-
PDF
- Split View
-
Views
-
Cite
Cite
Martine Claremont, Roland Houart, Suzanne T. Williams, David G. Reid, A molecular phylogenetic framework for the Ergalataxinae (Neogastropoda: Muricidae), Journal of Molluscan Studies, Volume 79, Issue 1, February 2013, Pages 19–29, https://doi.org/10.1093/mollus/eys028
Close - Share Icon Share
Abstract
The validity of the muricid subfamily Ergalataxinae has recently been confirmed with molecular data, but its composition and the relationships among its constituent genera remain unclear. In order to investigate this, we use four genes (28S rRNA, 12S rRNA, 16S rRNA and cytochrome c oxidase subunit I) to construct a Bayesian phylogeny of 52 ergalataxine species in 18 genera, representing c. 40% of the currently accepted species and 86% of the genera. This is the most complete phylogeny of this taxonomically confusing subfamily yet produced. Our results indicate the polyphyly of many traditional genera, including Morula, Pascula and Orania. In order to improve the correspondence between classification and phylogeny, we restrict the definition of Morula, resurrect Tenguella and elevate Oppomorus to full genus, but describe no new genera. Several species in this analysis could not be identified and may be new, but we do not describe them. Further molecular and morphological analyses, in the context of this framework, should help to resolve the remaining ambiguities in the classification of this subfamily. The oldest fossil member of the Ergalataxinae known to us is of Early Oligocene age.
INTRODUCTION
The Muricidae are a large, taxonomically complex family of neogastropods. Many species in this family are well known for their charismatic shells (Murex, Chicoreus), as predators (Ocenebra, Urosalpinx) of commercially important oysters, as an ancient source of purple dye (Bolinus, Plicopurpura) or as food items (Concholepas, Murex). However, until recently, the phylogeny of the family was poorly known; subfamilies have been erected based largely on superficial shell and radular resemblances rather than on rigorous phylogenetic analyses. One such subfamily is the Ergalataxinae, the diagnosis of which was based on somewhat vague characters of the shell, operculum, radula and egg capsule (Kuroda & Habe, 1971). It has been criticised as “the least well-defined [subfamily] in the entire family Muricidae” (Vokes, 1996a, 27). Ever since its introduction, the taxonomy of the group and its relationship to another muricid subfamily, the Rapaninae (=Thaidinae of authors), has been controversial. Some workers contended that there was little morphological support for the separation of the Rapaninae and Ergalataxinae (Tan, 1995, 2000). Others described new ergalataxine genera and transferred existing genera to the subfamily (e.g. Houart, 1995a, b, 1997; Vokes, 1996a, b). Recent molecular phylogenetic analyses have tested these opposing views, establishing the monophyly of the Ergalataxinae and supporting its recognition as a subfamily (Claremont, Reid & Williams, 2008; Barco et al., 2010; Claremont et al., in press).
Like the rest of the Muricidae, ergalataxines are carnivorous, preying upon a wide variety of organisms including corals, polychaetes, barnacles and other molluscs (e.g. Taylor, 1980; Tan, 1995; Ishida, 2004; Claremont, Reid & Williams, 2011a). Ergalataxines are mainly found in the tropical Indo-West Pacific and occupy a wide range of habitats, with some species common in the high intertidal, while others live more than 500 m deep (Kuroda & Habe, 1971; Houart, 1995b; Tan, 1995). The subfamily was initially established for only three genera (Ergalatax, Bedevina and Cytharomorula), but many traditionally ‘thaidid’ genera have been transferred to it subsequently (e.g. Morula, Spinidrupa, Cronia, Drupella; Tröndlé & Houart, 1992; Kool, 1993; Vokes, 1996a; Tan, 2000; Vermeij & Carlson, 2000; Houart, 2004; Claremont et al., 2011a). New ergalataxine genera have also been described, based on characters of the shell and radula (e.g. Habromorula; Houart, 1995a).
Characters of the shell, internal anatomy and radula have also been used to suggest affinities between various genera (e.g. Fujioka, 1985; Houart, 1995a, b, 2004), but cases of plasticity, polymorphism and of parallel and convergent evolution within the Ergalataxinae have made phylogenetic reconstruction based on morphology difficult. For example, radular morphology has been shown to vary within species according to sex, growth rate and season (Fujioka, 1985; Kool, 1993; Tan, 1995). Such is the prevalence of homoplasy in morphological character transformations that early cladistic analyses did not even find support for the monophyly of the Ergalataxinae (Kool, 1993; Vermeij & Carlson, 2000). Although molecular studies have confirmed its monophyly (see above), so far they have suffered from limited taxonomic sampling. A comprehensive molecular analysis of this important subfamily remains to be done.
Previous molecular phylogenetic analyses of the Muricidae have included only a few representatives of the Ergalataxinae (6 species in Claremont et al., 2008; 10 species in Barco et al., 2010; 6 species in Claremont et al., in press). Here, we aim to provide a more comprehensive phylogenetic framework of the Ergalataxinae for further taxonomic, ecological and phylogeographic work; to test monophyly of nominal genera currently defined by morphological characters and to provide a new phylogenetic classification. To do this, we construct a molecular phylogeny of 52 ergalataxine species using one nuclear and three mitochondrial genes.
MATERIAL AND METHODS
Taxon sampling and identification
A total of 109 ergalataxine specimens were obtained for molecular analysis (Supplementary Material, Table S1). These were identified morphologically by the authors (R.H., D.G.R. and M.C.) as belonging to 52 species in 18 genera, representing 40% of the c. 129 accepted species in the subfamily and all but three of the 21 currently accepted genera (Houart, 1986, 1987, 1991, 1995a, b, 1996a, b, 1997, 1998, 2000, 2002, 2004, 2008; Tröndlé & Houart, 1992; Tan, 1995; Vokes, 1996b; Vermeij & Carlson, 2000; Table 1). We have sequenced the type species of 15 genera (Houart, 1995b; Vermeij & Carlson, 2000; Tables 1, Supplementary Material Table S1). Based on morphology, nine additional genera have been included in the Ergalataxinae by some authors (Tröndlé & Houart, 1992; Houart, 1995b, 2004; Vokes, 1996b). Of these nine genera, one is unavailable; one is a possible synonym of another ergalataxine genus; three have been removed from the subfamily on the basis of their morphology (Ponder, 1972; Radwin & D'Attilio, 1976; R. Houart, 2010, unpubl.); and one has been removed based on molecular analyses (Barco et al., 2010; see Supplementary Material, Table S2). The three ergalataxine genera not included in our analysis (Daphnellopsis, Lindapterys, Uttleya) are poorly known and lack clear morphological similarities to other Ergalataxinae, so have been placed in the Ergalataxinae only tentatively (R. Houart, unpubl.; Supplementary Material, Table S2). Representative species of these genera should be included in future molecular analyses.
