-
PDF
- Split View
-
Views
-
Cite
Cite
J. Romeike, T. Friedl, G. Helms, S. Ott, Genetic Diversity of Algal and Fungal Partners in Four Species of Umbilicaria (Lichenized Ascomycetes) Along a Transect of the Antarctic Peninsula, Molecular Biology and Evolution, Volume 19, Issue 8, August 2002, Pages 1209–1217, https://doi.org/10.1093/oxfordjournals.molbev.a004181
Close - Share Icon Share
Abstract
Lichens from the genus Umbilicaria were collected across a 5,000-km transect through Antarctica and investigated for DNA sequence polymorphism in a region of 480–660 bp of the nuclear internal transcribed spacer region of ribosomal DNA. Sequences from both fungal (16 ascomycetes) and photosynthetic partners (22 chlorophytes from the genus Trebouxia) were determined and compared with homologs from lichens inhabiting more temperate, continental climates. The phylogenetic analyses reveal that Antarctic lichens have colonized their current habitats both through multiple independent colonization events from temperate embarkation zones and through recent long-range dispersal in the Antarctic of successful preexisting colonizers. Furthermore, the results suggest that relichenization—de novo establishment of the fungus-photosynthesizer symbiosis from nonlichenized algal and fungal cells—has occurred during the process of Antarctic lichen dispersal. Independent dispersal of algal and fungal cultures therefore can lead to a successful establishment of the lichen symbiosis even under harsh Antarctic conditions.
Introduction
The Antarctic region harbors the most extreme climate and the most narrow biodiversity of all continents. The eukaryotic flora of the Antarctic region consists of two flowering plants, 50 liverworts, 104 bryophytes (mosses), and roughly 427 lichens. Lichens thus dominate the Antarctic flora both in terms of species diversity and in terms of total biomass (Ochyra 1998 , pp. 278; Bednarek-Ochyra et al. 2000 , pp. 236; Øvstedal and Lewis Smith 2001 , pp. 411). How these lichens came to colonize and inhabit the Antarctic region is a matter of considerable debate. In principle, there are three possibilities and combinations thereof (1) relict endemism from warmer Antarctic palaeoclimates, (2) rare colonization events of specialized ecotypes particularly suited to the harsh Antarctic climate followed by speciation and spread on the continent, and (3) temporally continuous, including recent, multiple independent colonization events (Hertel 1984 ; Kappen 1993a,2000 ; Seppelt, Green, and Schroeter 1995 ; Castello and Nimis 1997 ). Studies of ecophysiological adaptations of Antarctic lichens, for example tolerance to high UV dosage, desiccation, and cold, have suggested that physiological plasticity may be the factor limiting the colonization success of Antarctic lichens (Kappen 1993b;Schroeter et al. 1997 ). This would favor the view that colonization events should be rare. However, molecular data that could help to uncover colonization history have not been reported for Antarctic lichens, and only a few reports of molecular data for any member of the Antarctic flora (mosses, lichens) are known (Skotnicki, Selkirk, and Beard 1998 ; Dyer and Murtagh 2001 ; Skotnicki, Ninham, and Selkirk 2000 ).
Unlike higher plants, lichens are not single organisms, rather they are two—a fungus, commonly an ascomycete (the mycobiont), and a photosynthesizing organism (the photobiont), which can either be a cyanobacterium or a eukaryotic green alga, usually a member of the genus Trebouxia (Trebouxiophyceae). Unlike phanerogams, which undergo long-range dispersal by seeds (seed plants) or spores (ferns and mosses), lichens have two fundamentally different mechanisms of long-range dispersal. The first of these is the joint dispersal of the mycobiont and photobiont, either by means of a vegetative fragment (simply a piece of the lichen thallus) or by means of specialized dispersal organs (soredia) which are small (∼100–150 μm in diameter) sporelike dispersal units that are produced in specialized cuplike structures called soralia. These can be carried easily by wind. The second mechanism of lichen dispersal is the independent dispersal of the mycobiont (as ascospores) and the photobiont (as single vegetative cells), which then grow individually in a new habitat, come into contact, and form a new lichen at that spot de novo (Ott 1987 ). Independent dispersal of the mycobiont can also occur by vegetative structures of fungal hyphae—so-called thalloconidia—which are characteristic of the genus Umbilicaria.
