 running on the web site of the database. Enzymes, whose functions have already been confirmed, were chosen for query sequences, and their conserved domains with high similarity were used for the calculation of e-values by the BLASTP program. The best matched genes are indicated in Table 1.
Acknowledgments
We wish to thank Dr. H. Sakurai (Waseda University) for the gift of A. variabilis ATCC 29413. This study was supported in part by a Grant-in-Aid for Scientific Research from JSPS to S.T. (16570038).
This paper is dedicated to the memory of Professor Ken-ichiro Takamiya, 1943–2005.
Fig. 1 HPLC elution profile of pigments extracted from Anabaena variabilis ATCC 29413. The eluent was methanol/water (9 : 1, v/v) for 20 min and then 100% methanol (2.0 ml/min). Absorbances at 475 (solid line) and 664 nm (dashed line) are shown. Peaks are referred to in the text and figures.
Fig. 2 Absorption spectrum of myxol (peak-2 in Fig. 1) in methanol/water (9 : 1, v/v).
Fig. 3 Structure of some carotenoids.
Fig. 4 Biosynthetic pathway of carotenoids and their enzymes in Anabaena variabilis ATCC 29413. Numbers in parentheses indicated the peak numbers in Fig. 1. See the text for details.
Presumed enzymes and the gene numbers of Anabaena variabilis ATCC 29413
| Gene | Enzyme | Query sequence for BLASTP | Contig-Gene no. | Identity (%)/e-value |
| crtE | Geranylgeranyl diphosphate synthase | Thermosynechococcus elongatus BP-1 CrtE a | 232-Ava2704 | 68/e-118 |
| crtB | Phytoene synthase | Synechocystis sp. PCC 6803 CrtB b | 232-Ava4794 | 70/e-128 |
| crtP | Phytoene desaturase | Synechocystis sp. PCC 6803 CrtP c | 232-Ava4795 | 77/0.0 |
| crtQ | ζ-Carotene desaturase | Synechocystis sp. PCC 6803 CrtQ d | 232-Ava0200 | 75/0.0 |
| crtH | cis-to-trans Carotene isomerase | Synechocystis sp. PCC 6803 CrtH e | 232-Ava3112 | 73/0.0 |
| cruA | Lycopene cyclase | Chlorobium tepidum TLS CruA f | 232-Ava3214 | 38/1e-68 |
| crtR | β-Carotene hydroxylase | Synechocystis sp. PCC 6803 CrtR g | 232-Ava1693 | 66/e-129 |
| crtW | β-Carotene ketolase | Anabaena sp. PCC 7120 CrtW h | 232-Ava3888 | 95/e-154 |
| crtO | β-Carotene ketolase | Anabaena sp. PCC 7120 CrtO i | 232-Ava1581 | 98/0.0 |
| crtD | Methoxyneurosporene desaturase | Synechocystis sp. PCC 6803 CrtD j | 232-Ava2342 | 67/0.0 |
| Gene | Enzyme | Query sequence for BLASTP | Contig-Gene no. | Identity (%)/e-value |
| crtE | Geranylgeranyl diphosphate synthase | Thermosynechococcus elongatus BP-1 CrtE a | 232-Ava2704 | 68/e-118 |
| crtB | Phytoene synthase | Synechocystis sp. PCC 6803 CrtB b | 232-Ava4794 | 70/e-128 |
| crtP | Phytoene desaturase | Synechocystis sp. PCC 6803 CrtP c | 232-Ava4795 | 77/0.0 |
| crtQ | ζ-Carotene desaturase | Synechocystis sp. PCC 6803 CrtQ d | 232-Ava0200 | 75/0.0 |
| crtH | cis-to-trans Carotene isomerase | Synechocystis sp. PCC 6803 CrtH e | 232-Ava3112 | 73/0.0 |