Status of genera of Ergalataxinae included in this study.
| Genus . | Type species . | First placed in Ergalataxinae by . | Type species analysed herein . | Validity . |
|---|---|---|---|---|
| Bedevina Habe, 1946 | Trophon birileffi Lishke, 1871 | Kuroda & Habe, 1971 | Yes | Valid; could include Spinidrupa |
| Cronia H. & A. Adams, 1853 | Purpura amygdala Kiener, 1835 | Tröndlé & Houart, 1992 | Yes | Valid; could include Maculotriton and Ergalatax |
| Cytharomorula Kuroda, 1953 | Cytharomorula vexillum Kuroda, 1953 | Kuroda & Habe, 1971 | Yes | Uncertain; not monophyletic in any analysis |
| Drupella Thiele, 1925 | Purpura elata Blainville, 1832 [=Drupa cornus Röding, 1798] | Claremont et al., 2011a | Yes | Valid |
| Ergalatax Iredale, 1931 | Ergalatax recurrens Iredale, 1931 [=Murex pauper Watson, 1883] | Kuroda & Habe, 1971 | No (although included E. contracta is possible synonym; Houart, 2008) | Uncertain; not monophyletic in any analysis. In clade with Cronia and Maculotriton; potential synonym of Cronia |
| Habromorula Houart, 1995 | Purpura biconica Blainville, 1832 | Houart, 2004 | Yes | Not a valid genus; within Morula clade; monophyly of Habromorula not strongly contradicted, so potentially valid subgenus |
| Lataxiena Jousseaume, 1883 | Lataxiena lataxiena Jousseaume, 1883 [=Trophon fimbriata Hinds, 1844] | Houart, 1995b | Yes | Valid; composition unclear due to uncertainty surrounding Orania |
| Maculotriton Dall, 1904 | Triton bracteata Hinds, 1844 [=Buccinum serriale Deshayes, 1834] | Tröndlé & Houart, 1992 | Yes | Valid; potential synonym of Cronia |
| Morula Schumacher, 1817 | Morula papillosa Schumacher, 1917 [=Drupa uva Röding, 1798] | Houart, 2004 | Yes | Valid; definition restricted (cf. Houart, 2002, 2004) |
| Muricodrupa Iredale, 1918 | Purpura fenestrata Blainville, 1832 | Tröndlé & Houart, 1992 | Yes | Valid; definition restricted (M. fiscella excluded) |
| Oppomorus Iredale, 1937 | Morula nodulifera Menke, 1829 | Houart, 2004 | Yes | Valid genus (subgenus of Houart, 2004) |
| Orania Pallary, 1900 | Pseudomurex spadae Libassi, 1859 [=Murex fusulus Brocchi, 1814] | Houart, 1995b | No | Uncertain; type species not included; species previously assigned to Orania form clades with members of Lataxiena, Usilla, Cytharomorula and Pascula. |
| Pascula Dall, 1908 | Trophon citricus Dall, 1908 | Tröndlé & Houart, 1992 | No | Uncertain; type species not included; paraphyletic as currently defined |
| Phrygiomurex Dall, 1904 | Triton sculptilis Reeve, 1844 | Tröndlé & Houart, 1992 | Yes | Valid |
| Spinidrupa Habe & Kosuge, 1966 | Murex euracantha A. Adams, 1851 | Houart, 1995a | Yes | Uncertain; some species in clade with Bedevina, which has priority |
| Tenguella Arakawa, 1965 | Purpura granulata Duclos, 1832 | This study | Yes | Valid; synonymized with Morula by Fujioka (1985), but shown here to be distinct |
| Trachypollia Woodring, 1928 | Trachypollia sclera Woodring, 1928 | Vokes, 1996a | No | Probably valid, although sampling limited and type species not included |
| Usilla H. Adams, 1861 | Vexilla nigrofusca Pease, 1860 [=Vexilla fusconigra Pease, 1860 = Purpura avenacea Lesson, 1842] | Vokes, 1996b | Yes | Valid, but type in clade with type of Lataxiena and some Orania; retained as genus because morphologically distinct (following Vokes,1996b; Houart & Tröndlé, 2008) |
| Genus . | Type species . | First placed in Ergalataxinae by . | Type species analysed herein . | Validity . |
|---|---|---|---|---|
| Bedevina Habe, 1946 | Trophon birileffi Lishke, 1871 | Kuroda & Habe, 1971 | Yes | Valid; could include Spinidrupa |
| Cronia H. & A. Adams, 1853 | Purpura amygdala Kiener, 1835 | Tröndlé & Houart, 1992 | Yes | Valid; could include Maculotriton and Ergalatax |
| Cytharomorula Kuroda, 1953 | Cytharomorula vexillum Kuroda, 1953 | Kuroda & Habe, 1971 | Yes | Uncertain; not monophyletic in any analysis |
| Drupella Thiele, 1925 | Purpura elata Blainville, 1832 [=Drupa cornus Röding, 1798] | Claremont et al., 2011a | Yes | Valid |
| Ergalatax Iredale, 1931 | Ergalatax recurrens Iredale, 1931 [=Murex pauper Watson, 1883] | Kuroda & Habe, 1971 | No (although included E. contracta is possible synonym; Houart, 2008) | Uncertain; not monophyletic in any analysis. In clade with Cronia and Maculotriton; potential synonym of Cronia |
| Habromorula Houart, 1995 | Purpura biconica Blainville, 1832 | Houart, 2004 | Yes | Not a valid genus; within Morula clade; monophyly of Habromorula not strongly contradicted, so potentially valid subgenus |
| Lataxiena Jousseaume, 1883 | Lataxiena lataxiena Jousseaume, 1883 [=Trophon fimbriata Hinds, 1844] | Houart, 1995b | Yes | Valid; composition unclear due to uncertainty surrounding Orania |
| Maculotriton Dall, 1904 | Triton bracteata Hinds, 1844 [=Buccinum serriale Deshayes, 1834] | Tröndlé & Houart, 1992 | Yes | Valid; potential synonym of Cronia |
| Morula Schumacher, 1817 | Morula papillosa Schumacher, 1917 [=Drupa uva Röding, 1798] | Houart, 2004 | Yes | Valid; definition restricted (cf. Houart, 2002, 2004) |
| Muricodrupa Iredale, 1918 | Purpura fenestrata Blainville, 1832 | Tröndlé & Houart, 1992 | Yes | Valid; definition restricted (M. fiscella excluded) |
| Oppomorus Iredale, 1937 | Morula nodulifera Menke, 1829 | Houart, 2004 | Yes | Valid genus (subgenus of Houart, 2004) |
| Orania Pallary, 1900 | Pseudomurex spadae Libassi, 1859 [=Murex fusulus Brocchi, 1814] | Houart, 1995b | No | Uncertain; type species not included; species previously assigned to Orania form clades with members of Lataxiena, Usilla, Cytharomorula and Pascula. |
| Pascula Dall, 1908 | Trophon citricus Dall, 1908 | Tröndlé & Houart, 1992 | No | Uncertain; type species not included; paraphyletic as currently defined |
| Phrygiomurex Dall, 1904 | Triton sculptilis Reeve, 1844 | Tröndlé & Houart, 1992 | Yes | Valid |
| Spinidrupa Habe & Kosuge, 1966 | Murex euracantha A. Adams, 1851 | Houart, 1995a | Yes | Uncertain; some species in clade with Bedevina, which has priority |
| Tenguella Arakawa, 1965 | Purpura granulata Duclos, 1832 | This study | Yes | Valid; synonymized with Morula by Fujioka (1985), but shown here to be distinct |
| Trachypollia Woodring, 1928 | Trachypollia sclera Woodring, 1928 | Vokes, 1996a | No | Probably valid, although sampling limited and type species not included |
| Usilla H. Adams, 1861 | Vexilla nigrofusca Pease, 1860 [=Vexilla fusconigra Pease, 1860 = Purpura avenacea Lesson, 1842] | Vokes, 1996b | Yes | Valid, but type in clade with type of Lataxiena and some Orania; retained as genus because morphologically distinct (following Vokes,1996b; Houart & Tröndlé, 2008) |
See Supplementary material, Table S2 for status of unsampled, excluded and doubtful genera. Valid genera are those confirmed as monophyletic, with type species included. Potential synonymy and other uncertainties should be resolved by further sampling and analysis.