Thus, the diversity and dispersal of lichens depends both on the capacity for joint dispersal and on the accessibility of individual partners in the new environment, an indispensable prerequisite for the disjoint dispersal mechanism. Disjoint dispersal of the separate bionts requires the process of relichenization. The success of the recognition process and the contact phase between the mycobiont and photobiont in the initial phase of relichenization depends upon the environmental and physiological factors, particularly on the selectivity of the partners to each other. Selectivity describes the process where bionts interact preferentially with particular species as partners (Galun and Bubrick 1984 ; Bubrick, Frensdorff, and Galun 1985 ). Lichens that depend on relichenization for their colonization may express a low selectivity toward their photobionts (Beck, Kasalicky, and Rambold 2002 ) because relichenization may be more successful if a broader range of respective algal strains were acceptable than a more narrow one. Lichens can even use different species of algae as photobionts during their life cycle (Friedl 1987 ). However, if no suitable photobiont is available, the mycobiont can survive in a loose association with other algae (Ott 1987 ) or behave as a saprotrophic fungus (Lawrey 1984 , pp. 407). Such fungi can then form lichen thalli only with certain algal species; therefore, they have a high selectivity toward their photobionts (Helms et al. 2001 ).
The morphology, physiology, and colonization behavior of Antarctic lichens have been characterized at selected sites forming a discrete geographical transect across the Antarctic Peninsula (Ott and Sancho 1991 , 1993 ; Ott et al. 1997 ; Lud, Huiskes, and Ott 2001 ). The major species of macrolichens in this Antarctic transect belong to the genera Usnea (subgenus Neuropogon) and Umbilicaria. Among the Antarctic macrolichens, the genus Umbilicaria is of interest because fewer of the species occur in the Antarctic than in similar harsh or high-mountain habitats (Sancho, Kappen, and Schroeter 1992 ), suggesting that the colonization history of the Antarctic Umbilicaria species differs from that of congeners in ecologically similar habitats.
The molecular systematics of many lichens (photobionts and mycobionts) from temperate climates has been characterized in some detail using the internal transcribed spacer (ITS) of the nuclear ribosomal DNA (Friedl and Rokitta 1997 ; Rambold, Friedl, and Beck 1998 ; Helms et al. 2001 ). Here we report DNA sequence polymorphism in the ITS region of mycobionts and photobionts of lichens from the genus Umbilicaria (mycobionts associated with species of the genus Trebouxia), collected across a transect of the Antarctic Peninsula (distance of 4,860 km), and a comparison of these sequences in the context of sequence diversity among lichenizing ascomycetes and chlorophytes from non-Antarctic accessions.
Materials and Methods
Lichen Samples and Research Areas
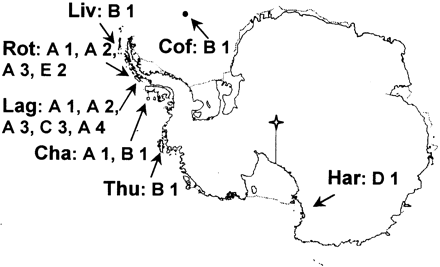
The photobiont and mycobiont ITS rDNA sequences were determined from lichen specimens as listed in table 1 . The lichen samples were collected along a transect from the northern maritime Antarctic to the continent (fig. 1 ). The transect starts on Coffer Island (South Orkney Islands, 60°45′S, 45°09′W) at the northernmost area of the maritime Antarctic at the borderline to the sub-Antarctic region and extends southwards along the west side of the Antarctic Peninsula to the west coast of Ellsworth Island (West Antarctica) and Victoria Land (East Antarctica). Following the transect south from Coffer Island at a distance of 830 km, Livingston Island (South Shetland Islands, 62°41′S, 60°23′W) is located in the middle of the maritime Antarctic, and Lagoon Island and Rothera Point (Adelaide Island, 67°34′S, 68°07′W, 650 km away from Livingston Island) are on the southernmost borderline between the maritime Antarctic and the continental Antarctic region. Rothera Point and Lagoon Island are only 5 km apart. The following three collection sites belong to the continental area of Antarctica. At a distance of 390 km southwest of Lagoon Island, Charcot Island (69°57′S,75°25′W) is placed at the west side of Alexander Island. At the transition from the Antarctic Peninsula to the continent, 840 km south of Charcot Island to the west of Ellsworth Land (West Antarctica) is the location of Thurston Island (72°28′S, 97°40′W). The southernmost sampling site is placed at Harrow Peaks (74°04′S, 164°45′E, Victoria Land) close to the west coast of East Antarctica, 2,780 km away from Thurston Island.
