-
PDF
- Split View
-
Views
-
Cite
Cite
Isabel Peixeiro, Ângela Inácio, Cristina Barbosa, Ana Luísa Silva, Stephen A. Liebhaber, Luísa Romão, Interaction of PABPC1 with the translation initiation complex is critical to the NMD resistance of AUG-proximal nonsense mutations, Nucleic Acids Research, Volume 40, Issue 3, 1 February 2012, Pages 1160–1173, https://doi.org/10.1093/nar/gkr820
Close - Share Icon Share
Abstract
Nonsense-mediated mRNA decay (NMD) is a surveillance pathway that recognizes and rapidly degrades mRNAs containing premature termination codons (PTC). The strength of the NMD response appears to reflect multiple determinants on a target mRNA. We have previously reported that mRNAs containing PTCs in close proximity to the translation initiation codon (AUG-proximal PTCs) can substantially evade NMD. Here, we explore the mechanistic basis for this NMD resistance. We demonstrate that translation termination at an AUG-proximal PTC lacks the ribosome stalling that is evident in an NMD-sensitive PTC. This difference is associated with demonstrated interactions of the cytoplasmic poly(A)-binding protein 1, PABPC1, with the cap-binding complex subunit, eIF4G and the 40S recruitment factor eIF3 as well as the ribosome release factor, eRF3. These interactions, in combination, underlie critical 3′–5′ linkage of translation initiation with efficient termination at the AUG-proximal PTC and contribute to an NMD-resistant PTC definition at an early phase of translation elongation.
INTRODUCTION
Nonsense-mediated decay (NMD) targets mRNAs harboring premature translation–termination codons (PTCs) for rapid decay. This surveillance pathway limits the synthesis of potentially deleterious C-terminally truncated proteins encoded by mutant mRNAs (1–4). As such, NMD serves as an important modifier of many genetic disorders (5–7). The NMD pathway also functions as an important determinant of wild-type gene expression with ∼10% of the mammalian transcriptome impacted by components of the NMD apparatus (8–10). A comprehensive description of the determinants and mechanisms of NMD is therefore central to the understanding of both normal and mutant gene expression.
A substantial body of evidence supports a role for the supramolecular exon junction complexes (EJC) in triggering NMD. The EJC is deposited 20- to 24-nt upstream of each exon–exon junction during transcript splicing in the nucleus (11). These complexes are subsequently displaced from the mature mRNA in the cytoplasm by the elongating ribosome (12) during the first round of translation [‘pioneer round of translation’; (13,14)]. If a PTC is located >50–54 nt 5′ to the last exon–exon junction, one or more EJCs will remain beyond the reach of the elongating ribosome and will be retained on the mRNA. The retained EJC(s) can interact with the translation termination complex via bridging interactions between the release complex-associated proteins, UPF1 and SMG-1 (15) and the EJC-associated factors, UPF2–UPF3 (11). This bridging interaction has been proposed to trigger accelerated decay (i.e. NMD) of the PTC-containing mRNA.
While multiple reports support a role for the EJC complex in NMD, an accumulating body of data indicates that additional determinants may play a significant role in this surveillance pathway. Converging lines of evidence from studies in Drosophila melanogaster, Caenorhabditis elegans and Saccharomyces cerevisiae reveal that NMD can be triggered independently of transcript splicing and EJC deposition (9,16). These studies further reveal that 3′-untranslated region (UTR) length and the proximity of the PTC to the cytoplasmic poly(A)-binding protein 1 (PABPC1) may constitute critical determinants of NMD. Studies in yeast support these findings by demonstrating that correct positioning of the termination codon relative to PABPC1 are necessary for efficient termination and NMD resistance (17). Recent studies in mammalian cell-culture support the conclusion that the strength of the NMD response is inversely related to the distance between the PTC and PABPC1 (18,19); shortening this distance by tethering PABPC1 in close proximity to an otherwise NMD-sensitive PTC suppresses NMD, even in the presence of a downstream EJC (18–21). In further support of this model, it has been demonstrated in vitro that PABPC1 can competitively block the association of UPF1 with eRF3 (19) with a corresponding blunting of UPF1 actions and the NMD response (19). Thus the impact of PABPC1 on NMD appears to reflect its ability to interact with, and alter the impact of, supramolecular interactions at the translation termination complex.
We have previously reported that mRNAs containing PTCs in close proximity to the translation initiation AUG codon (AUG-proximal PTCs) escape NMD. This was initially surprising as these mRNAs would be expected to contain residual EJCs and in addition would situate the PTC quite far, in a linear sense, from the poly(A) tail and PABP (22). Detailed analyses of AUG-proximal PTC mRNAs revealed that their observed NMD resistance did not reflect downstream translation reinitiation or extension of ribosome elongation 3′ of the PTC and was instead a direct effect of the termination event being located in close proximity to the AUG (23,24). Based on these studies, and on the observation by others that PABPC1 is able to bind simultaneously to the cap-binding complex subunit eIF4G and to the poly(A) tail, [‘closed-loop’ mRNP configuration (25)], we have proposed a model in which the short open reading frame on an AUG-proximal nonsense-mutated mRNA, situates PABPC1 and its associated cap-binding complex, in close proximity to the PTC. This proximity would allow PABPC1 to alter the structure and/or function of the translation termination complex with a consequent inhibitory effect on NMD (4,21).
In the present report, we further focus on the mechanism of NMD resistance of AUG-proximal PTCs. These studies support the model in which PABPC1 is brought into close proximity with an AUG-proximal PTC via interactions with the translation initiation complexes. This proximity of PABP to the AUG-proximal PTC allows PABP to interact with eRF3 with a consequent enhancement of the release reaction and repression of the NMD response.
MATERIALS AND METHODS
Plasmid constructs
The wild-type β-globin gene (βN), as well as the human β-globin variants β15 and β39 were cloned into the pTRE2pur vector (BD Biosciences) as previously described (24). The pDEST26-PABPC1delC construct was obtained by deleting the DNA fragment encoding the C-terminal portion of PABPC1 from pDEST26-PABPC1 plasmid (IOH13850-pDEST26; RZPD). To accomplish this deletion, the coding sequence of PABPC1, excluding the C-terminal domain (NM 002568, nucleotides 2140–2394, corresponding to C-terminal 85 amino acids), was amplified by PCR from pDEST26-PABPC1 using a pair of primers, one with an AgeI linker (primers #1 and #2; Table 1). After HindIII digestion, the PCR DNA product was inserted into HindIII/AgeI sites of pDEST26-PABPC1. The pUHD-eRF3 plasmid was a generous gift of Marla J. Berry. eRF3 cDNA sequence was then subloned into pcDNA3.1 (Invitrogen). To obtain pcDNA-eRF3delN, the coding sequence of eRF3, excluding the N-terminal domain (X17644, nucleotides 1–648, corresponding to N-terminal 138 amino acids), was amplified by PCR from pcDNAeRF3 using a pair of primers, one with an EcoRI linker and another with an EcoRV linker (primers #3 and #4; Table1) and inserted into EcoRI/EcorV sites on pcDNA3.1/hygro+ (Invitrogen). The plasmid encoding PAIP2, pDEST26-PAIP2 (o834B1117-pDEST26; RZPD) was purchased from BD Bioscience.
DNA oligonucleotides used in the current work
| Primer . | Sequence (5′ → 3′) . |
|---|---|
| #1 | GTGGAAAATCCAAAGGATTTGG |
| #2 | ACCGGTCAAAGGTTCCTGACCTTGTAC |
| #3 | CCGGAATTCGCCATGGAACTTTCAGAACC |
| #4 | AAAGATATCTTAGTCTTTCTCTGGAACCAG |
| #5 | GTGGATCCTGAGAACTTCAGGCT |
| #6 | CAGCACACAGACCAGCACGT |
| #7 | CGCAACCTCCCCTTCTACG |
