-
PDF
- Split View
-
Views
-
Cite
Cite
Kim Schwarze, Kevin L. Campbell, Thomas Hankeln, Jay F. Storz, Federico G. Hoffmann, Thorsten Burmester, The Globin Gene Repertoire of Lampreys: Convergent Evolution of Hemoglobin and Myoglobin in Jawed and Jawless Vertebrates, Molecular Biology and Evolution, Volume 31, Issue 10, October 2014, Pages 2708–2721, https://doi.org/10.1093/molbev/msu216
Close - Share Icon Share
Abstract
Agnathans (jawless vertebrates) occupy a key phylogenetic position for illuminating the evolution of vertebrate anatomy and physiology. Evaluation of the agnathan globin gene repertoire can thus aid efforts to reconstruct the origin and evolution of the globin genes of vertebrates, a superfamily that includes the well-known model proteins hemoglobin and myoglobin. Here, we report a comprehensive analysis of the genome of the sea lamprey (Petromyzon marinus) which revealed 23 intact globin genes and two hemoglobin pseudogenes. Analyses of the genome of the Arctic lamprey (Lethenteron camtschaticum) identified 18 full length and five partial globin gene sequences. The majority of the globin genes in both lamprey species correspond to the known agnathan hemoglobins. Both genomes harbor two copies of globin X, an ancient globin gene that has a broad phylogenetic distribution in the animal kingdom. Surprisingly, we found no evidence for an ortholog of neuroglobin in the lamprey genomes. Expression and phylogenetic analyses identified an ortholog of cytoglobin in the lampreys; in fact, our results indicate that cytoglobin is the only orthologous vertebrate-specific globin that has been retained in both gnathostomes and agnathans. Notably, we also found two globins that are highly expressed in the heart of P. marinus, thus representing functional myoglobins. Both genes have orthologs in L. camtschaticum. Phylogenetic analyses indicate that these heart-expressed globins are not orthologous to the myoglobins of jawed vertebrates (Gnathostomata), but originated independently within the agnathans. The agnathan myoglobin and hemoglobin proteins form a monophyletic group to the exclusion of functionally analogous myoglobins and hemoglobins of gnathostomes, indicating that specialized respiratory proteins for O2 transport in the blood and O2 storage in the striated muscles evolved independently in both lineages. This dual convergence of O2-transport and O2-storage proteins in agnathans and gnathostomes involved the convergent co-option of different precursor proteins in the ancestral globin repertoire of vertebrates.
Introduction
Transport and storage of molecular O2 in vertebrates is accomplished by proteins of the globin superfamily. Globins are small heme-containing proteins that reversibly bind O2 and other gaseous ligands, and which are present in essentially all animal phyla. The globin superfamily is a classical model system to study the function and evolution of proteins, genes, and gene families (Hardison 1996; Graur and Li 2000; Gillemans et al. 2003; Vinogradov et al. 2007). In addition to their role in O2 supply, globins may also be instrumental in the detoxification of reactive O2 and nitrogen species, regulation of apoptosis, and signal transduction (Dickerson and Geis 1983; Weber and Vinogradov 2001; Wittenberg and Wittenberg 2003; Burmester and Hankeln 2009).
The vertebrate globins provide a classic example of how gene duplication can facilitate the evolution of new protein functions. The globin repertoire of extant vertebrates is the product of successive genome and gene duplication events, followed by differential gene retention among lineages (Hoffmann et al. 2011; Storz et al. 2011, 2013; Hoffmann, Opazo, and Storz 2012). Eight distinct globin types have been identified in the jawed vertebrates (Gnathostomata). Hemoglobin (Hb), which transports O2 in red blood cells (Dickerson and Geis 1983), and myoglobin (Mb), which supplies O2 to the mitochondria of cardiac and striated muscle cells (Wittenberg and Wittenberg 1989), are the best known members of the globin superfamily and are among the most intensively investigated proteins in the biomedical sciences. Although Mb is a monomer, Hb from jawed vertebrates forms a heterotetramer of two α- and two β-chains (Dickerson and Geis 1983), with each of the latter belonging to distinct globin families that originated through gene duplication before the radiation of gnathostomes. Different α- and β-subtypes have evolved independently by gene duplication in the gnathostome classes (Hoffmann, Storz, et al. 2010; Schwarze and Burmester 2013). In many cases, the Hb subtypes are expressed at different stages of development. More recently, other globins with less well-defined functions have been discovered. These include neuroglobin (Ngb) (Burmester et al. 2000) and androglobin (Adgb) (Hoogewijs et al. 2012), which originated early in the evolution of metazoan animals and have orthologs in protostomes and deuterostomes. Adgb and cytoglobin (Cygb) (Kawada et al. 2001; Burmester et al. 2002; Trent and Hargrove 2002) appear to be present in all gnathostomes, whereas globin E (GbE) (Kugelstadt et al. 2004; Blank, Kiger, et al. 2011), globin X (GbX) (Roesner et al. 2005; Blank, Wollberg, et al. 2011), and globin Y (GbY) (Fuchs et al. 2006) have been secondarily lost in multiple vertebrate lineages independently (Hoffmann et al. 2011; Schwarze and Burmester 2013; Storz et al. 2013).