DNA Extraction, PCR, Sequencing
DNA was extracted from about 2 mm2 pieces of air-dried herbarium material of Umbilicaria species as previously described (Helms et al. 2001 ). About 500-bp-long DNA fragments extending from the 3′-end of the 18S to the 5′-end of the 26S rDNA were amplified using the biont-specific PCR primer combinations (for photobionts as in Helms et al. 2001 , for mycobionts My1200f+ and My1700f [unpublished data] combined with ITS4 [White et al. 1990] ). PCR conditions were as in Helms et al. (2001) , except that the annealing temperature was set to 51°C, and the extension was done for 1 min with an increment of 3 s after each cycle. PCR products were cleaned with the High Pure™ PCR Product Purification Kit (Roche) and sequenced directly using fluorescence-labeled primers (Helms et al. 2001 ). Reactions were run on an ALFexpress II automated sequencer (Amersham Pharmacia).
Phylogenetic Analyses
The ITS rDNA sequences determined in this study were manually aligned using the multiple sequence editor SeqEdit (from the Ribosomal Database Project; Maidak et al. 2001 ), the programs SeqPup (Gilbert 1995 ), and BioEdit (Hall 1999 ; http://www.mbio.ncsu.edu/RNaseP/info/programs/BIOEDIT/bioedit.html). The photobiont sequences were compared with all the available homologs from Trebouxia. Initial phylogenetic analyses revealed the same clades as found in previous analyses (Friedl et al. 2000 ; Helms et al. 2001 ); therefore, for the final analyses, only examples of taxa from the four different clades and sequences which were monophyletic with sister group to the newly determined sequences were considered. Regions of ambiguous alignment in both ITS regions and the 5.8S rDNA gene sequences were excluded from the phylogenetic analyses. The final photobiont ITS data set contained 27 taxa and 630 characters per site of which 280 were variable and 209 were parsimony-informative. The photobiont ITS alignment used in this study is available from TF and from Treebase (http://herbaria.harvard.edu/treebase/).
Phylogenetic analyses of the mycobiont ITS gene sequences were not done because three of the four newly determined fungal sequences appeared to be identical or very similar to sequences already available for Umbilicaria (Ivanova et al. 1999 ). Accession numbers for all ITS rDNA sequences determined in the present study are given in the note of table 1 . Three independent types of data analyses were used to assess the evolutionary relationships of the photobiont ITS sequences. All analyses were performed using PAUP* V4.0b8 (Swofford 2001 ). In maximum parsimony (MP) analyses, the sites were weighted (rescaled consistency index [RC] over an interval of 1–1,000 bp; Bhattacharya and Medlin 1995 ) and then used as input for bootstrap analyses (2,000 replications). The introduced gaps were treated as missing data. Heuristic search conditions were with starting trees built stepwise with 10 random additions of taxa, using the tree-bisection–reconnection branch-swapping algorithm to find the best tree. The best-scoring trees were held at each step. For distance and maximum likelihood (ML) analyses, the “GTR+G” (Rodriguez et al. 1990 ) model of DNA substitution was chosen on the basis of the likelihood ratio test statistic as implemented in the program MODELTEST 3.04 (Posada and Crandall 1998 ). Gamma shape parameters, substitution rate matrices, and nucleotide frequencies were also estimated using MODELTEST. Distance trees were constructed using both the minimum evolution criterion (ME; Rzhetsky and Nei 1995 ), with the same heuristic search procedure as in the MP analyses, and the Neighbor-Joining method (NJ; Saitou and Nei 1987 ). Bootstrap resampling (2,000 replications) was performed on both ME and NJ trees. ML searches were done under the same model (GRT+G) and settings as in the distance analyses. The initial ML tree (with a −ln likelihood of 4879.58832) was used as the input for subsequent ML runs to search for trees with better −ln likelihoods, but no better likelihood scores were obtained.