Status of genera of Ergalataxinae included in this study.
| Genus . | Type species . | First placed in Ergalataxinae by . | Type species analysed herein . | Validity . |
|---|---|---|---|---|
| Bedevina Habe, 1946 | Trophon birileffi Lishke, 1871 | Kuroda & Habe, 1971 | Yes | Valid; could include Spinidrupa |
| Cronia H. & A. Adams, 1853 | Purpura amygdala Kiener, 1835 | Tröndlé & Houart, 1992 | Yes | Valid; could include Maculotriton and Ergalatax |
| Cytharomorula Kuroda, 1953 | Cytharomorula vexillum Kuroda, 1953 | Kuroda & Habe, 1971 | Yes | Uncertain; not monophyletic in any analysis |
| Drupella Thiele, 1925 | Purpura elata Blainville, 1832 [=Drupa cornus Röding, 1798] | Claremont et al., 2011a | Yes | Valid |
| Ergalatax Iredale, 1931 | Ergalatax recurrens Iredale, 1931 [=Murex pauper Watson, 1883] | Kuroda & Habe, 1971 | No (although included E. contracta is possible synonym; Houart, 2008) | Uncertain; not monophyletic in any analysis. In clade with Cronia and Maculotriton; potential synonym of Cronia |
| Habromorula Houart, 1995 | Purpura biconica Blainville, 1832 | Houart, 2004 | Yes | Not a valid genus; within Morula clade; monophyly of Habromorula not strongly contradicted, so potentially valid subgenus |
| Lataxiena Jousseaume, 1883 | Lataxiena lataxiena Jousseaume, 1883 [=Trophon fimbriata Hinds, 1844] | Houart, 1995b | Yes | Valid; composition unclear due to uncertainty surrounding Orania |
| Maculotriton Dall, 1904 | Triton bracteata Hinds, 1844 [=Buccinum serriale Deshayes, 1834] | Tröndlé & Houart, 1992 | Yes | Valid; potential synonym of Cronia |
| Morula Schumacher, 1817 | Morula papillosa Schumacher, 1917 [=Drupa uva Röding, 1798] | Houart, 2004 | Yes | Valid; definition restricted (cf. Houart, 2002, 2004) |
| Muricodrupa Iredale, 1918 | Purpura fenestrata Blainville, 1832 | Tröndlé & Houart, 1992 | Yes | Valid; definition restricted (M. fiscella excluded) |
| Oppomorus Iredale, 1937 | Morula nodulifera Menke, 1829 | Houart, 2004 | Yes | Valid genus (subgenus of Houart, 2004) |
| Orania Pallary, 1900 | Pseudomurex spadae Libassi, 1859 [=Murex fusulus Brocchi, 1814] | Houart, 1995b | No | Uncertain; type species not included; species previously assigned to Orania form clades with members of Lataxiena, Usilla, Cytharomorula and Pascula. |
| Pascula Dall, 1908 | Trophon citricus Dall, 1908 | Tröndlé & Houart, 1992 | No | Uncertain; type species not included; paraphyletic as currently defined |
| Phrygiomurex Dall, 1904 | Triton sculptilis Reeve, 1844 | Tröndlé & Houart, 1992 | Yes | Valid |
| Spinidrupa Habe & Kosuge, 1966 | Murex euracantha A. Adams, 1851 | Houart, 1995a | Yes | Uncertain; some species in clade with Bedevina, which has priority |
| Tenguella Arakawa, 1965 | Purpura granulata Duclos, 1832 | This study | Yes | Valid; synonymized with Morula by Fujioka (1985), but shown here to be distinct |
| Trachypollia Woodring, 1928 | Trachypollia sclera Woodring, 1928 | Vokes, 1996a | No | Probably valid, although sampling limited and type species not included |
| Usilla H. Adams, 1861 | Vexilla nigrofusca Pease, 1860 [=Vexilla fusconigra Pease, 1860 = Purpura avenacea Lesson, 1842] | Vokes, 1996b | Yes | Valid, but type in clade with type of Lataxiena and some Orania; retained as genus because morphologically distinct (following Vokes,1996b; Houart & Tröndlé, 2008) |
| Genus . | Type species . | First placed in Ergalataxinae by . | Type species analysed herein . | Validity . |
|---|---|---|---|---|
| Bedevina Habe, 1946 | Trophon birileffi Lishke, 1871 | Kuroda & Habe, 1971 | Yes | Valid; could include Spinidrupa |
| Cronia H. & A. Adams, 1853 | Purpura amygdala Kiener, 1835 | Tröndlé & Houart, 1992 | Yes | Valid; could include Maculotriton and Ergalatax |
| Cytharomorula Kuroda, 1953 | Cytharomorula vexillum Kuroda, 1953 | Kuroda & Habe, 1971 | Yes | Uncertain; not monophyletic in any analysis |
| Drupella Thiele, 1925 | Purpura elata Blainville, 1832 [=Drupa cornus Röding, 1798] | Claremont et al., 2011a | Yes | Valid |
| Ergalatax Iredale, 1931 | Ergalatax recurrens Iredale, 1931 [=Murex pauper Watson, 1883] | Kuroda & Habe, 1971 | No (although included E. contracta is possible synonym; Houart, 2008) | Uncertain; not monophyletic in any analysis. In clade with Cronia and Maculotriton; potential synonym of Cronia |
| Habromorula Houart, 1995 | Purpura biconica Blainville, 1832 | Houart, 2004 | Yes | Not a valid genus; within Morula clade; monophyly of Habromorula not strongly contradicted, so potentially valid subgenus |
| Lataxiena Jousseaume, 1883 | Lataxiena lataxiena Jousseaume, 1883 [=Trophon fimbriata Hinds, 1844] | Houart, 1995b | Yes | Valid; composition unclear due to uncertainty surrounding Orania |
| Maculotriton Dall, 1904 | Triton bracteata Hinds, 1844 [=Buccinum serriale Deshayes, 1834] | Tröndlé & Houart, 1992 | Yes | Valid; potential synonym of Cronia |