| #8 | GGTGACGGTGAAGCCGAG |
| #9 | CTGCTCATTGCAGGCCAGAT |
| #10 | GAGCCTGGGCCATGAAGAG |
| #11 | CACCCAGTCATTTTGGCCTC |
| #12 | CGACAGTTCCCAACAGGGTC |
| #13 | GGACAAGCATGGTTTTAGGCA |
| #14 | TGCTGCTCCTGAGTAATTCCC |
| #15 | ATGGCGCAGACGCAGGG |
| #16 | CCGCACTAGGCTGGAACATC |
| #17 | FAM-GCAATGAAAATAAATGTTTTTTATT |
| #18 | FAM-CCCTTGAGGTTGTCCAGGT |
| Primer . | Sequence (5′ → 3′) . |
|---|---|
| #1 | GTGGAAAATCCAAAGGATTTGG |
| #2 | ACCGGTCAAAGGTTCCTGACCTTGTAC |
| #3 | CCGGAATTCGCCATGGAACTTTCAGAACC |
| #4 | AAAGATATCTTAGTCTTTCTCTGGAACCAG |
| #5 | GTGGATCCTGAGAACTTCAGGCT |
| #6 | CAGCACACAGACCAGCACGT |
| #7 | CGCAACCTCCCCTTCTACG |
| #8 | GGTGACGGTGAAGCCGAG |
| #9 | CTGCTCATTGCAGGCCAGAT |
| #10 | GAGCCTGGGCCATGAAGAG |
| #11 | CACCCAGTCATTTTGGCCTC |
| #12 | CGACAGTTCCCAACAGGGTC |
| #13 | GGACAAGCATGGTTTTAGGCA |
| #14 | TGCTGCTCCTGAGTAATTCCC |
| #15 | ATGGCGCAGACGCAGGG |
| #16 | CCGCACTAGGCTGGAACATC |
| #17 | FAM-GCAATGAAAATAAATGTTTTTTATT |
| #18 | FAM-CCCTTGAGGTTGTCCAGGT |
DNA oligonucleotides used in the current work
| Primer . | Sequence (5′ → 3′) . |
|---|---|
| #1 | GTGGAAAATCCAAAGGATTTGG |
| #2 | ACCGGTCAAAGGTTCCTGACCTTGTAC |
| #3 | CCGGAATTCGCCATGGAACTTTCAGAACC |
| #4 | AAAGATATCTTAGTCTTTCTCTGGAACCAG |
| #5 | GTGGATCCTGAGAACTTCAGGCT |
| #6 | CAGCACACAGACCAGCACGT |
| #7 | CGCAACCTCCCCTTCTACG |
| #8 | GGTGACGGTGAAGCCGAG |
| #9 | CTGCTCATTGCAGGCCAGAT |
| #10 | GAGCCTGGGCCATGAAGAG |
| #11 | CACCCAGTCATTTTGGCCTC |
| #12 | CGACAGTTCCCAACAGGGTC |
| #13 | GGACAAGCATGGTTTTAGGCA |
| #14 | TGCTGCTCCTGAGTAATTCCC |
| #15 | ATGGCGCAGACGCAGGG |
| #16 | CCGCACTAGGCTGGAACATC |
| #17 | FAM-GCAATGAAAATAAATGTTTTTTATT |
| #18 | FAM-CCCTTGAGGTTGTCCAGGT |
| Primer . | Sequence (5′ → 3′) . |
|---|---|
| #1 | GTGGAAAATCCAAAGGATTTGG |
| #2 | ACCGGTCAAAGGTTCCTGACCTTGTAC |
| #3 | CCGGAATTCGCCATGGAACTTTCAGAACC |
| #4 | AAAGATATCTTAGTCTTTCTCTGGAACCAG |
| #5 | GTGGATCCTGAGAACTTCAGGCT |
| #6 | CAGCACACAGACCAGCACGT |
| #7 | CGCAACCTCCCCTTCTACG |
| #8 | GGTGACGGTGAAGCCGAG |
| #9 | CTGCTCATTGCAGGCCAGAT |
| #10 | GAGCCTGGGCCATGAAGAG |
| #11 | CACCCAGTCATTTTGGCCTC |
| #12 | CGACAGTTCCCAACAGGGTC |
| #13 | GGACAAGCATGGTTTTAGGCA |
| #14 | TGCTGCTCCTGAGTAATTCCC |
| #15 | ATGGCGCAGACGCAGGG |
| #16 | CCGCACTAGGCTGGAACATC |
| #17 | FAM-GCAATGAAAATAAATGTTTTTTATT |
| #18 | FAM-CCCTTGAGGTTGTCCAGGT |
Cell culture, plasmid and siRNA transfection
HeLa cells, stably expressing the tet transactivator (HeLa/tTA) (26), were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. siRNA duplexes (Table 2) were designed as 19-mers with 3′-dTdT overhangs and purchased from Thermo. Transfections of cells with siRNAs were carried out using Lipofectamine 2000 Transfection Reagent (Invitrogen), following the manufacturer's instructions, in 35-mm plates using 200 pmol of siRNA oligonucleotides and 4 µl of transfection reagent. Twenty-four hours later, cells were transfected with an additional 50 pmol of siRNAs along with 300 ng of the test construct DNA and 1700 ng of pEGFP vector (BD Biosciences) to control for transfection efficiency. Where specified, 1700 ng of pDEST26-PABPC1, pDEST26-PABPC1delC, pcDNA-eRF3delN or 1000 ng of pDEST26-PAIP2 [since in the study of Khalegpour et al. (27), 5000 ng of PAIP2 DNA resulted in a decrease of ∼30% of the reporter translation in HeLa cells] were co-transfected with 300 ng of the test β-globin construct DNA.
Sequences of the siRNAs used in the current work
| siRNA . | Sequence (5′ → 3′) . |
|---|---|
| PABPC11 | UCACUGGCAUGUUGUUGGA |
| PAB3UTR | UUGAUCAGGGACCAUGAAA |
| eRF3a1 | GAGGAACAGUCAUUGUGUG |
| eRF3a2 | UCCUCUCAAGAGGAACAGU |
| UPF1a | AAGAUGCAGUUCCGCUCCAUU |
| UPF1b | GCAGCCACAUUGUAAAUCAUU |
| eIF3h1 | ACUGCCCAAGGAUCUCUCU |
| eIF3h2 | GAUCGGCUUGAAAUUACCA |
| eIF3f1 | GUGAAGGAGAAAUGGGUUU |
| eIF3f2 | AUACGCGUACUACGACACU |
| eIF3e1 | CCAGGGAUGGUAGGAUGCU |
| eIF3e2 | UGCAGAAUUGGGAUGCAGC |
| Luc | CGUACGCGGAAUACUUCGA |
| siRNA . | Sequence (5′ → 3′) . |
|---|---|
| PABPC11 | UCACUGGCAUGUUGUUGGA |
| PAB3UTR | UUGAUCAGGGACCAUGAAA |
| eRF3a1 | GAGGAACAGUCAUUGUGUG |
| eRF3a2 | UCCUCUCAAGAGGAACAGU |
| UPF1a | AAGAUGCAGUUCCGCUCCAUU |
| UPF1b | GCAGCCACAUUGUAAAUCAUU |
| eIF3h1 | ACUGCCCAAGGAUCUCUCU |
| eIF3h2 | GAUCGGCUUGAAAUUACCA |
| eIF3f1 | GUGAAGGAGAAAUGGGUUU |
| eIF3f2 | AUACGCGUACUACGACACU |
| eIF3e1 | CCAGGGAUGGUAGGAUGCU |
| eIF3e2 | UGCAGAAUUGGGAUGCAGC |
| Luc | CGUACGCGGAAUACUUCGA |
Sequences of the siRNAs used in the current work
| siRNA . | Sequence (5′ → 3′) . |
|---|---|
| PABPC11 | UCACUGGCAUGUUGUUGGA |
| PAB3UTR | UUGAUCAGGGACCAUGAAA |
| eRF3a1 | GAGGAACAGUCAUUGUGUG |
| eRF3a2 | UCCUCUCAAGAGGAACAGU |
| UPF1a | AAGAUGCAGUUCCGCUCCAUU |
| UPF1b | GCAGCCACAUUGUAAAUCAUU |
| eIF3h1 | ACUGCCCAAGGAUCUCUCU |
| eIF3h2 | GAUCGGCUUGAAAUUACCA |
| eIF3f1 | GUGAAGGAGAAAUGGGUUU |
| eIF3f2 | AUACGCGUACUACGACACU |
| eIF3e1 | CCAGGGAUGGUAGGAUGCU |
| eIF3e2 | UGCAGAAUUGGGAUGCAGC |
| Luc | CGUACGCGGAAUACUUCGA |
| siRNA . | Sequence (5′ → 3′) . |
|---|---|
| PABPC11 | UCACUGGCAUGUUGUUGGA |
| PAB3UTR | UUGAUCAGGGACCAUGAAA |
| eRF3a1 | GAGGAACAGUCAUUGUGUG |
| eRF3a2 | UCCUCUCAAGAGGAACAGU |
| UPF1a | AAGAUGCAGUUCCGCUCCAUU |
| UPF1b | GCAGCCACAUUGUAAAUCAUU |
| eIF3h1 | ACUGCCCAAGGAUCUCUCU |
| eIF3h2 | GAUCGGCUUGAAAUUACCA |
| eIF3f1 | GUGAAGGAGAAAUGGGUUU |
| eIF3f2 | AUACGCGUACUACGACACU |
| eIF3e1 | CCAGGGAUGGUAGGAUGCU |
| eIF3e2 | UGCAGAAUUGGGAUGCAGC |
| Luc | CGUACGCGGAAUACUUCGA |
RNA isolation
Total RNA from transfected cells was prepared using the Nucleospin RNA extraction II (Marcherey-Nagel) following the manufacturer's instructions.
Reverse transcription-coupled quantitative PCR
Synthesis of cDNA was carried out using 2 µg of total RNA and Superscript II Reverse Transcriptase (Invitrogen), according to the manufacturer's instructions. Real-time PCR was performed in ABI Prism 7000 Sequence Detection System, using SybrGreen Master Mix (Applied Biosystems). Primers specific for β-globin (primers #5 and #6; Table 1) and for puromycin (primers #7 and #8; Table 1) were designed using the ABI Primer Express software. Quantification was performed using the relative standard curve method (ΔΔCt, Applied Biosystems). The following cycling parameters were used: 10 min at 95°C and 40 cycles of 15 s at 95°C and 1 min at 60°C. Technical triplicates from three to four independent experiments were assessed in all cases.
Semiquantitative RT–PCR
A total of 1500 ng of total mRNA were reverse-transcribed (RT) with Superscript II Reverse Transcriptase (Invitrogen) according to the manufacturer's standard protocol and using 250 ng of Random Primers (Invitrogen) in a final volume of 20 µl. The PCR reactions for eIF3h, eIF3f or eIF3e and histone deacetylase 1 (HDAC1) cDNAs were performed in parallel at similar conditions: 3 µl of the RT product was amplified in a 50-µl reaction volume using 0.2 mM dNTPs, 1.5 mM MgCl2, 15 pmol of each primer (primers #9 and #10 for eIF3h, primers #11 and #12 for eIF3f, primers #13 and #14 for eIF3e and primers #15 and #16 for HDAC1; Table 1), 0.75 U of Amplitaq (Promega) and 1 × PCR buffer (Promega). Thermocycler conditions were 95°C for 4 min followed by 26 cycles of 95°C for 45 s, 56°C for 45 s and 72°C for 45 s followed by a final extension of 72°C for 10 min. Ten-microliter aliquots from each RT–PCR sample were analyzed by electrophoresis on 1.8% agarose gels.
SDS–PAGE and western blotting
Protein lysates were resolved, according to standard protocols, in 10% SDS–PAGE and transferred to PVDF membranes (Bio-Rad). Membranes were probed using mouse monoclonal anti-α-tubulin (Sigma) at 1:10 000 dilution, goat polyclonal anti-hUPF1 (Bethyl Labs) at 1:500 dilution, rabbit polyclonal anti-PABPC1 N-terminal domain (Cell Signaling) at 1:500 dilution, rabbit polyclonal anti-eRF3 (Abcam) at 1:500 dilution, goat polyclonal anti-PAIP2 (Santa Cruz) at 1:250 dilution or anti-GFP (Abcam) at 1:5000 dilution. Detection was carried out using secondary peroxidase-conjugated anti-mouse IgG (Bio-Rad), anti-rabbit IgG (Bio-Rad) or anti-goat IgG (Sigma) antibodies followed by chemiluminescence.