Results
Photobionts
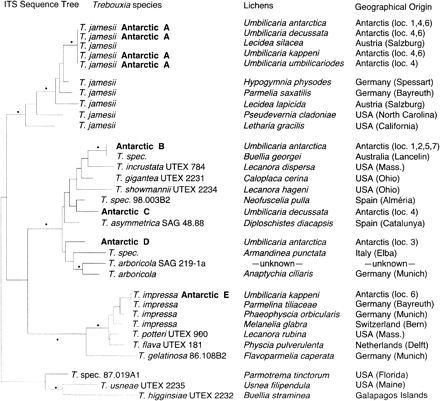
A total of 22 photobiont ITS sequences was determined from U. antartica, U. decussata, U. kappeni, and U. umbilicarioides in this study (table 1 ). Five distinct Trebouxia ITS rDNA sequences were found among these species which, for convenience, are referred to as algal variants Antarctic A, B, C, D, and E. Whereas variants Antarctic A and B were found 15 and 4 times, respectively, Antarctic C, D, and E occurred only once in the investigated samples. Among the 15 sequences of algal variant A, six sequences showed up to 11 different positions (fig. 3 ). Seven sequences from the Umbilicaria species were identical, although the samples were taken from three different localities (Charcot Island, Rothera Point, Lagoon Island). The absolute pairwise differences among the five ITS Antarctic types were 79.6 positions on average with a range from 52 to 119 positions. The algal variants Antarctic B, C, and D were most closely related to each other. There was an average difference of 54 positions among these sequences, whereas their absolute pairwise sequence differences with the variants Antarctic A and E ranged from 90 to 119 positions. These differences are also reflected in the ITS rDNA phylogeny (fig. 2 ). Antarctic A and E were each in a distinct clade of Trebouxia, whereas Antarctic B, C, and D were united in one clade (fig. 2 ). The monophyletic origin of each of these clades was highly supported in bootstrap tests (fig. 2 ). Antarctic A, on the basis of the sequence from a cultured algal strain of Lecidea silacea, has been identified as T. jamesii using morphological characters (Beck 1999 ). Antarctic B had only two positions that were different from an unidentified Trebouxia sequence determined from Buellia georgei. Antarctic E was identical with a cultured T. impressa strain isolated from Parmelina tiliacea, except for one base substitution within the ITS rDNA sequence. The algal variants Antarctic C and D differed greatly from the available Trebouxia ITS sequences; therefore, phylogenetic analyses were needed to infer their phylogenetic relationships. In the ITS phylogeny, however, their exact positions were unclear. Both algal variants were within a clade representing an array of different species (T. arboricola, T. asymmetrica, T. gigantea, T. incrustata, and T. showmanii) and two Trebouxia strains whose species assignment is unclear (Trebouxia sp. 98.003B2 and Trebouxia sp. from Amandinea punctata). The exact phylogenetic position of each of these species or strains within the clade was ambiguous (fig. 2 ). In distance analyses, there was moderate bootstrap support (68%–71% in NJ-ME) for an alliance of the algal variant Antarctic C with T. asymmetrica and Trebouxia sp. 98.003B2, but this relationship was not resolved in the ML analysis. For the variant Antarctic D, each of the different analyses methods revealed different positions within the same clade, and none was supported in bootstrap analyses.