| Morula Schumacher, 1817 | Morula papillosa Schumacher, 1917 [=Drupa uva Röding, 1798] | Houart, 2004 | Yes | Valid; definition restricted (cf. Houart, 2002, 2004) |
| Muricodrupa Iredale, 1918 | Purpura fenestrata Blainville, 1832 | Tröndlé & Houart, 1992 | Yes | Valid; definition restricted (M. fiscella excluded) |
| Oppomorus Iredale, 1937 | Morula nodulifera Menke, 1829 | Houart, 2004 | Yes | Valid genus (subgenus of Houart, 2004) |
| Orania Pallary, 1900 | Pseudomurex spadae Libassi, 1859 [=Murex fusulus Brocchi, 1814] | Houart, 1995b | No | Uncertain; type species not included; species previously assigned to Orania form clades with members of Lataxiena, Usilla, Cytharomorula and Pascula. |
| Pascula Dall, 1908 | Trophon citricus Dall, 1908 | Tröndlé & Houart, 1992 | No | Uncertain; type species not included; paraphyletic as currently defined |
| Phrygiomurex Dall, 1904 | Triton sculptilis Reeve, 1844 | Tröndlé & Houart, 1992 | Yes | Valid |
| Spinidrupa Habe & Kosuge, 1966 | Murex euracantha A. Adams, 1851 | Houart, 1995a | Yes | Uncertain; some species in clade with Bedevina, which has priority |
| Tenguella Arakawa, 1965 | Purpura granulata Duclos, 1832 | This study | Yes | Valid; synonymized with Morula by Fujioka (1985), but shown here to be distinct |
| Trachypollia Woodring, 1928 | Trachypollia sclera Woodring, 1928 | Vokes, 1996a | No | Probably valid, although sampling limited and type species not included |
| Usilla H. Adams, 1861 | Vexilla nigrofusca Pease, 1860 [=Vexilla fusconigra Pease, 1860 = Purpura avenacea Lesson, 1842] | Vokes, 1996b | Yes | Valid, but type in clade with type of Lataxiena and some Orania; retained as genus because morphologically distinct (following Vokes,1996b; Houart & Tröndlé, 2008) |
See Supplementary material, Table S2 for status of unsampled, excluded and doubtful genera. Valid genera are those confirmed as monophyletic, with type species included. Potential synonymy and other uncertainties should be resolved by further sampling and analysis.
We used 66 previously published ergalataxine sequences (Supplementary Material, Table S1; Claremont et al., 2008, 2011a; Barco et al., 2010). Outgroup species were selected from the Rapaninae, as this is the sister subfamily to the Ergalataxinae in some analyses (Claremont et al., 2008; but see Barco et al., 2010). All rapanine outgroup sequences have previously been published (Claremont et al., 2008; Barco et al., 2010). New sequences were submitted to GenBank (Supplementary Material, Table S1). Location of voucher material is noted in Supplementary Material, Table S1. Generic assignments in the following text and figures are valid, based on the conclusions of this study (summarized in Table 1). Where generic assignments remain unclear (because of polyphyly and/or lack of type species in the analysis), we have followed those of Houart (e.g. Houart, 1986, 1987, 1991, 1995a, b, 1996a, b, 1997, 1998, 2000, 2002, 2004, 2008), except for ‘Thais’ castanea (where we have followed Steyn & Lussi, 1998). Single quotation marks have been used to indicate these cases of uncertainty.
DNA sequencing and alignment
For all samples, three mitochondrial genes [cytochrome c oxidase subunit I (COI), 16S rRNA and 12S rRNA] and one nuclear gene (28S rRNA), known to be informative for phylogenetic analysis in the Muricidae (Claremont et al., 2008; Barco et al., 2010), were sequenced, following the protocols of Claremont et al. (2011b). Polymerase chain reactions (PCRs) amplified c. 1500 bp of 28S, 750 bp of 16S, 700 bp of COI and 650 bp of 12S. Primers and PCR conditions for all genes were as described by Barco et al. (2010), except for some forward fragments of 16S that were obtained using a new internal primer, 16S-Int56F (5′-AAC RGC CGC GGT ACT CTG-3′) and some COI sequences that were obtained using primers COIF and COI-MUR (Claremont et al., 2011b). We were unable to amplify all genes for all specimens. Thus, the 12S alignment consisted of 115 sequences, the 16S alignment of 106 sequences, the 28S alignment of 95 sequences and the COI alignment of 78 sequences (Supplementary Material, Table S1).
Sequences were assembled and edited with Sequencher (v. 4.8; GeneCodes Corporation, Ann Arbor, MI). Clear heterozygous peaks in both the forward and reverse sequence of 28S were coded as polymorphisms. After removal of primer regions, 28S sequences were 1497 bp and 12S sequences were 573 bp. COI sequences obtained with COIF/COI-MUR were 703 bp; those sequences obtained using universal primers (Folmer et al., 1994) were 658 bp. Sequences of 16S obtained with CGLeuR (Hayashi, 2005) and 16S-Int56F (this paper) were 705 bp, while those obtained with CGLeuR (Hayashi, 2005) and 16SA (Palumbi et al., 1991) were 825 bp.