Toeprinting assay
The #17 or #18 primers (Table 1) were synthesized and end labeled with 5′-FAM (Invitrogen). Twenty picomoles of primer and 200 ng of capped and polyadenylated mRNA were combined in 50 mM Tris–HCl, pH 7.5, heated to 68°C for 2 min and cooled to 37°C for 8 min and immediately added to the translation mixture, comprising 60% rabbit reticulocyte lysate (RRL) (Promega), 28 mM AcK, 180 μM MgCl2, 102 μg/ml creatine phosphokinase (Sigma), 6 mM creatine phosphate (Sigma), 24 mM KCl, 15 mM Hepes, 27 μM amino acid mixture (Promega) and 0.6 U/μl RNase inhibitor (Invitrogen). Where appropriated, 500 μg/ml cycloheximide (Sigma) was added before the reaction. The reactions were incubated at 30°C for 20 min. For the primer extension stage, 4 μl of the translation reaction were supplemented with 50 mM Tris–HCl, pH 7.5, 40 mM KCl, 3.5 mM MgCl2, 5 mM DTT, 0.8 mM each dNTP, 500 μg/ml cycloheximide, 1.5 U/μl ribonuclease inhibitor and 5 U/μl Superscript II reverse transcriptase (Invitrogen) in a final volume of 20 μl and placed at 37°C for 10 min. Primer extension products were extracted with phenol, and the ethanol-precipitated pellets were resuspended in 3 μl of Hi-Di Formamide (Applied Biosystems) and brought to a total volume of 12 μl aliquot with Hi-Di Formamide, which included 0.5 μl ROX 500 or 2500 size standard (Applied Biosystems). The products were separated by electrophoresis using standard GeneScan conditions and analyzed using the GeneMapper Software—version 3.7 (Applied Biosystems).
RESULTS
Translation termination at an AUG-proximal PTC obviates ribosome stalling characteristic of NMD-sensitive PTCs
Given the demonstrated linkage between NMD and defects in translation termination (17,19,28), we hypothesized that translation termination at an NMD-resistant AUG-proximal PTC might be more efficient than that at a more distal NMD-sensitive PTC. This possibility was explored by an in vitro ‘toeprinting’ analysis. We compared the residency of terminating ribosomes at the termination codon of the wild-type β-globin mRNA (βN), at a NMD resistant ‘AUG-proximal’ stop codon (β15), and at a more distal NMD sensitive PTC (β39). Capped, polyadenylated βN, β15 and β39 mRNAs were each incubated in translationally active RRL followed by primer extension using a fluorescently labeled primer (#17 primer; Figure 1A and Table 1) and analysis of reverse transcription products by capillary electrophoresis (29,30) (Figure 1B). Parallel controls included incubation of the mRNAs in RRL containing cycloheximide (CHX) and incubation in buffers in the absence of RRL. CHX at the levels used, blocks peptidyl transferase and should therefore ablate toeprinting signals corresponding to the terminating ribosome and accentuate signals corresponding to ribosomes blocked at the initiation AUG codon (‘AUG toeprint’). mRNA incubated in the absence of RRL was used to distinguish genuine ribosome-generated toeprints (TP) from primer extension ‘short-stops’ due to mRNA secondary structures.
Translation termination at the AUG-proximal β15 PTC occurs in the absence of the ribosome stalling evident at the more distal β39 PTC. (A) Diagram of the human β-globin mRNA showing positions of initiation and termination codons [native (N) or premature (at position 15 or 39)]. The arrow indicates the position and orientation of the #17 primer (Table 1) used in the primer-extension (toeprinting) assays. The lines below the mRNA diagram represent the length in nucleotides of the synthesized cDNA from the #17 primer to the 5′-end of the mRNA, the initiation AUG codon, or to the βN, β15 and β39 translation termination codons. (B) Electropherograms of βN, β15 and β39 toeprint assays using the fluorescently labeled #17 primer. The toeprint reactions were performed in the presence or absence of cycloheximide (+CHX or −CHX, respectively). A control reaction was carried out in the absence of added rabbit reticulocyte lysate (−RRL). Primer extension products were suspended in a mix containing ROX 2500 molecular weight marker (Applied Biosystems) and subjected to capillary electrophoresis. Size standard peaks are shown below and sequence position is indicated in nucleotides. The toeprint peaks at 626 nt represent the full-length transcript; the peaks at 620 nt correspond to the cap-binding complex-bound full-length transcript; the peaks at 555-nt map 18-nt downstream the AUG codon (AUG TP). The peak at 440-nt maps to a position 16 nt downstream the β39 stop codon (PTC TP). (C) The arrow below the mRNA diagram represents position and orientation of #18 primer (Table 1) used in the second, high-resolution toeprinting assays. The lines below the mRNA diagram represent the length in nucleotides of the primer-extended cDNA to the 5′-end of the mRNA, to the initiation AUG, as well as to the translation termination codons at positions 15 or 39. (D) Electropherograms of β15 and β39 toeprint assays using the fluorescently labeled #18 primer. A parallel sequencing reaction on the βN cDNA, performed with the same primer is shown on the top. Underlined sequences indicate codon position: 0 (AUG), 15 and 39. The toeprint reactions were performed in the presence or absence of cycloheximide (+CHX or −CHX, respectively). A control reaction lacking −RRL is shown. Primer extension and sequencing products were resuspended in a mix containing ROX 500 (Applied Biosystems) molecular weight standard and separated by capillary electrophoresis. Sequence position is indicated in nucleotides. Peaks at 230 nt correspond to the initiation AUG toeprint that maps 18-nt downstream the AUG codon. The peak at 115 nt, present uniquely in the β39 analysis, maps 16-nt downstream of the PTC.
The toeprinting analyses of βN, β15 and β39 mRNAs under each of the three incubation conditions are summarized in Figure 1B. The predominant peak at 626 nt observed on each of the three mRNAs represents the full-length transcripts. A second peak corresponding to the initiation AUG toeprint (at 555 nt, AUG TP) is seen for all three mRNAs (βN, β15 and β39 mRNAs), and in each case it is appropriately enhanced by CHX. A toeprint at 440 nt is specifically observed on the β39 mRNA incubated in the RRL. This band, mapping to a position 16 nt 3′ to codon 39, is ablated in the presence of CHX and is not generated in the –RRL incubation. The position of this toeprint is consistent with the presence of a stalled ribosome at the β39 termination codon (PTC TP; Figure 1B). The parallel analysis of the βN and β15 mRNAs failed to reveal and extension product of 116 or 512 nt that could be ascribed to ribosome stalling at the normal stop codon or at codon 15, respectively.
To confirm and extend the initial toeprinting study, a reanalysis of the β15 and β39 transcripts was performed with a primer positioned closer to the PTCs to achieve higher resolution (#18 primer; Figure 1C and Table 1). The β15 and β39 mRNAs both generate a peak at 230 nt (Figure 1D) that is accentuated in the presence of CHX (AUG TP; Figure 1D). This product maps to a position 18 nt 3′ the AUG codon, as confirmed by direct sequencing with the same primer (Figure 1D). Additional CHX-independent peaks, observed for both transcripts in the presence but not in the absence of RRL most likely represent mRNA regions where mRNA binding proteins or structures impede cDNA synthesis. The analysis of the β39 mRNA generated an additional peak at 115 nt that was not seen in the presence of CHX; this signal corresponds to a toeprint at 16 nt downstream the PTC at codon 39 (PTC TP; Figure 1D). No corresponding band was observed immediately 3′ to codon 15 of the β15 mRNA. It is unlikely that the absence of a toeprint corresponding to the β15 PTC is due to inefficient translation because these transcripts show a normal initiator AUG toeprint that responds positively to CHX (Figure 1B and D). Together, these in vitro studies reveal aberrant ribosome termination/release at the β39 PTC that is not seen at the β15 PTC.
PABPC1 plays an essential role in establishing NMD resistance of an AUG-proximal nonsense-mutated mRNA
Prior studies have demonstrated that PABPC1 can associate simultaneously with the cap-associated eIF4G and the poly(A) tail (25). This dual binding has the potential to bridge the 3′ and 5′ termini of an mRNA resulting in a circularized or ‘closed loop’ conformation (25). Such a conformation would bring PABPC1 into close proximity with 5′-terminal translation complexes. In related studies, PABPC1 has been shown to inhibit NMD when located close to the PTC (18–21) and this effect is dependent on its C-terminal domain (21). With these observations in mind, we hypothesized that PABPC1 might be brought into the vicinity of the AUG-proximal PTC during cap-dependent translation by binding to and tracking with the initiation complex. This would allow it to interact with eRF3 and repress NMD. To test this model, HeLa cells were transfected with a siRNA targeting PABPC1. Twenty-four hours later, these cells were transiently co-transfected with a plasmid-encoding PABPC1 lacking the 85 residues of the C-terminal segment critical to eRF3 interactions (31), (PABPdelC, see ‘Materials and Methods’ section) and with plasmid encoding βN, β15 or β39 mRNAs (Figure 2A). Levels of endogenous PABPC1 and exogenous PABPdelC proteins were monitored by western blot (Figure 2B, lanes 4–6) and the impact of altering the levels and structure of the PABPC1 on the expression of the various β-globin mRNAs was monitored by RT–quantitative PCR (RT–qPCR) (Figure 2C). The studies were controlled by a parallel study in cells treated with a control (Luciferase; LUC) siRNA (Figure 2B, lanes 1–3). The data revealed that expression of the C-terminally deleted PABPC1 in PABPC1-depleted cells (Figure 2B, Lanes 4–6) results in a significant decrease on β15 mRNA (from 103% to 70% of βN). In contrast, the same alteration in PABP constitution fails to alter β39 mRNA expression levels (Figure 2C). These observations are consistent with the model that NMD evasion of β15 transcripts is dependent on PABPC1, and specifically on a function mediated by its C-terminal domain. Efforts at testing the single effect of endogenous PABPC1 depletion on the NMD resistance of β15 transcripts failed because HeLa cells became detached from plates after the second transfection. The apparent toxicity of the siRNA-mediated PABPC1 depletion suggests that the knockdown of this critical factor was effective (our unpublished data).