| cruA | Lycopene cyclase | Chlorobium tepidum TLS CruA f | 232-Ava3214 | 38/1e-68 |
| crtR | β-Carotene hydroxylase | Synechocystis sp. PCC 6803 CrtR g | 232-Ava1693 | 66/e-129 |
| crtW | β-Carotene ketolase | Anabaena sp. PCC 7120 CrtW h | 232-Ava3888 | 95/e-154 |
| crtO | β-Carotene ketolase | Anabaena sp. PCC 7120 CrtO i | 232-Ava1581 | 98/0.0 |
| crtD | Methoxyneurosporene desaturase | Synechocystis sp. PCC 6803 CrtD j | 232-Ava2342 | 67/0.0 |
The Contig-Gene numbers were cited from the Anabaena variabilis ATCC 29413 genome database Ver. 26jun05 produced by JGI. Database searches were carried out with the BLAST program. Enzymes, whose functions have already been confirmed, were chosen for the query sequences. Accesssion numbers are as follows: a AB016093 (GenBank) (Ohto et al. 1999), 99% identical to tll0020 (Cyanobase), b S45360 (PIR) (Martínez-Férez et al. 1994), 100% identical to slr1255 (Cyanobase), cslr1254 (Cyanobase), S74886 (PIR) (Martínez-Férez and Vioque 1992), dslr0940 (Cyanobase), S76141 (PIR) (Breitenbach et al. 1998), esll0033 (Cyanobase), S75951 (PIR) (Masamoto et al. 2001), fCT0456 (Cyanobase), Q8KF75(TrEMBL) (Maresca et al. 2005), gsll1468 (Cyanobase), S77365 (PIR) (Masamoto et al. 1998, Lagarde and Vermass, 1999), halr3189 (Cyanobase) (Mochimaru et al. 2005), iall3744 (Cyanobase) (Mochimaru et al. 2005), jslr1293 (Cyanobase), S74689 (PIR) (Mohamed and Vermaas 2004).
Presumed enzymes and the gene numbers of Anabaena variabilis ATCC 29413
| Gene | Enzyme | Query sequence for BLASTP | Contig-Gene no. | Identity (%)/e-value |
| crtE | Geranylgeranyl diphosphate synthase | Thermosynechococcus elongatus BP-1 CrtE a | 232-Ava2704 | 68/e-118 |
| crtB | Phytoene synthase | Synechocystis sp. PCC 6803 CrtB b | 232-Ava4794 | 70/e-128 |
| crtP | Phytoene desaturase | Synechocystis sp. PCC 6803 CrtP c | 232-Ava4795 | 77/0.0 |
| crtQ | ζ-Carotene desaturase | Synechocystis sp. PCC 6803 CrtQ d | 232-Ava0200 | 75/0.0 |
| crtH | cis-to-trans Carotene isomerase | Synechocystis sp. PCC 6803 CrtH e | 232-Ava3112 | 73/0.0 |
| cruA | Lycopene cyclase | Chlorobium tepidum TLS CruA f | 232-Ava3214 | 38/1e-68 |
| crtR | β-Carotene hydroxylase | Synechocystis sp. PCC 6803 CrtR g | 232-Ava1693 | 66/e-129 |
| crtW | β-Carotene ketolase | Anabaena sp. PCC 7120 CrtW h | 232-Ava3888 | 95/e-154 |
| crtO | β-Carotene ketolase | Anabaena sp. PCC 7120 CrtO i | 232-Ava1581 | 98/0.0 |
| crtD | Methoxyneurosporene desaturase | Synechocystis sp. PCC 6803 CrtD j | 232-Ava2342 | 67/0.0 |
| Gene | Enzyme | Query sequence for BLASTP | Contig-Gene no. | Identity (%)/e-value |
| crtE | Geranylgeranyl diphosphate synthase | Thermosynechococcus elongatus BP-1 CrtE a | 232-Ava2704 | 68/e-118 |
| crtB | Phytoene synthase | Synechocystis sp. PCC 6803 CrtB b | 232-Ava4794 | 70/e-128 |
| crtP | Phytoene desaturase | Synechocystis sp. PCC 6803 CrtP c | 232-Ava4795 | 77/0.0 |