Ribosomal (28S, 16S and 12S) sequences were aligned using the Q-INS-i method of MAFFT (Multiple Alignment using Fast Fourier Transform; v. 6.847b; Katoh & Toh, 2008). Based on preliminary analyses, we did not expect long gaps in our alignments, so the offset value was set to 0.1. The resulting alignments were adjusted by eye in MacClade (v. 4.06 OSX; Maddison & Maddison, 2003). Gblocks (v. 0.91 beta; Castresana, 2000) was then used to remove poorly aligned sites (minimum number of sequences for a conserved position: 70%; minimum number of sequences for a flanking position: 90%; maximum number of contiguous non-conserved positions: 3; minimum length of a block: 5; all gap positions allowed). Elimination of ambiguous regions reduced the 28S alignment by 5% to 1432 bp, the 12S alignment by 10% to 518 bp and the 16S alignment by 14% to 620 bp. COI sequences were aligned by eye in MacClade. There were 91 phylogenetically informative base pairs in 28S, 232 bp in 12S, 173 bp in 16S and 272 bp in COI.
For each gene partition, 24 different models of nucleotide substitution were tested with MrModelTest (v. 2.2; Nylander, 2004). Additionally, we tested a site-specific model of evolution of COI using PAUP* (v. 4.0b10; Swofford, 2002). The model chosen by both the hierarchical ratio test and Akaike's Information Criterion in MrModelTest was GTR + I + G for 28S, 16S and 12S, and HKY + I + G for COI. Comparison of log-likelihood values indicated that HKY + I + G was a significantly better fit to the data than a model that allowed site-specific rate variation across codon partitions.
Phylogenetic analysis (single gene)
All single-gene alignments were analysed using Bayesian inference and the Markov Chain Monte-Carlo Method (MCMC; MrBayes v. 3.1; Huelsenbeck & Ronquist, 2001). Model parameters for each gene were set according to the model selected by Mr Model Test and were free to vary among gene partitions. There were two independent runs of the MCMC analysis, each using four chains. Convergence between the independent MCMC runs was tested by examining the average deviations of split frequencies and the potential scale-reduction factor (PSRF), as well as the traces in Tracer (v. 1.5; Drummond & Rambaut, 2007). The MCMC analysis was run for five million generations until the MCMC runs converged. In all cases, we used a sample frequency of 1,000 and a burn-in of 1,501; other parameters were default values. Clades with posterior probability (PP) >95% were considered well supported, while PP of 90–95% was taken as marginal support. Branches in the consensus tree supported by PP <50% were collapsed. All PSRF values for MrBayes analyses were 1.00, while average deviations of split frequencies converged on zero, indicating that all trees had reached stationarity.
Construction and analysis of concatenated alignments
Inspection of individual gene trees did not reveal any well-supported clades (PP > 95%) in conflict. Conflict among strongly supported clades (PP > 95%) can be interpreted as evidence of genetic incongruence and divergent phylogenetic histories, while conflict among weakly supported clades (PP < 50%) may be due to stochastic error (Wiens, 1998; Reeder, 2003; Williams & Ozawa, 2006). We therefore created concatenated datasets. As more sequences were available for 28S and 12S than for 16S and COI, we constructed two datasets: a four-gene dataset (28S + 12S + 16S + COI) and a two-gene dataset (28S + 12S). The two-gene dataset consisted of 94 sequences, while the four-gene alignment consisted of 59 sequences.
Bayesian analysis of the two-gene and four-gene datasets was performed exactly as above; MCMC analyses of both datasets were run until stationarity was reached at nine million generations.
RESULTS
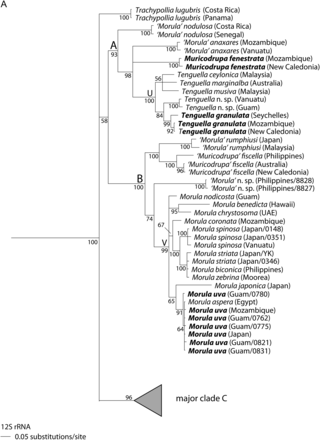
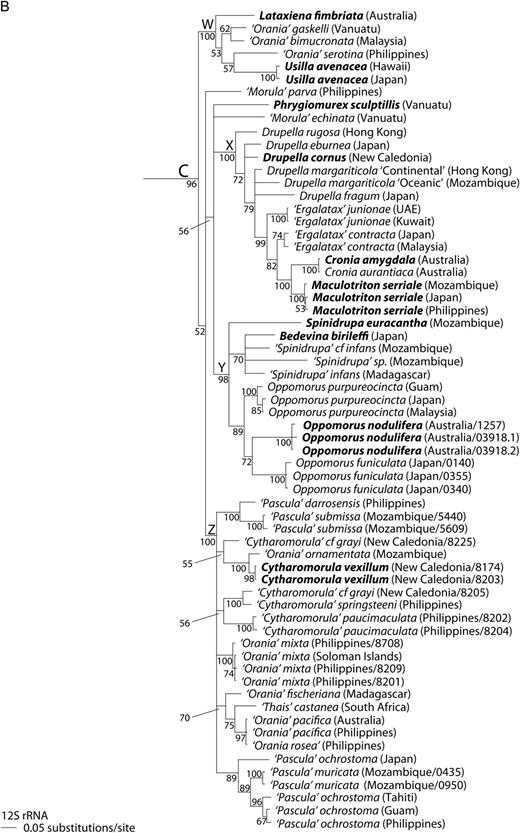
In general, the single-gene analyses were less well resolved than two-gene and four-gene analyses (Figs 1–3), with 28S being particularly poorly resolved at the suprageneric level (Supplementary Material, Fig. S1). We found that the two-gene and the four-gene analyses were most useful for determining the relationships among genera (Figs 1 and 2) while, because it had the most available sequences, the 12S analysis was most useful for determining relationships among species (Fig. 3). For this reason, we will not discuss the single-gene trees other than 12S in detail; none of the remaining single-gene trees have any well-supported branches in conflict with the trees discussed (Supplementary Material Fig. S1).