PABPC1 plays an essential role in NMD resistance of an AUG-proximal PTC. (A) Diagram representing βN, the NMD-resistant β-globin mRNA with an AUG-proximal nonsense mutation at codon 15 (UGA; β15) and the NMD-sensitive β39 mRNA. The positions of initiation (AUG) and termination (native and premature) codons are indicated. (B) Representative western blot analysis of HeLa cells extracts transfected with human PABPC1 siRNA (lanes 4–6) or with a control Luciferase siRNA target (LUC siRNA; lanes 1–3). Twenty-four hours after siRNA treatment, cells were transfected with the β-globin reporter constructs (βN, β15 or β39) with or without a plasmid expressing PABPdelC mutant protein (pDESTPABPC1delC plasmid); lanes 4–6 or 1–3, respectively. Twenty-four hours post-transfection, protein and RNA were isolated from the cells for analysis. Immunoblotting to confirm PABPC1 knockdown was carried out with anti-PABPC1 (specific for the N-terminal domain) and with anti-α-tubulin antibodies as a loading control (lanes 4–6 versus lanes 1–3). Identification of each band is on the right. (C) Depletion of endogenous PABPC1 in conjunction with expression of exogenous PABPdelC protein represses β15 mRNA levels. Relative β-globin mRNA levels under control conditions (LUC siRNA-treated HeLa cells; dark bars) and at conditions of PABPdelC expression in PABPC1-depleted HeLa cells (PABPC1 siRNA + PABPdelC overexpression; light bars), normalized to the levels of puromycin resistance mRNA (Puror; plasmids carrying the reporter β-globin gene also contain the Puror gene), were determined by quantitative RT–qPCR and compared to the corresponding βN mRNA levels (defined as 100%). Average and standard deviation (SD) of three independent experiments corresponding to three independent transfections are shown in the histogram. (D) Representative western blot analysis of HeLa cells extracts transfected with control Luciferase siRNA (LUC siRNA; lanes 1–3) or with siRNA targeting the human PABPC1 3′-UTR (PABPC1 3′-UTR siRNA; lanes 4–9). After siRNA treatment, cells were transfected with the plasmids expressing βN, β15 or β39 mRNAs (lanes 1–3) or cotransfected with these plasmids in combination with a plasmid expressing PABPdelC mutant protein [pDESTPABPC1delC plasmid as above in (B); lanes 4–6], or with a plasmid expressing wild-type PABPC1 (encoded by an mRNA with an heterologous 3′-UTR resistant to the PABPC1 siRNA; lanes 7–9). Protein levels present in the cell extracts were analyzed by western blot for PABPC1 and α-tubulin (loading control) as in (B), to monitor endogenous PABPC1 knockdown (lanes 4–9 versus lanes 1–3) and exogenous expression of mutant PABPdelC protein (lanes 4–6) or PABPC1 expression rescue (lanes 7–9). Identification of each band is indicated to the right of the gel image. The histogram below the immunoblot shows the average and SD values of three independent experiments for quantification of relative PABPC1 protein levels. (E) Expression of wild-type PABPC, but not PABPdelC, restores β15 expression in PABPC1 depleted cells. Relative β-globin mRNA levels for the control conditions (LUC siRNA-treated cells; dark bars), at conditions of PABPdelC expression in PABPC1-depleted HeLa cells (PABPC1 siRNA + PABPdelC overexpression; light bars), and at conditions of PABPC1 rescue (PABPC1 siRNA-treated cells plus expression of exogenous PABPC1; light bars) were determined by RT–qPCR and compared to the corresponding βN mRNA levels as in (C). Average values and SD of three independent experiments corresponding to three independent transfections are shown in the histogram. All values are represented as a percentage (%) of the corresponding βN mRNA (defined as 100%).
The preceding experiment was repeated by directly comparing the impact of the native PABPC1 versus the PABPdelC mutant in the cells depleted of endogenous PABPC1 (Figure 2D and E). HeLa cells were transfected with a siRNA targeting the 3′-UTR region of PABPC1-encoding mRNA (PAB3UTR; Table 2). Twenty-four hours later, the PABPC1-depleted HeLa cells were co-transfected with plasmids encoding the β-globin reporters and with a plasmid-encoding PABPdelC protein (Figure 2D, lanes 4–6) or encoding exogenous PABPC1 (with a 3′-UTR resistant to PABPC1-directed siRNA, Figure 2D, lanes 7–9). Levels of β-globin mRNAs were measured by RT–qPCR (Figure 2E). As before, we observed that depletion of PABPC1, followed by expression of PABPdelC (Figure 2D, lanes 4–6 versus lanes 1–3), decreased β15 mRNA levels while the corresponding levels of the β39 mRNA remained unaltered (Figure 2F). In contrast, repletion of the cells with wild-type PABPC1 from ∼40–50% to ∼75–95% of the normal protein levels (Figure 4D, lanes 7–9 versus lanes 4–6) fully restored β15 mRNA levels so that they were comparable to βN mRNA levels, while β39 mRNA remained at low levels (Figure 2E). Together, these results demonstrate that the NMD resistance of the β15 RNA is dependent on the action of PABPC1 and in particular on the function of its C-terminal domain.
Interaction of PABPC1 with the ribosome release factor, eRF3, is critical to NMD evasion by an AUG-proximal nonsense-mutated mRNA
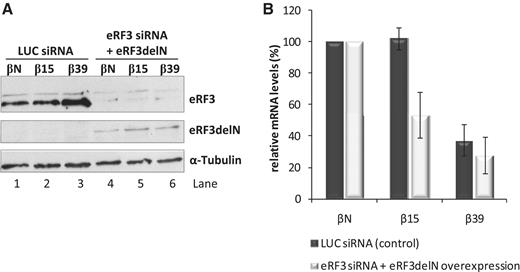
Prior studies have revealed that PABPC1 can bind to eRF3 (15,31–36). This binding blocks UPF1 binding to the release complex. PABPC1 is thus able to antagonize the UPF1–eRF3 interaction involved in NMD (19). PABPC1 interacts with the N-terminal domain of eRF3 (33,37), while UPF1 interacts with the non-overlapping eRF3 GTPase domain (20). To further test the model that NMD resistance of the AUG-proximal nonsense-mutated transcripts relies on the PABPC1–eRF3 interaction, we depleted HeLa cells of eRF3 by siRNA treatment and subsequently co-transfected these cells with plasmids bearing the β-globin reporters genes (βN, β15 or β39) and with a plasmid encoding an eRF3 mutant protein lacking its N-terminal domain [residues 1–183 (eRF3delN)]. Importantly, this deletion does not impair eRF3–eRF1 or eRF3–UPF1 interactions (20,34). eRF3 levels as monitored by western blot were depleted to ∼25% of wild-type levels (Figure 3A, lanes 4–6 versus lanes 1–3). Under these eRF3-depleted conditions, the expression eRF3delN resulted in a significant repression of β15 mRNAs to 55% of βN mRNA levels, while levels of β39 transcript were not appreciably altered (Figure 3B). These results, along with those summarized in Figure 2, lead us to conclude that an AUG-proximal nonsense-mutated mRNA can evade NMD through a mechanism that is dependent on the interaction of PABPC1 with eRF3.
The N-terminal domain of eRF3 is critical to NMD evasion by an AUG-proximal nonsense-mutated mRNA. (A) Western blot analysis of HeLa cells extracts transfected with human eRF3 siRNA (lanes 4–6) or with a control Luciferase siRNA target (LUC siRNA; lanes 1–3). After siRNA treatment, cells were transfected with plasmids expressing βN, β15 or β39 mRNAs with or without a plasmid expressing eRF3delN mutant protein (pcDNAeRF3delN plasmid); lanes 4–6 or 1–3, respectively. Twenty-four hours post-transfection, protein and RNA were isolated from the cells. Immunoblotting was carried out with anti-eRF3 to monitor endogenous eRF3 knockdown (lanes 4–6 versus lanes 1–3) and expression of mutant eRF3delN protein (lanes 4–6). Detection of α-tubulin served as a loading control. Identification of each band is on the right. (B) Depletion of endogenous eRF3 in combination with expression of exogenous eRF3delN represses β15 mRNA levels. Relative levels of β-globin mRNA under control conditions (LUC siRNA-treated cells; dark bars) and in eRF3-depleted cells expressing exogenous eRF3delN protein (eRF3 siRNA + eRF3delN overexpression; light bars), normalized to the levels of Puror mRNA expressed from the β-globin plasmids, were determined by quantitative RT–PCR and compared to the corresponding βN mRNA levels (defined as 100%). Average and SD values of four independent experiments corresponding to three independent transfections are shown in the histogram.
Inhibition of PABPC1 function by PAIP2 sensitizes an AUG-proximal nonsense-mutated mRNA to NMD
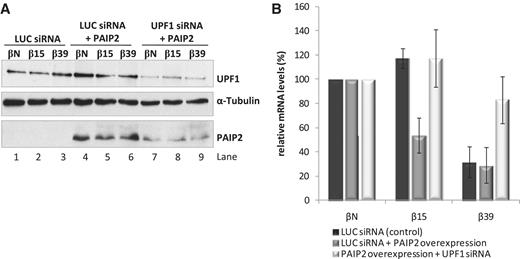
The closed loop configuration of a capped mRNA is dependent on the PABPC1–eIF4G interaction. The PABP-interacting protein 2, PAIP2, competitively blocks this interaction via direct binding to PABPC1 (38). We therefore reasoned that over-expression of PAIP2 might sensitize an AUG-proximal nonsense-mutated mRNA to NMD by inhibition of the closed loop conformation. To ensure that the PAIP2 over-expression was not having a secondary effect on NMD efficiency via a repression of translation, we limited our studies to low concentrations of PAIP2 DNA (see ‘Materials and Methods’ section). HeLa cells were transiently co-transfected with the plasmids encoding βN, β15, or β39 mRNAs along with the plasmid-encoding PAIP2. PAIP2 protein levels were monitored by western blot (Figure 4A; lanes 1–3 versus lanes 4–9) and levels of β-globin mRNAs were quantified by RT–qPCR (Figure 4B). PAIP2 expression induced a substantial decrease in β15 mRNA levels from 117% to 53% of the corresponding βN mRNA levels, without altering the expression of the β39 mRNA (Figure 4B). To confirm that the decrease of β15 mRNA levels seen in the PAIP2 treated cells reflected NMD, the experiment was repeated in UPF1-depleted HeLa cells. The UPF1 depletion resulted in the expected increase in β39 mRNA and, in addition, reversed the repressive effect of the PAIP2 over-expression on β15 mRNA (Figure 4A and B). These data lead us to conclude that an AUG-proximal nonsense-mutated mRNA can be sensitized to NMD by blocking the interaction of PABPC1 with eIF4G.