Three of the four investigated Umbilicaria species were found to contain more than one algal partner. In U. antarctica, out of 10 samples studied, the algal variant Antarctic A (= T. jamesii) was found in five, Antarctic B in four, and Antarctic D in one (table 1 ). From U. decussata, three out of four specimens contained Antarctic A (= T. jamesii), and one had Antarctic C. Similarly, in U. kappeni Antarctic A (= T. jamesii) was found in three of the four available samples, and Antarctic E (= T. impressa) was found in one. However, in U. umbilicarioides all the four studied samples contained the algal variant Antarctic A (= T. jamesii).
Mycobionts
In contrast to the photobionts, almost no variation was found in the ITS rDNA sequences of the mycobionts (fig. 4 ). No sequence variation in mycobiont ITS rDNA was found among the three samples of U. kappeni. The ITS rDNA of that species differed in only a single position from the corresponding sequence of U. antartica AF096213; i.e., a G to A change at position 587 relative to the sequence AF096213. This base change was in a rather conserved position because none of the other available Umbilicaria fungal ITS sequences showed a variation there. Out of the 11 samples of U. antartica from which the mycobiont ITS rDNA were determined in this study, 6 had sequences identical with the U. antartica AF096213 sequence, 2 were incomplete sequences, and 2 differed from it in a single position: a C to T change at position 275 relative to the AF096213 sequence (fig. 4 ). This change also occurred in sequences of U. decussata, U. kappeni, and U. umbilicarioides. The sequence of one sample was left out because of too many gaps. For U. decussata and U. umbilicarioides, only a single mycobiont sequence each could be determined. The U. umbilicarioides sequence differed from the already available sequence from that species (AF096210) which was from another locality outside of Antarctica (Ivanova et al. 1999 ) for a total of three positions (positions 527, 714, 715). Between the U. decussata ITS sequence determined here and that from another sample of the same species (AF096214, Ivanova et al. 1999 ) 13 sequence positions were different (fig. 4 ). In phylogenetic analyses both sequences were each other's closest neighbors.
Discussion
Genetic Diversity of the Photobionts
The diversity of photobionts in the investigated Umbilicaria species was higher than that in previous studies of other lichens as for example in species of Physciaceae (Helms et al. 2001 ) and Cladoniaceae (DePriest et al. 2000 ). The unexpectedly high algal diversity reflects a low selectivity toward the algal partner in these lichen species. A similar high degree of photobiont diversity within a given species of lichens has not been reported to date. Friedl (1989) found six species of Parmeliaceae from tree barks of different localities in Europe; each was found to be associated with two or three different species of Trebouxia on the basis of morphological studies of cultured algal strains. In contrast, a rather high selectivity toward the photobiont was detected in species of foliose Physciaceae (e.g., Physcia, Physconia, Phaeophyscia) using algal-specific PCR and ITS rDNA sequence comparisons (Helms et al. 2001 ). Species of foliose Physciaceae had ITS algal variants that were closely related within the same clade in the Trebouxia ITS phylogeny and exhibited very little sequence differences.
In the 11 samples taken from Lagoon Island, three algal variants Antarctic A, C, and E were found (table 1 , fig. 1 ). At Charcot Island, from which two samples were taken, two algal variants were found. In the four samples taken from Rothera Point, two algal variants were detected. From all other studied localities only one sample each was available. Variant diversity of the photobiont in certain Umbilicaria species may not decrease with smaller sampling areas or sample sizes. In U. decussata and U. kappeni different algal variants were found in samples of each of these lichens at the same locality, Lagoon Island. The collection sites for the different samples from both lichens at that locality were at a distance of approximately 50 m from each other. Similarly, U. antarctica algal variants Antarctic A and B were found at the same locality, Charcot Island, with the collection sites of the samples only 40 m away from each other. The samples of U. kappeni that contained different algal variants were collected at two sites at Lagoon Island which were only about 30 m away from each other. Small sampling areas as well as small distances do not automatically mean that the samples belong to the same populations. In U. decussata, even one and the same specimen contained two different algal variants in different parts of the thallus. A third alga, T. impressa, has been reported for U. decussata by Aoki et al. (1998) , but this identification was not confirmed using DNA sequence data.