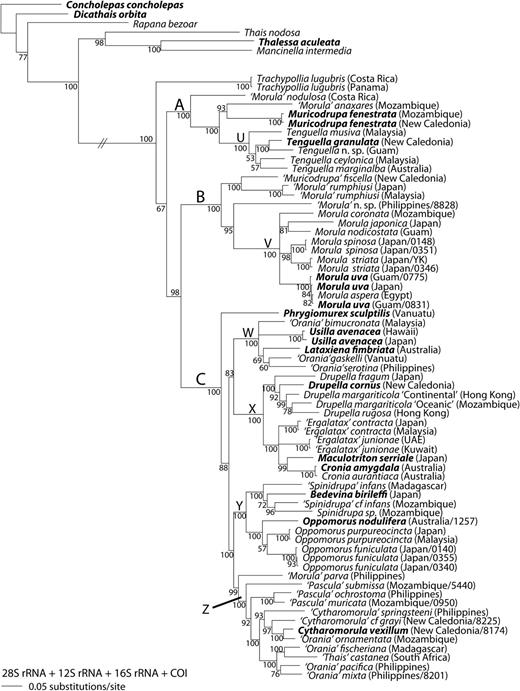
Bayesian phylogeny of the Ergalataxinae and rapanine outgroups, based on the four-gene analysis of 28S rRNA, 12S rRNA, 16S rRNA and cytochrome c oxidase subunit I (COI) sequences. Posterior probabilities ≥50 are shown at the nodes. Generic assignments are based on our revised classification (Table 1); uncertain assignments are indicated by single quotation marks. Type species of valid genera are indicated in bold. Locality is shown in parentheses after species name; when more than one sample of a species is from a given location, the last four digits of the voucher numbers are given (see Supplementary material Table S1). Major clades are indicated by letters A–C and subclades by U–Z.
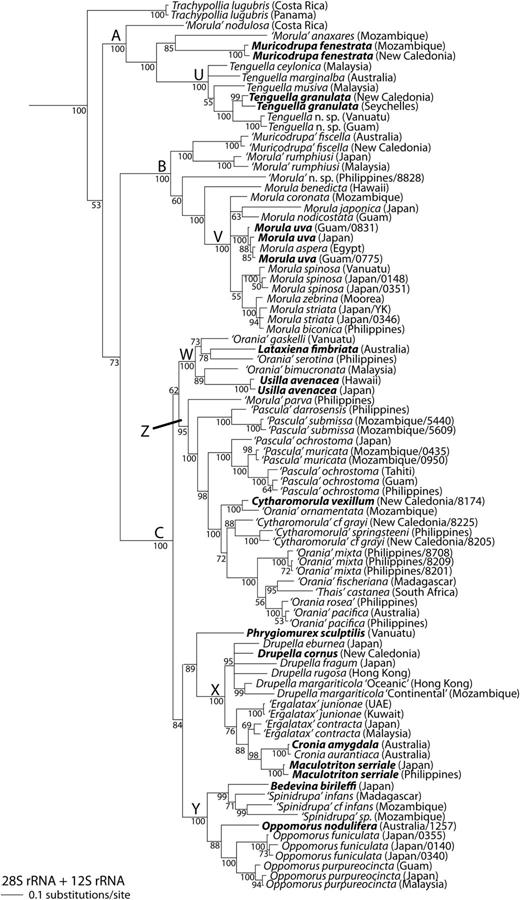
Bayesian phylogeny of the Ergalataxinae, based on the two-gene analysis of 28S and 12S rRNA. Rapanine outgroups not shown. Conventions as in Figure 1.
Suprageneric relationships
Three well-supported suprageneric clades were found (Figs 1 and 2; Clades A–C; PP = 100%), with a single lineage (Trachypollia lugubris) in an ambiguous position (Figs 1–3). Relationships among the major clades were not well supported in most trees, although B and C formed a clade in the all-gene tree (Fig. 1; PP = 98%).
Within clade A, we recovered species that have been previously assigned to the genera Morula sensu stricto, Muricodrupa and Tenguella (Figs 1 and 2; PP = 100%). Included in clade B were species that have been previously assigned to Morula s. s., Muricodrupa, Morula (Habromorula) and Cytharomorula (Figs 1–3; PP = 100%). Within clade C fell species that have previously been assigned to Orania, Cronia s. s., Cronia (Usilla), Lataxiena, Drupella, Ergalatax, Maculotriton, Bedevina, Spinidrupa, Morula s. s., Morula (Oppomorus), Pascula, Cytharomorula and Phrygiomurex (Figs 1–3; PP > 95%).
Genus-level clades
Tenguella granulata, T. ceylonica, T. musiva and T. marginalba formed a well-supported subclade U within major clade A, together with an apparently undescribed species (Tenguella n. sp.; Figs 1–3A; PP = 100%). Muricodrupa fenestrata and ‘Morula’ anaxares formed a poorly supported clade in the two-gene and four-gene analyses (Fig. 1, PP = 93%; Fig. 2, PP = 85%). A sister relationship between ‘Morula’ nodulosa and all other species in major clade A was well supported (Figs 1–3A; PP > 98%).
Within major clade Ba well-supported subclade V consisted of Morulauva and 10 other Morula species (M. aspera, M. benedicta, M. biconica, M. chrysostoma, M. coronata, M. japonica, M. nodicostata, M. spinosa, M. striata and M. zebrina; Figs 1–3A; PP > 99%). ‘Morula’ rumphiusi and ‘Morula’ fiscella formed a well-supported clade in all analyses (Figs 1–3A; PP = 100%). An unidentified species (‘Morula’ n. sp.) fell within major clade B, but its position was not resolved (Figs 1–3A).
Four well-supported subclades W–Z were observed within major clade C (Figs 1, 2 and 3B; PP > 95%; but Z unsupported in 12S analysis Fig. 3B). Usilla avenacea, Lataxiena fimbriata, ‘Orania’ bimucronata, ‘O.’ gaskelli and ‘O.’ serotina formed subclade W (Figs 1, 2 and 3B; PP = 100%). Within subclade × Drupella cornus, D. eburnea, D. fragum, D. rugosa, D. margariticola ‘Continental’ and D. margariticola ‘Oceanic’ formed a well-supported clade in the two-gene and four-gene analyses (Figs 1 and 2; PP > 95%). A sister relationship between this clade and one consisting of Ergalatax contracta, ‘E.’ junionae, Cronia amygdala, C. aurantiaca and Maculotriton serriale was well supported in the four-gene analysis (Fig. 1; PP = 100%). Within subclade Y, a clade of Oppomorus nodulifera, O. purpureocincta and O. funiculata was well supported in the four-gene analysis (Fig. 1; PP = 100%). Also within subclade Y, a clade consisting of ‘Spinidrupa’ infans, ‘S.’ cf. infans, ‘Spinidrupa' sp. and Bedevina birileffi was well supported in both the two-gene and four-gene analyses (Figs 1 and 2; PP > 98%), but the position of Spinidrupa euracantha was not resolved within subclade Y (Fig. 3B). Within subclade Z, a clade consisting of Cytharomorula vexillum, ‘C.’ cf. grayi, ‘C.’ springsteeni, ‘Orania’ ornamentata, ‘O.’ mixta, ‘O.’ pacifica, ‘O.’ fischeriana, ‘O.’ cf. rosea and ‘Thais’ castanea was well supported in the two-gene and four-gene analyses (Figs 1 and 2; PP = 100%). ‘Pascula’ ochrostoma and ‘P.’ muricata formed a clade (Figs 1 and 2; PP = 100%), as did ‘P.’ darrosensis and ‘P.’ submissa (Figs 2 and 3B; PP = 100%). ‘Morula’ parva was sister to all the other members of subclade Z in the two-gene and four-gene analyses (Figs 1 and 2; PP > 95%). The position of ‘Cytharomorula’ paucimaculata in subclade Z was unresolved (Fig. 3B). The positions of Phrygiomurex sculptilis and ‘Morula’ echinata (only 12S sequence available for the latter) within major clade C were not resolved (Figs 1, 2 and 3B).