Overexpression of PAIP2 induces NMD sensitivity in the β15 nonsense-mutated mRNA. (A) Western blot analysis of HeLa cells treated with siRNAs specific to Luciferase (LUC) (lanes 1–6), or siRNA targeting UPF1 (lanes 7–9). After siRNA treatment, cells were transfected with plasmids expressing βN, β15 or β39 mRNAs with or without a plasmid expressing PAIP2 protein (pDEST26PAIP2 plasmid); lanes 4–9 or 1–3, respectively. Twenty-four hours post-transfection, protein and RNA were isolated from the cells for analysis. Immunoblotting was carried out with anti-UPF1, anti-PAIP2 and anti-α-tubulin antibodies. Detection of α-tubulin served as a loading control. Identification of each band is indicated to the right of the gel image. (B) Overexpression of PAIP2 represses β15 mRNA levels in a UPF1-sensitive manner. Relative β-globin mRNA levels under control conditions (dark bars), PAIP2 overexpression (dark grey bars), and in conditions of PAIP2 overexpression co-existing with UPF1 depletion (light bars) are shown. All values determined by RT–qPCR are normalized to the mRNA levels of Puror, and compared to the corresponding βN mRNA levels. Average values and SD of four independent experiments corresponding to four independent transfections are shown. All values are represented as a percentage (%) of the corresponding βN mRNA (defined as 100%).
The eIF3 subunits that tether the 40S ribosomal subunit to eIF4G are required for NMD resistance of an AUG-proximal nonsense-mutated mRNA
Interactions between mRNA 5′-end and the poly(A) tail are thought to be established during the initial round of translation. These interactions involve interactions of PABPC1 with the initiation factor eIF4G. eIF4G can simultaneously interact with PABPC1 and with the 40S recruitment factor, eIF3 (39). Mammalian eIF3 is composed of 13 subunits designated eIF3a to eIF3m (40–42). The structure and organization of this multisubunit has not been complete elucidated, although crystallography, mass spectrometry and immunoprecipitation studies allows the prediction of an interaction map of the free eIF3 as it is represented in Figure 5A [adapted from (40–42)]. The eIF3–eIF4G interaction is established via eIF3e and eIF3f subunits (40,43). This interaction of eIF4G with eIF3 has the potential to retain the eIF4G-bound PABPC1 on the scanning 40S ribosome and on the 80S ribosome during the early phase of elongation. Besides, in plants, the eIF3h subunit helps to prevent the permanent loss of reinitiation competence (44), suggesting that this subunit might also be involved in tethering 40S to eIF4G. Such an activity could blunt NMD of an AUG-proximal nonsense-mutated mRNA by bringing the PABPC1 into close proximity with the PTC. To test this hypothesis, we assessed the impact of depleting eIF3 subunits on the expression of an AUG-proximal PTC mRNA. Efficient knockdown was confirmed at the mRNA level for each respective eIF3 subunit mRNA, by normalization to mRNA levels of the histone deacetylase 1 (HDAC1) (Figure 5B–D). HDAC1 mRNA was chosen as an internal control for these analyses since it is constitutively expressed (45). We further demonstrated that under the experimental conditions used in the study, these depletions did not alter cellular translation as assessed by GFP reporter expression (Figure 5E, lanes 3–5 versus lane 2 and 1).
The translation initiation factors eIF3h and eIF3f are required for the NMD-resistant phenotype of the β15 nonsense-mutated mRNA. (A) Representation of the predicted organization of the mammalian eIF3 subunits [adapted from (40–42)]. Free eIF3 complex can be assigned into three stable modules. One module consists of eIF3a, b, g and i, and it interacts with a second module composed by eIF3c, d, e, l and k. eIF3b functions as a scaffold protein connecting eIF3a, c, i and g subunits. The third subcomplex is composed by eIF3f, h and m. Subunits eIF3f and m bind to the subcomplex eIF3 c:d:e:l:k through subunit eIF3h. The remaining subunit eIF3j, a labile subunit, attaches to the complex via eIF3b (40–42). (B–D) Representative RT–PCR analyses of RNAs extracted from untreated (Ctl; lane 1), Luciferase (LUC; lane 2) or eIF3h, eIF3f and eIF3e (lane 3) siRNAs-treated HeLa cells, respectively, in panels (B–D). RT–PCRs were carried out with eIF3h, eIF3f or eIF3e mRNA specific primers to monitor endogenous eIF3h, eIF3f or eIF3e knockdown, respectively (lanes 3 versus lanes 1–2). The eIF3h, eIF3f and eIF3e mRNA levels were normalized to those of histone deacetylase 1 (HDAC1) mRNA level. In each panel, the right three lanes correspond to serial dilutions of RNA, demonstrating semiquantitative conditions used for RT–PCR. (E) Representative western blot analysis of HeLa cells extracts, untreated (Ctl, lane 1) or treated with Luciferase (LUC; lane 2), eIF3h (lane 3), eIF3f (lane 4) or eIF3e (lane 5) specific siRNAs (see ‘Material and Methods’ section). After siRNA treatment, cells were transiently transfected with the pEGFP plasmid (BD Biosciences) expressing green fluorescent protein (GFP). Protein lysates were analyzed by immunoblotting using anti-GFP and anti-α-tubulin (loading control) antibodies to monitor GFP protein expression. Identification of each band is indicated to the right of the gel image. (F, H and J) Semiquantitative RT–PCR analyses of RNAs extracted from HeLa cells transfected with a Luciferase (LUC) siRNA target (lanes 1–3) or human eIF3h, eIF3f or eIF3e siRNAs, respectively, at panels (F, H or J) (lanes 4–6 of each panel), using the same experimental settings as in (B). Twenty-four hours after siRNA-treatment, cells were transfected with plasmids expressing βN, β15 or β39 mRNAs. Twenty-four hours after constructs transfection, total RNA was isolated from the cells. RT–PCR was carried out as in (B) with eIF3h, eIF3f or eIF3e mRNA specific primers, respectively, to monitor endogenous eIF3h, eIF3f or eIF3e knockdown (lanes 4–6 versus lanes 1–3). (G) Knockdown of eIF3h represses β15 mRNA levels. Relative β-globin mRNA levels for the control conditions (LUC siRNA; dark bars) and at conditions of eIF3h depletion (eIF3h siRNA; light bars), normalized to the levels of puromycin resistance mRNA encoded from the β-globin gene plasmid, were determined by quantitative RT–PCR (RT–qPCR) and compared to the corresponding βN mRNA levels (defined as 100%). (I) Knockdown of eIF3f represses β15 mRNA levels. Relative β-globin mRNA levels for the control conditions (LUC siRNA; dark bars) and at conditions of eIF3f depletion (eIF3f siRNA; light bars), normalized to the levels of puromycin resistance mRNA were determined by RT–qPCR and compared to the corresponding βN mRNA levels (defined as 100%). (K) Knockdown of eIF3e fails to repress β15 mRNA expression. Relative β-globin mRNA levels for the control conditions (LUC siRNA; dark bars) and at conditions of eIF3e depletion (eIF3e siRNA; light bars), normalized to the levels of puromycin resistance mRNA were determined by RT–qPCR as in (G) and compared to the corresponding βN mRNA levels (defined as 100%). (G, I and K) histograms show average and SD values of three independent experiments corresponding to three independent transfections.
We next evaluated the effect of individually depleting each of the three eIF3 subunits on NMD. Genes encoding the β15, β39 and βN mRNAs were each transiently expressed in eIF3h-, eIF3f- or eIF3e-depleted HeLa cells. β-globin mRNA levels, quantified by RT–qPCR, revealed that eIF3h depletion (Figure 5F, lanes 4–6 versus lanes 1–3) results in a decrease of β15 mRNA level from 113% to 79% of the corresponding normal control without a corresponding effect on the β39 mRNA (Figure 5G). A similar selective repression of β15 mRNA was observed in eIF3f-depleted cells (Figure 5H and I). In contrast, depletion of eIF3e (Figure 5J) had no appreciable effect on β15 NMD-resistant transcript levels (Figure 5K). Instead, this depletion results in an increase in β39 mRNA levels [from 37% to 59% of βN; (Figure 5K)]. This later result is consistent with prior observations that the eIF3e subunit may be required, at least in part, for NMD-commitment (46). The destabilizing effect of eIF3h and eIF3f depletions on the β15 mRNA, together with the suggestion that eIF3 remains bound to the translating ribosome for a few peptide bonds (47,48) are consistent with the model where eIF3h and eIF3f have a role in bringing PABPC1 in association with eIF4G into the vicinity of the PTC of an mRNA with a short open reading frame. A schematic of this model is shown in Figure 6.
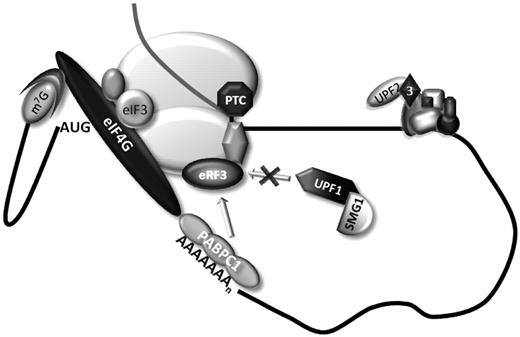
A model for NMD resistance of AUG-proximal nonsense-mutated mRNAs. The current and prior data supports the model shown in this figure. During cap-mediated translation initiation, PABPC1 interacts with the initiation factor eIF4G. This interaction indirectly tethers PABPC1 to the 40S ribosomal subunit via the interaction of eIF4G with eIF3 subunits. The resulting configuration brings PABPC1 into in the vicinity of the AUG initiation codon as a consequence of 43S scanning and the maintenance of eIF4G–PABPC1 association with the 40S during the initial phase of translation elongation brings it into close contact with an AUG-proximal PTC in a transcript where the ORF is quite short. This proximity to the PTC allows PABPC1 to interact with the release factor eRF3 at the termination complex, thus impairing the association of UPF1 to the ribonucleoprotein complex, resulting in efficient translation termination and inhibition of NMD.