Though a low selectivity toward the photobiont was observed, algal variants seem to be preferred by the investigated Umbilicaria species. Algal variant Antarctic A (T. jamesii) was shared by all four investigated Umbilicaria species. Three out of four samples from U. decussata and U. kappeni contained algal variant Antarctic A. It was also present in all the three studied specimens of U. umbilicarioides and in half of all the samples from U. antarctica. For U. antarctica the preference for one particular Trebouxia ITS algal variant cannot be clearly stated. Among the 10 samples one half had algal variant Antarctic A, whereas the other half had algal variant Antarctic B. All samples of U. umbilicarioides were from a rich population on a flat beach terrace, Lagoon Island. In contrast, algal variants Antarctic B–E were each found in only one species of Umbilicaria. Variant Antarctic A (T. jamesii) may be preferred because of the selectivity of the Umbilicaria mycobionts toward a certain photobiont. As an alternative, the algal variant Antarctic A may be simply the most abundant photobiont species in the studied area, i.e., it may be provided by lichenized propagules from other lichens or it may occur free-living there.
Although U. kappeni is dispersed with soredia that contained both partners, lichen thalli from close sampling sites contained different algal variants. This might be the result of genetically different populations or the populations that have emerged from different geohistorical periods depending on deglaciation processes. The degree of photobiont diversity may be a feature that is differently expressed in different taxonomic groups of lichenized ascomycetes and may therefore be phylogenetically significant (Rambold, Friedl, and Beck 1998 ). In the Antarctic species of Umbilicaria studied here, low selectivity toward the photobiont may also function as a strategy for survival under harsh environmental conditions. Algal variants Antarctic C and D appear to be novel, i.e., they have not been found in any other lichens and not in any other localities except Antarctica so far.
Beck (1999) also found variant Antarctic A (T. jamesii) in U. cylindrica on siliceous rock at a locality in southern Germany and in other lichens on heavy-metal–containing rock (e.g., Lecidea silacea). Conspicuously, the lichens containing algal variant Antarctic A (T. jamesii) in this study were all from localities that were rich in iron. Therefore, environmental conditions may contribute to a preference toward a particular photobiont. Most likely Trebouxia is distributed via the air within dry diaspores of lichens, such as soredia, which may be well suited for long-distance dispersal because of their low weight. Birds may also be involved in the distribution of these diaspores as well as clusters of free-living Trebouxia cells. It is still not known whether Trebouxia algae are free living and it cannot be concluded that Trebouxia are wind-distributed from other continents to the Antarctic area in the same way as discussed for vegetative diaspores. The different algal variants may be spread throughout the different Antarctic localities and can thereby be utilized by the different Umbilicaria species. Variant Antarctic B was found at four of the seven studied localities but was not detected at Adelaide Island (Lagoon Island and Rothera Point) from which the great majority (15 out of 21) of all samples was taken. Algal variant Antarctic A (T. jamesii) was found at both localities from Adelaide Island (Lagoon Island and Rothera Point) and at Charcot Island (Alexander Island, distance 400 km).
Mycobiont Genetic Diversity
In contrast to the photobionts, almost no genetic variation was found among the mycobionts with ITS rDNA sequences, despite the fact that considerable morphological differences were expressed among the Umbilicaria species. Our findings thus contrast with previous studies of mycobiontal genetic variation within lichen populations (Zoller, Lutzoni, and Scheidegger 1999 ). The studied species of Umbilicaria may also have diverged from each other only relatively short time ago in evolution. Our finding of high polymorphism in U. decussata mycobiont ITS rDNA, but almost no polymorphisms in other Antarctic species of Umbilicaria, may be caused either by unequal rates of evolution among these species or by recent divergence of the Antarctic forms.