Species-level relationships
This study was not designed to test species boundaries or to resolve relationships between species comprehensively. Nevertheless, some results suggest the need for further investigation. We found evidence indicating two possible new species (Tenguella n. sp. and ‘Morula’ n. sp.; Figs 1–3; and Supplementary material, Fig. S1). Small genetic distances are suggestive of possible synonymy of species; for example, there was <1% divergence among COI sequences for the pair Morula uva and M. aspera, and similarly for the pair Cronia amygdala and C. aurantiaca. In two cases, morphospecies were not monophyletic (‘Cytharomorula’ cf. grayi and ‘Pascula ochrostoma’; Figs 2 and 3B).
DISCUSSION
Phylogenetic relationships and classification of the ergalataxine genera
Our results indicate polyphyly in nearly all the ergalataxine genera, as hitherto defined by morphological characters. For example, species previously assigned to Morula are found in each of the three major clades, while species of Orania are found in each subclade within major clade C. Here, we use a molecular dataset to construct a new phylogenetic classification that reflects evolutionary relationships rather than superficial similarities in shell morphology. A phylogenetic classification requires that genera are clades that contain their designated type species, but some decisions about naming and ranking remain subjective. When making such decisions, we have preserved current usage (based on morphological definitions of genera) where possible. We emphasize that our study provides only an initial framework which should be refined and tested by further molecular and morphological analyses and consequently we name no new genera at this stage.
The genus Trachypollia is here represented by only one of its four currently recognized members, all of which occur in the eastern Pacific and Atlantic Oceans (EPA) (Figs 1–3, Table 1; Vokes, 1996a; Houart, 1997). Further species, including the type, should be investigated with molecular techniques, not only to test the monophyly of the genus, but also to resolve its position within the Ergalataxinae. If the included species T. lugubris is representative of the genus, it is clear that the Atlantic ‘Morula’ nodulosa must be excluded from the genus, in agreement with Vokes (1996a).
Within major clade A, we propose that three or four genera can be recognized: Tenguella, Muricodrupa and one or two undescribed genera. Subclade U (Figs 1–3A) contains the type species of Tenguella, T. granulata, and we therefore assign to Tenguella all species in subclade U (Figs 1–3A). Tenguella has previously been synonymized with Morula (Fujioka, 1985) and its species have not previously been recognized as a morphologically distinctive group. Included in this group is an unidentified species (Tenguella n. sp. in Figs 1–3A); this has been illustrated as ‘Azumamorula sp.’ (Dharma, 2005) and as ‘Morula mutica’ (Wilson, 1994). The shell shows some similarities to M. mutica, which is the type species of Azumamorula, and it is therefore possible that this genus may prove to be a synonym of Tenguella (Table 1). The other species in clade A are morphologically dissimilar to species of Tenguella. We recommend that Muricodrupa be retained for its type species, M. fenestrata. In the all-gene analysis (Fig. 1), ‘Morula’ anaxares forms a marginally significant clade with M. fenestrata, which is not contradicted in other analyses. Unworn shells of both species show similar sculpture and, if future studies confirm their sister relationship, ‘Morula’ anaxares may be transferred to Muricodrupa. A new genus is required for ‘Morula’ nodulosa, the sole representative of clade A in the Atlantic, well supported as sister to the remaining Indo-West Pacific (IWP) members (Figs 1–3A). The pattern of one or more EPA lineages sister to a more diverse IWP clade is common in tropical gastropod groups (e.g. Williams & Reid, 2004; Claremont et al., in press).
Eleven species that are commonly assigned to Morula form a monophyletic group within major clade B. Since this includes M. uva, the type species of Morula, we propose that the generic name should be restricted to this subclade (Figs 1–3A: subclade V). This clade also contains M. biconica, the type species of Habromorula, but species previously assigned to this group [as a subgenus of Morula: M. (H.) biconica, M. (H.) coronata, M. (H.) japonica, M. (H.) spinosa, M. (H.) striata; Houart, 2004] do not form a clade in any analysis (Figs 1–3A). However, monophyly of this morphologically distinctive group is not strongly contradicted, so that further sampling and analysis could yet support Habromorula as a monophyletic subgenus of Morula. Because they are morphologically dissimilar to species of Morula s. s., other species in clade B (‘Morula’ rumphiusi and ‘Morula’ fiscella) should be assigned to a new genus. The placement of an unidentified species (‘Morula’ n. sp.) is not certain.
Major clade C is the largest and most morphologically disparate of the ergalataxine subgroups and our sampling includes the type species of 10 described genera. Most of these are poorly defined morphologically and some (Phrygiomurex, Usilla, Maculotriton, Bedevina) may be monotypic. The affinities of Phrygiomurex sculptilis and ‘Morula’ echinata are not resolved.
Species in subclade W (Figs 1–3B) are morphologically heterogeneous, including both the smooth-shelled Usilla and foliose Lataxiena, together with some Orania species. The genus Orania is strongly polyphyletic in our analyses, with members in subclades W and Z (Figs 1–3B). Since the type species of Orania (O. fusulus from the Mediterranean; Table 1) was not included, it is impossible to determine to which of the three to five clades of ‘Orania’ the name should be applied. Clearly a revision of the genus is necessary. Meanwhile, in view of the morphological disparity in this group, we suggest that Orania, Usilla and Lataxiena should be retained according to their accustomed usage.
Within subclade X (Figs 1–3B) the genus-level classification of Drupella has been investigated in detail elsewhere (Claremont et al., 2011a). The genus is well supported (Figs 1 and 2), consisting of D. cornus, D. fragum, D. eburnea, D. rugosa and two cryptic species of D. margariticola sensu lato (D. margariticola ‘Oceanic’ and D. margariticola ‘Continental’). Other species in subclade X form a well-supported clade in the all-gene analysis (Fig. 1), including the type species of Cronia and Maculotriton. The type species of Ergalatax, E. pauper, was not sampled, but the included E. contracta is probably close to it and has been considered synonymous (Houart, 2008). Of these three generic names, Cronia has priority (Table 1). Although Ergalatax is not monophyletic in any analysis (Figs 1–3B), our sampling of this genus was limited. Therefore, we conservatively suggest that all three genera are retained until relationships among them can be more thoroughly investigated with increased species-level sampling.