DISCUSSION
We have previously reported that mammalian mRNAs bearing AUG-proximal PTCs evade NMD (22,24). This immunity to NMD is independent of sequence context of the open reading frame and PTC, independent of transcript identity, and appears to be a general attribute of the mammalian NMD pathway (24). Extensive mapping studies, using the human β-globin gene as a primary model, have revealed that this effect reflects a close proximity of a PTC to the translation initiation codon (23,24). In the case of the β-globin mRNA, PTCs that occur up to 23 codons into the open reading frame effectively evade NMD while mRNAs with PTCs at a greater distance from the AUG are targeted for decay. We have further demonstrated that NMD evasion by AUG-proximal PTCs does not, as a rule, reflect defects in transcript splicing, impaired translation or reinitiation of translation 3′ to the PTC (23,24). Thus, the mechanism underlying the NMD resistance of the AUG-proximal nonsense-mutated mRNA remains to be more clearly delineated.
The potential role of translation reinitiation in NMD resistance of AUG-proximal PTCs has been the subject of a recent report. In that study (49) the NMD resistance of β-globin AUG-proximal nonsense-mutated mRNAs was attributed to translation reinitiation 3′ to the PTC. It may be of importance, however, to note that the interpretation of these studies is complicated by the use of a β-globin gene in which exon 1 was transformed into a functional exon 2 by the introduction of an exogenous intron into the 5′-UTR. The expression of this recombinant β-globin gene, even lacking a PTC, was repressed when compared to the normal β-globin transcript. In contrast, we have found using globin genes with native structures, that even in cases in which we do detect some level of reinitiation 3′ to the AUG-proximal PTC, site-specific elimination of the reinitiation sites fails to fully restore NMD sensitivity (24). Thus, while a contribution of translation reinitiation in NMD evasion cannot be completely ruled out in any particular circumstance, our detailed analyses of the human β-globin gene lead us to conclude that AUG proximity is a major inhibitor of the NMD pathway, but other mechanisms can be considered.
The present data points to a major role for PABPC1 in protecting mRNAs harboring AUG-proximal PTCs from the NMD surveillance pathway (4,21). A critical contribution of interactions between PABPC1 and components of translation initiation complexes is highlighted by the dependence of the NMD evasion on interactions of PABPC1 with the eIF4G and most probably of eIF4G with eIF3 subunits. The data further reveal that these interactions are linked to the establishment of an efficient translation termination reaction at the AUG-proximal PTC. Toeprinting analysis of capped and polyadenylated human β-globin transcripts reveals substantial ribosome stalling at the NMD sensitive β39 stop codon that is absent at the termination codon of the β15 mRNA, as well as at the stop codon of the normal transcript (Figure 1). The results of these studies lead us to conclude that the interactions of PABPC1 with the pre-initiation complex and 40S ribosome place it in a favorable context to enhance translation termination efficiency.
How does the positioning of the PABPC1 at the 5′-end of the mRNA specifically blunt the NMD response on an AUG-proximal nonsense-mutated mRNA? Our current observations lead us to propose that this repositioned PABPC1 can enhance the efficiency of the translation termination at the AUG-proximal PTC and in so doing represses NMD. It has been demonstrated by others that PABPC1 can compete with UPF1 for binding to eRF3 with a consequent antagonistic effect on NMD response (19,20). Recruitment of PABPC1 to the 5′-end of the mRNA, with retention of ribosome association during the early phase of elongation, would position PABPC1 in close proximity to an early PTC. An impact on the termination reaction is supported by our prior demonstration that AUG-proximal nonsense-mutated transcripts fail to bind UPF1 as efficiently as NMD-sensitive transcripts (21). The interactions that support the repositioning of PABPC1 close to an AUG-proximal PTC are further defined by the in vitro observation that interactions of PABPC1 with eIF4G, eIF4E and eIF3, and the termination factors eRF1 and eRF3 can form a closed loop structure during translation of short mRNAs (39). The proposed role for PABPC1 in blunting NMD on AUG-proximal nonsense-mutated mRNAs is also consistent with the observation by ourselves and others of an inverse relationship between the distance among the PTC and an artificially tethered PABPC1 and the strength of NMD [for review, see (2,4)]. It is worth noting that the destabilization of transcripts bearing AUG-proximal PTCs obtained by the impairment of PABPC1–eRF3 interaction corroborates the importance of the AUG-proximity effect in NMD-evasion of such transcripts.
In the case of the AUG-proximal nonsense-mutated mRNA, our data support the conclusion that the PABPC1–eIF4G interaction enables PABPC1 to travel with the eIF4F/43S complex, as it scans from the cap to the AUG. The dependence of the AUG-proximity effect on the function of eIF3 (Figure 5), which bridges 43S complex to eIF4G (48), further supports this model. Thus, we provide evidence that eIF4G and eIF3 initiation factors are both implicated in PABPC1 inhibitory effect on NMD. This model is further supported by the observation that the interruption of the PABPC1–eIF4G interaction by over-expression of PAIP2 leads to NMD of transcripts bearing an AUG-proximal PTC (Figure 4). The observations that eIF3 can remain bound to the translating ribosome during the initial phase of elongation (47,48) and that it can function as the link between the eIF4F-bound mRNA (through eIF4G) and the 40S ribosomal subunit (43), supports the notion that eIF3 subunits might be involved in the delivery of eIF4G-associated PABPC1 to the vicinity of the AUG-proximal PTC. The strength of the NMD effect decay would then be determined by the physical distance between PABPC1 at the AUG vicinity and the PTC; the closer this distance, the more effectively PABPC1 could interact with eRF3, diminishing the binding of UPF1, with the consequent enhancement of translation termination and blunting of the NMD response (Figure 6).
Considering that mRNA circularization (25,50) and PABPC1 function as NMD inhibitor when located in the PTC proximity (17–21) are common features in all eukaryotes, the model for the AUG-proximity effect that we propose here might be also a general attribute for these organisms. Here, we show that the human eIF3h and eIF3f subunits, which are not conserved in S. cerevisiae, have a role in bringing PABPC1 in association with eIF4G into the vicinity of an AUG-proximal PTC. While the individual functions of the different mammalian subunits are not yet well established, many of the eIF3 subunits are thought to be conserved (40–42,48). Indeed, the five subunits that constitute the yeast eIF3—eIF3a, b, c, g and i—are conserved in mammalian (48,51,52). However, mammalian eIF3 has evolved to include additional subunits, which are likely to function as specific regulatory factors and some extra structural motifs provide the capacity to mediate extra protein-protein or protein–RNA interactions. Accordingly, eIF3e possesses a PCI (proteosome-COP9-initiation factor) domain, and eIF3f and eIF3h have a MPN (Mpr1-Pad1-N-terminal) domain. PCI or MPN domains are found in components of large protein complexes and have been implicated in protein–protein interactions (52). In fact, several results have indicated that the non-core subunits of the mammalian eIF3 regulate specific mRNAs (53–55). Also, one attractive hypothesis is that the presence of a particular non-core subunit, e.g. eIF3h in the eIF3 protein complex, may in turn, govern the efficiency with which the 43S pre-initiation complex is able to scan through the 5′-UTR of a particular set of mRNAs (56). In addition, the non-core subunits may serve to stabilize the eIF3 complex (40) and/or may have redundant functions (55). It has been shown that mammalian subunits eIF3f:h:m constitute a stable module that is located on the periphery of the complex and is not involved in interactions between other subunits (41). Also, it is known that subunit eIF3h is essential for binding the trimer f:h:m to the core eIF3c subunit (41). Based on these data, the possibility exists that in yeast, the model for the AUG-proximity effect may be dependent on the eIF3c, which may not evolved in order to become as specialized as its mammalian ortholog and thus it performs different biological functions, among which, those attributable to the mammalian eIF3f and h subunits. The relevance of the mammalian non-conserved eIF3f and h subunits in bridging eIF4G in association with PABPC1 to the ribosome during the process of translation initiation, which seem to be maintained during the first steps of elongation, might be linked to the need of a tighter regulation of translation in higher eukaryotes.
In conclusion, the present data support a role for PABPC1 and associated translation initiation factors in NMD evasion of AUG-proximal nonsense-mutated transcripts. Future efforts addressing the biochemical mechanism by which PABPC1 is involved in cap-dependent translation initiation and how it participates in the eIF4F/43S scanning will contribute to our understanding of mRNA translation.
FUNDING
Fundação Luso-Americana para o Desenvolvimento and Center for Biodiversity, Functional and Integrative Genomics (BioFIG) and Fundação para a Ciência e a Tecnologia (SFRH/BD/35962/2007 to I.P., SFRH/BD/63581/2009 to C.B., SFRH/BPD/34583/2007 to A.L.S. and SFRH/BPD/20644/2004 to A.I.). Funding for open access charge: Nucleic Acids Research funding.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Marla J. Berry (University of Hawaii, Manoa, HI) for providing the pUHD-eRF3 plasmid. The authors also thank João Lavinha for comments on the manuscript.