| crtQ | ζ-Carotene desaturase | Synechocystis sp. PCC 6803 CrtQ d | 232-Ava0200 | 75/0.0 |
| crtH | cis-to-trans Carotene isomerase | Synechocystis sp. PCC 6803 CrtH e | 232-Ava3112 | 73/0.0 |
| cruA | Lycopene cyclase | Chlorobium tepidum TLS CruA f | 232-Ava3214 | 38/1e-68 |
| crtR | β-Carotene hydroxylase | Synechocystis sp. PCC 6803 CrtR g | 232-Ava1693 | 66/e-129 |
| crtW | β-Carotene ketolase | Anabaena sp. PCC 7120 CrtW h | 232-Ava3888 | 95/e-154 |
| crtO | β-Carotene ketolase | Anabaena sp. PCC 7120 CrtO i | 232-Ava1581 | 98/0.0 |
| crtD | Methoxyneurosporene desaturase | Synechocystis sp. PCC 6803 CrtD j | 232-Ava2342 | 67/0.0 |
The Contig-Gene numbers were cited from the Anabaena variabilis ATCC 29413 genome database Ver. 26jun05 produced by JGI. Database searches were carried out with the BLAST program. Enzymes, whose functions have already been confirmed, were chosen for the query sequences. Accesssion numbers are as follows: a AB016093 (GenBank) (Ohto et al. 1999), 99% identical to tll0020 (Cyanobase), b S45360 (PIR) (Martínez-Férez et al. 1994), 100% identical to slr1255 (Cyanobase), cslr1254 (Cyanobase), S74886 (PIR) (Martínez-Férez and Vioque 1992), dslr0940 (Cyanobase), S76141 (PIR) (Breitenbach et al. 1998), esll0033 (Cyanobase), S75951 (PIR) (Masamoto et al. 2001), fCT0456 (Cyanobase), Q8KF75(TrEMBL) (Maresca et al. 2005), gsll1468 (Cyanobase), S77365 (PIR) (Masamoto et al. 1998, Lagarde and Vermass, 1999), halr3189 (Cyanobase) (Mochimaru et al. 2005), iall3744 (Cyanobase) (Mochimaru et al. 2005), jslr1293 (Cyanobase), S74689 (PIR) (Mohamed and Vermaas 2004).
Abbreviations
References
Breitenbach, J., Fernández-González, B., Vioque, A. and Sandmann, G. (
Englert, G. (
Fernández-González, B., Sandmann G. and Vioque A. (
Frigarrd, N.-U., Chew, A.G.M., Li, H., Maresca, J.A. and Bryant, D.A. (
Happe, T., Schütz, K. and Böhme, H. (
Lagarde, D. and Vermaas, W. (
Landrum, J.T. and Bone, R.A. (
Linden, H., Misawa, N., Saito, T. and Sandmann, G. (
Maoka, T. and Matsuno, T. (
Maresca, J.A., Frigaard, N.-F. and Bryant, D.A. (
Martínez-Férez, I. and Vioque, A. (
Martínez-Férez, I., Fernández-González, B., Sandmann, G. and Vioque, A. (
Masamoto, K., Misawa, N., Kaneko, T., Kikuno, R. and Toh, H. (
Masamoto, K., Wada, H., Kaneko, T. and Takaichi, S. (
Matsuno, T., Maoka, T., Sakaguchi, S. and Nishizawa, T. (
Mochimaru, M., Masukawa, H. and Takaichi, S. (
Mohamed, H.E. and Vermaas, W. (
Ohto, C., Ishida, C., Nakane, H., Muramatsu, M., Nishino, T. and Obata, S. (
Sandmann, G. (
Steiger, S. and Sandmann, G. (
Takaichi, S. (
Takaichi, S., Maoka, T. and Masamoto, K. (
Takaichi, S., Mochimaru, M., Maoka, T. and Katoh, H. (
Takaichi, S. and Shimada, K. (
Thiel, T. and Pratte, B. (
Tsuchiya, T., Takaichi, S., Misawa, N., Maoka, T., Miyashita, H. and Mimuro, M. (
Tsushima, M., Maoka, T. and Matsuno, T. (