Subclade Y (Figs 1–3B) also contains morphologically diverse species. In the all-gene analysis (Fig. 1), the type species of Oppomorus, O. nodulifera, forms a well-supported clade with O. funiculata and O. purpureocincta and we therefore propose that Oppomorus should be accorded full generic rank (hitherto a subgenus of Morula, e.g. Houart, 2004). Of the remaining taxa in subclade Y, three species previously assigned to Spinidrupa form a clade (‘S.’ infans, ‘S.’ cf. infans and ‘S.’ sp.), but their relationship with the type species, S. euracantha, is unresolved in the only analysis in which the latter was included (Fig. 3B). Furthermore, in the all-gene analysis the ‘Spinidrupa’ clade also includes Bedevina birileffi, the type species of Bedevina, which has priority. Pending the inclusion of additional species, we conservatively recommend the retention of both Spinidrupa and Bedevina.
Subclade Z (Figs 1–3B) contains species of Pascula, Orania and Cytharomorula, but includes the type species of only the last of these. Based on the included species, all three genera, as currently defined, are polyphyletic. It may be that the entire clade should be assigned to one genus. Of these three generic names, Orania has priority (Table 1), but its members also appear in clade W and the type species was not included. Pending a more complete analysis, including the relevant type species, we have conservatively retained all species in the genera to which they have been previously assigned. Also included here is the enigmatic South African species ‘Thais’ castanea; clearly, this is not a member of the rapanine genus Thais, but its appropriate generic assignment remains to be determined. A temporary placement in Orania is suggested. In the two-gene and four-gene analyses (Figs 1 and 2), ‘Morula’ parva is sister to all other members of subclade Z, suggesting that a new genus may be required for this species.
Fossil record and age of the Ergalataxinae
Owing to similarity and convergence of shell form among ergalataxine and rapaninemuricids and lack of unambiguous synapomorphies for many clades (Vermeij & Carlson, 2000), unequivocal identification of fossils can be problematic. Vermeij & Carlson (2000) gave the earliest occurrences of several ergalataxine genera as Late Miocene, while Landau, Houart & Da Silva (2007) reported that the group appeared in the Atlantic in the Early Miocene. The oldest certain ergalataxine known to us is a specimen of ‘Taurasia’ sacyi Cossmann & Peyrot, 1923 from the Early Oligocene (Gaas, Espibos, France; Rupelian stage, 28.4–33.9 Ma; Collection of R. Houart). This fossil resembles Recent species of Pascula, Ergalatax and Cytharomorula, implying a minimum age for major clade C. This, however, is insufficient for a reliable time calibration of our phylogeny. Palaeontological evidence suggests a Cretaceous origin for the Muricidae as a whole (Vermeij, 1996). Some recent molecular studies have attempted to estimate ages of ergalataxine clades, giving figures of 5 Ma for Drupella (Claremont et al., 2011a) and 82 Ma for the separation of the Ergalataxinae and Rapaninae (Claremont et al., in press). Detailed study of ergalataxine fossils is required before these estimates can be refined.
CONCLUSIONS
Although this framework represents the most complete phylogeny of the Ergalataxinae yet constructed, much work remains to be done to achieve a robust phylogenetic classification of this taxonomically difficult subfamily. We have clarified the classification of ‘Morula’ s. l., defining as distinct clades the genera Morula s. s., Tenguella and Oppomorus, and have confirmed the validity of Muricodrupa, Phrygiomurex and Drupella. The definition and validity of several traditional genera, including Orania, Pascula, Cytharomorula, Cronia, Ergalatax, Maculotriton, Usilla, Lataxiena, Spinidrupa and Bedevina remain to be confirmed (see summary in Table 1). At least three new genera (for ‘Morula’ nodulosa; for ‘M.’ fiscella and ‘M.’ rumphiusi; and for ‘M.’ parva) appear to be required. Several living genera that have been included in Ergalataxinae by some authors were not available for inclusion in our analyses and their placement should be tested by molecular analysis (Azumamorula, Daphnellopsis, Lindapterys, Uttleya; Supplementary material: Table S2).
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of Molluscan Studies online.
ACKNOWLEDGEMENTS
We are very grateful for the assistance and loans provided by the staff of museums and research institutions, especially P. Bouchet, V. Héros, B. Buge and N. Puillandre of Museum Nationale d'Histoire Naturelle, Paris (MNHN); G. Paulay, J. Slapcinsky and M. Bemis of the Florida Museum of Natural History; E. Naranjo of the Universidad Nacional Autónoma de México; J.E. Michel-Morfin and V. Landa-Jaime of Universidad de Guadalajara; Y. Camacho of the University of Costa Rica; S. Slack-Smith and C. Whisson of the Western Australian Museum, Perth; M. Oliverio and A. Barco of ‘La Sapienza’ University of Rome; A. Kerr and B. Smith of the University of Guam Marine Laboratory; G. Williams of the Swire Marine Lab, Hong Kong; and I. Loch and A. Miller of the Australian Museum, Sydney. We would also like to thank many others who provided specimens or who assisted us in the field including L. Alsayegh, C. Bennett, C. Bird, C. Carlson, T. Haga, T. Hamada, G. Herbert, Y. Ito, R. Jones, Y. Kano, P. Kuklinski, T. Nakano, B. Ng, S. Nielson, N. Razalli, K.S. Tan, S.H. Tan, J.D. True, R.C. Willan, and Z. Yasin. The MNHN material used in this study was collected during the following expeditions: PANGLAO 2004 Marine Biodiversity Project [a joint project between University of San Carlos, Cebu City (USC; co-PI Danilo Largo) and MNHN (co-PI Philippe Bouchet), funded by the Total Foundation and the French Ministry of Foreign Affairs]; the AURORA 2007 cruise [M/V DA-BFAR associated with the National Museum of the Philippines (NMP, co-PI Marivene Manuel), MNHN (co-PI Philippe Bouchet) and BFAR, funded through a grant from the Lounsbery Foundation]; the MNHN-IRD-PNI Santo 2006 expedition (funded by grants from, among others, the Total Foundation and the Stavros Niarchos Foundation); EBISCO, NORFOLK 2 and CONCALIS cruises [R/V Alis deployed from Nouméa by the Institut de Recherche pour le Développement (IRD)]. M.C. was supported by a studentship from the Natural History Museum, London and by an Imperial College Deputy Rector's studentship. We thank the Acting Editor Ellen Strong, Marco Oliverio and one anonymous reviewer for helpful comments on a previous version of this manuscript.