Umbilicaria kappeni is regarded as a species distinct from U. antarctica, although both share a similar external morphology and sometimes grow intermixed. In contrast to U. antarctica, U. kappeni produces neither thalloconidia nor ascospores but exhibits a variety of vegetative propagules e.g., soredia, adventive lobes, so-called thallyles (Sancho, Schroeter, and Valladares 1998 ). Umbilicaria kappeni is the only Umbilicaria species known to reproduce by soredia that completely cover the upper thallus surface. The soredia are never developed in sorelia as is described for U. soralifera (Sancho, Schroeter, and Valladares 1998 ). The very similar mycobiont ITS sequences of U. antarctica support the view that both species are very closely related. However, there is one sequence position in a rather conserved region of the ITS 2 rDNA, different between both species; this character may be regarded as an autapomorphy to distinguish both species, in addition to differences in their reproduction biology. For such closely related species, ITS rDNA may not be evolving fast enough to uncover colonization history, and other molecular markers may be more useful.
Conclusions
The diversity and distribution patterns of lichen communities on terrestrial sites of the Antarctic mirror a combination of relict flora, long-distance dispersal, and recent colonization. It seems unlikely that a whole population will be derived from a single fungal spore and a single existing algal cell but the gene pool should be smaller than in less isolated habitats on temperate continents. The occurrence of algal species may also depend on environmental conditions. In iron-rich habitats, the studied Umbilicaria species were mainly associated with a certain strain of T. jamesii, a Trebouxia species described by Beck (1999) from iron-rich habitats. A high rate of occurrence of this species in the investigated Umbilicaria could mainly result from its availability and less from the selectivity of the mycobiont.
Concerning the particular environmental conditions of Antarctic habitats, the relatively low Degree of selectivity found in the mycobionts of the Umbilicaria species can be interpreted as highly advantageous for colonization and adaptation and therefore for competition with other organisms in extreme habitats. If a mycobiont is less selective in its photobiont choice it enlarges its possibilities to colonize a broader range of habitats successfully. Under the peculiar environmental conditions of the Antarctic, the relatively low degree of selectivity in lichens as the investigated Umbilicaria species might be interpreted as an adaptation mechanism that improves the success of the mycobiont. This may also be true for the harsh environmental conditions of high mountain regions. This is the first insight into the diversity of photobionts of lichen species in the Antarctic region on the basis of molecular data. A comparison of the population structures and colonization strategies of lichens from the temperate regions and from this extreme environment could provide insights into the evolutionary history of the dominant Antarctic lichen flora.
William Martin, Reviewing Editor
Keywords: Antarctica genetic diversity selectivity lichen photobiont mycobiont
Address for correspondence and reprints: S. Ott, Botanisches Institut, Heinrich-Heine-Universität, Universitätsstr. 1, D-40225 Düsseldorf, Germany. otts@uni-duesseldorf.de
Fig. 1.—Sampling sites of the four species of the genus Umbilicaria. The following locations represent the sampling sites along a transect from the northern maritime Antarctic to the continent: Cof: Coffer Island, Liv: Livingston Island, Rot: Rothera Point, Lag: Lagoon Island, Cha: Charcot Island, Thu: Thurston Island, Har: Harrow Peak. See Materials and Methods for details on the precise coordinates. A–E represent the algal ITS variants and the four different mycobiont species are numbered 1–4. South Pole.