REFERENCES
Author notes
Present address: Ângela Inácio, Centro de Biologia Ambiental, Departamento de Biologia Animal, Faculdade de Ciências, Universidade de Lisboa, Campo Grande C2 – Piso 3, 1749-016 Lisboa, Portugal Ana Luísa Silva, Centro de Investigação em Patobiologia Molecular, Instituto Português de Oncologia de Francisco Gentil, Rua Professor Lima Basto, 1099-023 Lisboa, Portugal



![Translation termination at the AUG-proximal β15 PTC occurs in the absence of the ribosome stalling evident at the more distal β39 PTC. (A) Diagram of the human β-globin mRNA showing positions of initiation and termination codons [native (N) or premature (at position 15 or 39)]. The arrow indicates the position and orientation of the #17 primer (Table 1) used in the primer-extension (toeprinting) assays. The lines below the mRNA diagram represent the length in nucleotides of the synthesized cDNA from the #17 primer to the 5′-end of the mRNA, the initiation AUG codon, or to the βN, β15 and β39 translation termination codons. (B) Electropherograms of βN, β15 and β39 toeprint assays using the fluorescently labeled #17 primer. The toeprint reactions were performed in the presence or absence of cycloheximide (+CHX or −CHX, respectively). A control reaction was carried out in the absence of added rabbit reticulocyte lysate (−RRL). Primer extension products were suspended in a mix containing ROX 2500 molecular weight marker (Applied Biosystems) and subjected to capillary electrophoresis. Size standard peaks are shown below and sequence position is indicated in nucleotides. The toeprint peaks at 626 nt represent the full-length transcript; the peaks at 620 nt correspond to the cap-binding complex-bound full-length transcript; the peaks at 555-nt map 18-nt downstream the AUG codon (AUG TP). The peak at 440-nt maps to a position 16 nt downstream the β39 stop codon (PTC TP). (C) The arrow below the mRNA diagram represents position and orientation of #18 primer (Table 1) used in the second, high-resolution toeprinting assays. The lines below the mRNA diagram represent the length in nucleotides of the primer-extended cDNA to the 5′-end of the mRNA, to the initiation AUG, as well as to the translation termination codons at positions 15 or 39. (D) Electropherograms of β15 and β39 toeprint assays using the fluorescently labeled #18 primer. A parallel sequencing reaction on the βN cDNA, performed with the same primer is shown on the top. Underlined sequences indicate codon position: 0 (AUG), 15 and 39. The toeprint reactions were performed in the presence or absence of cycloheximide (+CHX or −CHX, respectively). A control reaction lacking −RRL is shown. Primer extension and sequencing products were resuspended in a mix containing ROX 500 (Applied Biosystems) molecular weight standard and separated by capillary electrophoresis. Sequence position is indicated in nucleotides. Peaks at 230 nt correspond to the initiation AUG toeprint that maps 18-nt downstream the AUG codon. The peak at 115 nt, present uniquely in the β39 analysis, maps 16-nt downstream of the PTC.](https://oup.silverchair-cdn.com/oup/backfile/Content_public/Journal/nar/40/3/10.1093/nar/gkr820/2/m_gkr820f1a.jpeg?Expires=1716343847&Signature=hKFyO7K2RjW8o-G3Wd2xLA4TtW-SVfQxbXF71olE4EV0oaoxALEln8omw6GKYvzHKnlcn6ZpXRaYjxN2fhubBv59ru1ijtKzwfhmIKGPfopDYgvzYVMTAlVeJpGt9uvcqHZbcfNxrhLwE6By-scSFxWww7oSuqosNcujmYM8Dj7HSmGK9d011LmXo-J96xF6bVUiSl7U2AY8r8Ej3kBiVNPE8FroY2rJpQ4nLm~~6CtOoPDM3-ld6DxDBHrzhdV05HcF9JhEnZjI0BPTxLkyT~gpzVcR-sjw2ACzfB01SSp~fp1Ia8ziEgaK65tmZEF0T01rrdMOPsgEqYeI3rdG4A__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Translation termination at the AUG-proximal β15 PTC occurs in the absence of the ribosome stalling evident at the more distal β39 PTC. (A) Diagram of the human β-globin mRNA showing positions of initiation and termination codons [native (N) or premature (at position 15 or 39)]. The arrow indicates the position and orientation of the #17 primer (Table 1) used in the primer-extension (toeprinting) assays. The lines below the mRNA diagram represent the length in nucleotides of the synthesized cDNA from the #17 primer to the 5′-end of the mRNA, the initiation AUG codon, or to the βN, β15 and β39 translation termination codons. (B) Electropherograms of βN, β15 and β39 toeprint assays using the fluorescently labeled #17 primer. The toeprint reactions were performed in the presence or absence of cycloheximide (+CHX or −CHX, respectively). A control reaction was carried out in the absence of added rabbit reticulocyte lysate (−RRL). Primer extension products were suspended in a mix containing ROX 2500 molecular weight marker (Applied Biosystems) and subjected to capillary electrophoresis. Size standard peaks are shown below and sequence position is indicated in nucleotides. The toeprint peaks at 626 nt represent the full-length transcript; the peaks at 620 nt correspond to the cap-binding complex-bound full-length transcript; the peaks at 555-nt map 18-nt downstream the AUG codon (AUG TP). The peak at 440-nt maps to a position 16 nt downstream the β39 stop codon (PTC TP). (C) The arrow below the mRNA diagram represents position and orientation of #18 primer (Table 1) used in the second, high-resolution toeprinting assays. The lines below the mRNA diagram represent the length in nucleotides of the primer-extended cDNA to the 5′-end of the mRNA, to the initiation AUG, as well as to the translation termination codons at positions 15 or 39. (D) Electropherograms of β15 and β39 toeprint assays using the fluorescently labeled #18 primer. A parallel sequencing reaction on the βN cDNA, performed with the same primer is shown on the top. Underlined sequences indicate codon position: 0 (AUG), 15 and 39. The toeprint reactions were performed in the presence or absence of cycloheximide (+CHX or −CHX, respectively). A control reaction lacking −RRL is shown. Primer extension and sequencing products were resuspended in a mix containing ROX 500 (Applied Biosystems) molecular weight standard and separated by capillary electrophoresis. Sequence position is indicated in nucleotides. Peaks at 230 nt correspond to the initiation AUG toeprint that maps 18-nt downstream the AUG codon. The peak at 115 nt, present uniquely in the β39 analysis, maps 16-nt downstream of the PTC.](https://oup.silverchair-cdn.com/oup/backfile/Content_public/Journal/nar/40/3/10.1093/nar/gkr820/2/m_gkr820f1b.jpeg?Expires=1716343847&Signature=rF8fzmIOLIM2Uc86IDepazATK4qssctgWC7p8PupgyvS6K5MGDmD2oXiZ6RtcgFG78rZ-K3L-gyrMrx--lti~Ev5ZN9DdoIo8Rzby2HktUHr4SrO1j6Zqife27mmP1YGxemZje~3b2QSeRhxh7Nv0~k4~mkRVvqAVoBKxahHOBLZ0ndeaSkyVGJeegwh~e-y1ctrAR2mLtckMCiI7FzE6ehVq2uB67ENci4D3Rtl2nKkC-xI2XTnqHBGj63hEg72gCuI77bFhEc8Z~wniTUvmAnL6i2SBaKLbP~nXKX4j7Zn6ruUh27AzHRQ7z4johG7nzCOMtWJQYwttVuWI7~BJA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![PABPC1 plays an essential role in NMD resistance of an AUG-proximal PTC. (A) Diagram representing βN, the NMD-resistant β-globin mRNA with an AUG-proximal nonsense mutation at codon 15 (UGA; β15) and the NMD-sensitive β39 mRNA. The positions of initiation (AUG) and termination (native and premature) codons are indicated. (B) Representative western blot analysis of HeLa cells extracts transfected with human PABPC1 siRNA (lanes 4–6) or with a control Luciferase siRNA target (LUC siRNA; lanes 1–3). Twenty-four hours after siRNA treatment, cells were transfected with the β-globin reporter constructs (βN, β15 or β39) with or without a plasmid expressing PABPdelC mutant protein (pDESTPABPC1delC plasmid); lanes 4–6 or 1–3, respectively. Twenty-four hours post-transfection, protein and RNA were isolated from the cells for analysis. Immunoblotting to confirm PABPC1 knockdown was carried out with anti-PABPC1 (specific for the N-terminal domain) and with anti-α-tubulin antibodies as a loading control (lanes 4–6 versus lanes 1–3). Identification of each band is on the right. (C) Depletion of endogenous PABPC1 in conjunction with expression of exogenous PABPdelC protein represses β15 mRNA levels. Relative β-globin mRNA levels under control conditions (LUC siRNA-treated HeLa cells; dark bars) and at conditions of PABPdelC expression in PABPC1-depleted HeLa cells (PABPC1 siRNA + PABPdelC overexpression; light bars), normalized to the levels of puromycin resistance mRNA (Puror; plasmids carrying the reporter β-globin gene also contain the Puror gene), were determined by quantitative RT–qPCR and compared to the corresponding βN mRNA levels (defined as 100%). Average and standard deviation (SD) of three independent experiments corresponding to three independent transfections are shown in the histogram. (D) Representative western blot analysis of HeLa cells extracts transfected with control Luciferase siRNA (LUC siRNA; lanes 1–3) or with siRNA targeting the human PABPC1 3′-UTR (PABPC1 3′-UTR siRNA; lanes 4–9). After siRNA treatment, cells were transfected with the plasmids expressing βN, β15 or β39 mRNAs (lanes 1–3) or cotransfected with these plasmids in combination with a plasmid expressing PABPdelC mutant protein [pDESTPABPC1delC plasmid as above in (B); lanes 4–6], or with a plasmid expressing wild-type PABPC1 (encoded by an mRNA with an heterologous 3′-UTR resistant to the PABPC1 siRNA; lanes 7–9). Protein levels present in the cell extracts were analyzed by western blot for PABPC1 and α-tubulin (loading control) as in (B), to monitor endogenous PABPC1 knockdown (lanes 4–9 versus lanes 1–3) and exogenous expression of mutant PABPdelC protein (lanes 4–6) or PABPC1 expression rescue (lanes 7–9). Identification of each band is indicated to the right of the gel image. The histogram below the immunoblot shows the average and SD values of three independent experiments for quantification of relative PABPC1 protein levels. (E) Expression of wild-type PABPC, but not PABPdelC, restores β15 expression in PABPC1 depleted cells. Relative β-globin