Fig. 2.—ML phylogeny of ITS rDNA sequences from photobionts of four Antarctic Umbilicaria species and other strains of Trebouxia. The origin of the photobiont sequences is given to the right of the species names. The five algal ITS variants detected in the Antarctic Umbilicaria species are named Antarctic A–E. The Antarctic localities for the algal ITS variants are given in brackets (see fig. 1 ). The sequences T. jamesii-Lecidea silacea and T. impressa-Parmelina tiliacea were identical with sequences determined in this study; they were simply added to the figure. Dots mark internal nodes which were defined by bootstrap support above 70% in NJ, ME, and weighted parsimony analyses and were also shared with the ML tree. The tree was rooted by the branch leading to T. sp.-T. usneae-T. higginsiae. (Photobiont accession numbers: AJ249565, Z68705, AJ293770, AF242467, Z68697, AJ249577, AJ249574, AJ007388, AJ007386, AJ249576, AJ293795, AF128270, Z68700, Z68701, AF128271, AF242466, AF242460, AF242469, AJ249573, AJ293783, AJ249572, AJ293780, Z68702)
Fig. 3.—Polymorphisms of the entire internal transcribes spacers (ITS I and ITS II) and the intervening 5.8S ribosomal DNA gene are shown for the photobiont Trebouxia jamesii sila of four Umbilicaria species investigated. Sample number see table 1 , Trebouxia jamesii sila*: accession number AF128270. Sequences were determined on both strands. Only polymorphisms that were unambiguous on both strands are shown. Nucleotide differences to the uppermost sequence are shown, dots indicate identity with the uppermost sequence, except for gaps, all of which are indicated as (“–”)
Fig. 4.—Polymorphisms of the entire internal transcribes spacers (ITS I and ITS II) and the intervening 5.8S ribosomal DNA gene are shown for the mycobionts of the four Umbilicaria species. Sample numbers see table 1 , Umbilicaria antarctica: AF096213, Umbilicaria decussata: AF096210, Umbilicaria umbilicarioides: AF096214. The incomplete sequence 1 (9) is not shown. Sequences were determined on both strands. Only polymorphisms that were unambiguous on both strands are shown. Nucleotide differences to the uppermost sequence are shown, dots indicate identity with the uppermost sequence, except for gaps, all of which are indicate as (“–”)
J.R. and S.O. thank Dr. R. I. L. Smith (Herbarium of British Antarctic Survey) for providing Antarctic samples from South Orkney Islands, Thurston Island, Victoria Land, and one sample from Charcot Island. We thank the British Antarctic Survey, UK, the people at Rothera Research Station, and Base Antàrtica Española Juan Carlos I for generous logistic support. This research was supported by grants of the DFG extended to S.O. (Ot 96/6-1/2/3/4) and T.F. (Fr 905/7-1,2) which is gratefully acknowledged. J.R. and T.F. gratefully acknowledge the help of E. Zufall-Roth in the laboratory. These results are included in the doctoral thesis of J.R.
References
Aoki M., T. Nakano, H. Kanda, H. Deguchi,
Beck A.,
Beck A., T. Kasalicky, G. Rambold,
Bednarek-Ochyra H., J. Vána, R. Ochyra, R. I. Lewis Smith,
Bhattacharya D., L. K. Medlin,
Bubrick P., A. Frensdorff, M. Galun,
Castello M., P. L. Nimis,
DePriest P. R., M. Piercey-Nomore, M. Sikaroodi, K. Kärkkäinen, I. Oksanen, R. Yahr, T. Ahti,
Dyer P., G. Murtagh,
Friedl T.,
Friedl T.,
Friedl T., A. Besendahl, P. Pfeiffer, D. Battacharya,
Friedl T., C. Rokitta,
Galun M., P. Bubrik,
Hall T. A.,
Helms G., T. Friedl, G. Rambold, H. Mayrhofer,
Ivanova N. V., P. DePriest, V. K. Bobrovas, A. V. Troitsky,
Kappen L.,
Lud D., A. Huiskes, S. Ott,
Maidak B. L., J. R. Cole, T. G. Lilburn, C. T. Parker Jr., P. R. Saxman, R. J. Farris, G. M. Garrity, G. J. Olsen, T. M. Schmidt, J. M. Tiedje,
Ochyra R.,
Ott S., T. Meier, H. M. Jahns,
Ott S., R. Przeswosnik, F. Sojo, H. M. Jahns,
Ott S., L. G. Sancho,
———.
Øvstedal D. O., R. I. Lewis Smith,
Posada D., K. A. Crandall,
Rambold G., T. Friedl, A. Beck,
Rodriguez F., J. F. Oliver, A. Marín, J. R. Medina,
Rzhetsky A., M. Nei,
Saitou N., M. Nei,
Sancho L. G., L. Kappen, B. Schroeter,
Sancho L. G., B. Schroeter, F. Valladares,
Schroeter B., L. Kappen, T. G. A. Green, R. D. Seppelt,
Seppelt R. D., T. G. A. Green, B. Schroeter,
Skotnicki M. L., J. A. Ninham, P. M. Selkirk,
Skotnicki M. L., P. M. Selkirk, C. Beard,
Swofford D. L.,
White T. J., T. Burns, S. Lee, J. Taylor,