mRNA levels for the control conditions (LUC siRNA-treated cells; dark bars), at conditions of PABPdelC expression in PABPC1-depleted HeLa cells (PABPC1 siRNA + PABPdelC overexpression; light bars), and at conditions of PABPC1 rescue (PABPC1 siRNA-treated cells plus expression of exogenous PABPC1; light bars) were determined by RT–qPCR and compared to the corresponding βN mRNA levels as in (C). Average values and SD of three independent experiments corresponding to three independent transfections are shown in the histogram. All values are represented as a percentage (%) of the corresponding βN mRNA (defined as 100%).](https://oup.silverchair-cdn.com/oup/backfile/Content_public/Journal/nar/40/3/10.1093/nar/gkr820/2/m_gkr820f2.jpeg?Expires=1716343847&Signature=3ROYVWNWSEOBkKVz5HIAYSYGnAlXH~OGLbgWB5sRjmpmv~WK4Ro-wjDjw5lZrCO2Qax-ERWXKXphdY9BtRr~lP1ekI9QxQgefsSJe2rlLnO~WbLouV0o~50VvVS6bA3PacG~MBNCQ0aY1bIOlEO~fAcdIWSJjZFj9UDrPpMomjRnLRkj5luiJe0p7jlZuJf68AXeLGMVYQynhAAp3w9NFTjVT684p7bKuBnMq6~SseaoKVTVBW2q4K-iNAHU~eHVLG2Zs~yoDfAmkM8CBF--5NTi0YGWPDFisUuFLJ56QEnZvC5769p7omhudEuuwGW54yEheHJqCIe9XhaT9VPK6Q__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![The translation initiation factors eIF3h and eIF3f are required for the NMD-resistant phenotype of the β15 nonsense-mutated mRNA. (A) Representation of the predicted organization of the mammalian eIF3 subunits [adapted from (40–42)]. Free eIF3 complex can be assigned into three stable modules. One module consists of eIF3a, b, g and i, and it interacts with a second module composed by eIF3c, d, e, l and k. eIF3b functions as a scaffold protein connecting eIF3a, c, i and g subunits. The third subcomplex is composed by eIF3f, h and m. Subunits eIF3f and m bind to the subcomplex eIF3 c:d:e:l:k through subunit eIF3h. The remaining subunit eIF3j, a labile subunit, attaches to the complex via eIF3b (40–42). (B–D) Representative RT–PCR analyses of RNAs extracted from untreated (Ctl; lane 1), Luciferase (LUC; lane 2) or eIF3h, eIF3f and eIF3e (lane 3) siRNAs-treated HeLa cells, respectively, in panels (B–D). RT–PCRs were carried out with eIF3h, eIF3f or eIF3e mRNA specific primers to monitor endogenous eIF3h, eIF3f or eIF3e knockdown, respectively (lanes 3 versus lanes 1–2). The eIF3h, eIF3f and eIF3e mRNA levels were normalized to those of histone deacetylase 1 (HDAC1) mRNA level. In each panel, the right three lanes correspond to serial dilutions of RNA, demonstrating semiquantitative conditions used for RT–PCR. (E) Representative western blot analysis of HeLa cells extracts, untreated (Ctl, lane 1) or treated with Luciferase (LUC; lane 2), eIF3h (lane 3), eIF3f (lane 4) or eIF3e (lane 5) specific siRNAs (see ‘Material and Methods’ section). After siRNA treatment, cells were transiently transfected with the pEGFP plasmid (BD Biosciences) expressing green fluorescent protein (GFP). Protein lysates were analyzed by immunoblotting using anti-GFP and anti-α-tubulin (loading control) antibodies to monitor GFP protein expression. Identification of each band is indicated to the right of the gel image. (F, H and J) Semiquantitative RT–PCR analyses of RNAs extracted from HeLa cells transfected with a Luciferase (LUC) siRNA target (lanes 1–3) or human eIF3h, eIF3f or eIF3e siRNAs, respectively, at panels (F, H or J) (lanes 4–6 of each panel), using the same experimental settings as in (B). Twenty-four hours after siRNA-treatment, cells were transfected with plasmids expressing βN, β15 or β39 mRNAs. Twenty-four hours after constructs transfection, total RNA was isolated from the cells. RT–PCR was carried out as in (B) with eIF3h, eIF3f or eIF3e mRNA specific primers, respectively, to monitor endogenous eIF3h, eIF3f or eIF3e knockdown (lanes 4–6 versus lanes 1–3). (G) Knockdown of eIF3h represses β15 mRNA levels. Relative β-globin mRNA levels for the control conditions (LUC siRNA; dark bars) and at conditions of eIF3h depletion (eIF3h siRNA; light bars), normalized to the levels of puromycin resistance mRNA encoded from the β-globin gene plasmid, were determined by quantitative RT–PCR (RT–qPCR) and compared to the corresponding βN mRNA levels (defined as 100%). (I) Knockdown of eIF3f represses β15 mRNA levels. Relative β-globin mRNA levels for the control conditions (LUC siRNA; dark bars) and at conditions of eIF3f depletion (eIF3f siRNA; light bars), normalized to the levels of puromycin resistance mRNA were determined by RT–qPCR and compared to the corresponding βN mRNA levels (defined as 100%). (K) Knockdown of eIF3e fails to repress β15 mRNA expression. Relative β-globin mRNA levels for the control conditions (LUC siRNA; dark bars) and at conditions of eIF3e depletion (eIF3e siRNA; light bars), normalized to the levels of puromycin resistance mRNA were determined by RT–qPCR as in (G) and compared to the corresponding βN mRNA levels (defined as 100%). (G, I and K) histograms show average and SD values of three independent experiments corresponding to three independent transfections.](https://oup.silverchair-cdn.com/oup/backfile/Content_public/Journal/nar/40/3/10.1093/nar/gkr820/2/m_gkr820f5a.jpeg?Expires=1716343847&Signature=uo0YNveyZxOpFOEvCCj2xf6HUuxavJiIf9Jc-hXh6Wph0Do09arkTh0AkJ4B3FuPOYprPzmgoZBA2-3R0rOcDzxDNNLPfm0cehbywLDl8QEMuQZvP7qx5~hQL4HfT0PuzC6ibdYPu5Qd0tI8VMMEzp~7fIFH7S6W1bOQY0MXBhVtGXu3u0kJpoSEklh5-FgIVqPDC5m6ldD9PnBp1-HWesZ8AUjxY7atazoiWmeMZBMNur49M5tZktlFbhKcYHJjjo4e~zUk5nUJFBnf3TdUgXPPYVMM632t2KfoUH-hqDFyfSZ6PtcJR-JHo4RSI8JoVs8uNoa0W-v7Ew~onuLmGw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![The translation initiation factors eIF3h and eIF3f are required for the NMD-resistant phenotype of the β15 nonsense-mutated mRNA. (A) Representation of the predicted organization of the mammalian eIF3 subunits [adapted from (40–42)]. Free eIF3 complex can be assigned into three stable modules. One module consists of eIF3a, b, g and i, and it interacts with a second module composed by eIF3c, d, e, l and k. eIF3b functions as a scaffold protein connecting eIF3a, c, i and g subunits. The third subcomplex is composed by eIF3f, h and m. Subunits eIF3f and m bind to the subcomplex eIF3 c:d:e:l:k through subunit eIF3h. The remaining subunit eIF3j, a labile subunit, attaches to the complex via eIF3b (40–42). (B–D) Representative RT–PCR analyses of RNAs extracted from untreated (Ctl; lane 1), Luciferase (LUC; lane 2) or eIF3h, eIF3f and eIF3e (lane 3) siRNAs-treated HeLa cells, respectively, in panels (B–D). RT–PCRs were carried out with eIF3h, eIF3f or eIF3e mRNA specific primers to monitor endogenous eIF3h, eIF3f or eIF3e knockdown, respectively (lanes 3 versus lanes 1–2). The eIF3h, eIF3f and eIF3e mRNA levels were normalized to those of histone deacetylase 1 (HDAC1) mRNA level. In each panel, the right three lanes correspond to serial dilutions of RNA, demonstrating semiquantitative conditions used for RT–PCR. (E) Representative western blot analysis of HeLa cells extracts, untreated (Ctl, lane 1) or treated with Luciferase (LUC; lane 2), eIF3h (lane 3), eIF3f (lane 4) or eIF3e (lane 5) specific siRNAs (see ‘Material and Methods’ section). After siRNA treatment, cells were transiently transfected with the pEGFP plasmid (BD Biosciences) expressing green fluorescent protein (GFP). Protein lysates were analyzed by immunoblotting using anti-GFP and anti-α-tubulin (loading control) antibodies to monitor GFP protein expression. Identification of each band is indicated to the right of the gel image. (F, H and J) Semiquantitative RT–PCR analyses of RNAs extracted from HeLa cells transfected with a Luciferase (LUC) siRNA target (lanes 1–3) or human eIF3h, eIF3f or eIF3e siRNAs, respectively, at panels (F, H or J) (lanes 4–6 of each panel), using the same experimental settings as in (B). Twenty-four hours after siRNA-treatment, cells were transfected with plasmids expressing βN, β15 or β39 mRNAs. Twenty-four hours after constructs transfection, total RNA was isolated from the cells. RT–PCR was carried out as in (B) with eIF3h, eIF3f or eIF3e mRNA specific primers, respectively, to monitor endogenous eIF3h, eIF3f or eIF3e knockdown (lanes 4–6 versus lanes 1–3). (G) Knockdown of eIF3h represses β15 mRNA levels. Relative β-globin mRNA levels for the control conditions (LUC siRNA; dark bars) and at conditions of eIF3h depletion (eIF3h siRNA; light bars), normalized to the levels of puromycin resistance mRNA encoded from the β-globin gene plasmid, were determined by quantitative RT–PCR (RT–qPCR) and compared to the corresponding βN mRNA levels (defined as 100%). (I) Knockdown of eIF3f represses β15 mRNA levels. Relative β-globin mRNA levels for the control conditions (LUC siRNA; dark bars) and at conditions of eIF3f depletion (eIF3f siRNA; light bars), normalized to the levels of puromycin resistance mRNA were determined by RT–qPCR and compared to the corresponding βN mRNA levels (defined as 100%). (K) Knockdown of eIF3e fails to repress β15 mRNA expression. Relative β-globin mRNA levels for the control conditions (LUC siRNA; dark bars) and at conditions of eIF3e depletion (eIF3e siRNA; light bars), normalized to the levels of puromycin resistance mRNA were determined by RT–qPCR as in (G) and compared to the corresponding βN mRNA levels (defined as 100%). (G, I and K) histograms show average and SD values of three independent experiments corresponding to three independent transfections.](https://oup.silverchair-cdn.com/oup/backfile/Content_public/Journal/nar/40/3/10.1093/nar/gkr820/2/m_gkr820f5b.jpeg?Expires=1716343847&Signature=xQndVsMA2RfNsHU2ldaz4PfSMiKQndBHphRTLMGeIVclb89YfhGBIYNi2hMOI57MjVeYs7Lkfhxfu~GdLPL8HyL57s6zh6e-UULUfCXBfqns1uyRSN~Oc4-5e02QhhmCYGGNYK3eWVKo3fzkVWCjP6A4nIqbCD73TpNc7jO1ydBui-ZuEXHkb0p1M-pgvrsXAS1v8lHQAcXHj7zLBs2QF2nne0uFIabiqVhgt2d45zHm72TjwesevcuJwLP5IAzznQzuyxGokcqmd210W-hz358i22fIrQVAohGpfwwi9ks7kQ5X28QqvoyIxZt1IQFqXV-QZ3h0XmZkuZBdIei0EQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

Comments