Agnatha, that is lampreys (Petromyzontiformes) and hagfishes (Myxiniformes), are jawless vertebrates that diverged from the ancestor of gnathostomes (jawed vertebrates) about 500 Ma (Kuraku and Kuratani 2006; Smith et al. 2013). Extant agnathans are most likely monophyletic and are referred to as Cyclostomata (Kuraku et al. 1999; Kuratani and Ota 2008). Despite their common name and analogous respiratory functions, O2-transport Hbs of jawed and jawless vertebrates are structurally distinct and are not orthologous. Previous phylogenetic analyses suggested that agnathan Hbs share a closer affinity to gnathostome Cygb than to red blood cell Hbs in gnathostomes that share an analogous function in circulatory O2 transport (Katoh and Miyata 2002; Hoffmann, Opazo, et al. 2010). Agnathan hemoglobins (aHbs) are usually monomers in the oxygenated state and polymerize to dimers or tetramers when deoxygenated (Fago et al. 2001). The O2-binding properties of lamprey aHbs resemble those of gnathostome Hbs, with both exhibiting low O2 affinities and large Bohr effects (Wald and Riggs 1951; Fago et al. 2001). As in the case of gnathostome Hbs, lamprey aHbs consist of multiple distinct subunit isoforms, which may be differentially expressed during ontogeny. For example, Lanfranchi et al. (1994) demonstrated a switch in aHb expression during development of the Lombardy brook lamprey (Lampetra zanandreai), which can be considered analogous to the switch of embryonic to adult Hb chains in gnathostomes.
Lampreys occupy a key phylogenetic position for illuminating the origins and evolution of vertebrate globins. The recent availability of the genomic sequences of the sea lamprey Petromyzon marinus (Smith et al. 2013) and the Arctic lamprey Lethenteron camtschaticum (Mehta et al. 2013) provides the opportunity to characterize the globin repertoire of these species. Genome mining and expression analyses show the presence of functional Hb-like, Mb-like, and Cygb genes in these species, and suggest that Ngb has been lost. Comparisons with the globins of jawed vertebrates by phylogenetic reconstructions and gene synteny analyses further allowed us to unravel the evolution of the vertebrate globin family. The results suggest that jawed and jawless vertebrates convergently evolved “Hbs” and “Mbs” with O2 transport/storage functions in blood and striated muscle, respectively, from a common globin ancestor.
Results
The Globin Gene Repertoire of the Sea Lamprey and the Arctic Lamprey
In silico analyses of the genome assembly (Smith et al. 2013) and transcriptome data sets from the sea lamprey (P. marinus) identified 25 globin genes (supplementary table S1, Supplementary Material online). At least partial sequences of these genes were found in the genome data and are distributed on 14 scaffolds. Two of the globin genes (aHb ps1 and aHb ps2), which are located on scaffolds GL476413 (739,046–740,131) and GL480013 (24,143–25,251), respectively, harbored premature stop codons and splice site mutations. No corresponding expressed sequence tags (ESTs) were found for these genes, consistent with the interpretation that they are pseudogenes. Two other genes (sea lamprey GbX2 and aHb5c; see below for the nomenclature) were only partially represented in the genome and transcriptome assemblies. The open reading frames of the other globin genes are composed of either three or five exons and range from 426 to 669 bp (141–222 amino acids) in length.
Analyses of the genome of the Arctic lamprey (L. camtschaticum) (Mehta et al. 2013) revealed 23 globin genes (supplementary table S2, Supplementary Material online), which are distributed on seven scaffolds. In addition, six contigs were found that include at least fragments of globin genes. By comparison with the globin repertoire of the sea lamprey and with other globin sequences, the full length open reading frames of 14 globin genes could be deduced. Two globin genes, which were not fully represented in the Arctic lamprey genome assembly, have full-length cDNA sequences at EMBL/GenBank (aHb1 and aHb7). Thus 16 globin genes of Arctic lamprey were full length. Ten other globin genes were likely incomplete in the current genome assembly (or represent truncated pseudogenes) and full-length coding sequences could not be determined by combining other data.
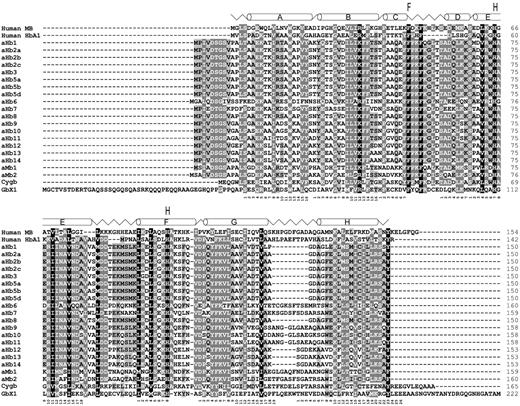
An alignment of the deduced amino acid sequences of all globins from the sea lamprey (fig. 1 and supplementary fig. S1, Supplementary Material online) and the Arctic lamprey (supplementary fig. S1, Supplementary Material online) revealed conservation of the typical residues required for heme-coordination and ligand stabilization, particularly the proximal and distal histidines at positions F8 (eighth amino acid of helix F) and E7, respectively, and the highly conserved phenylalanine at CD1.
Alignment of 23 sea lamprey globin proteins with human myoglobin (MB) and α-hemoglobin (HbA1). Incomplete sea lamprey sequences are not shown. The α-helical structure of sea lamprey aHb5 is shown on top of the alignments. Amino acids strictly conserved between the globins are shaded (black: 100% conservation, dark gray: 80%, light gray: 60%). The functionally important phenylalanine (F) at CD1 and the distal and proximal histidines (H) at E7 and E8 are indicated. The globin consensus numbering is given below the sequences.
In both lamprey species, partial sequences of putative Adgb orthologs (Hoogewijs et al. 2012) were detected (P. marinus: scaffold GL476565; L. camtschaticum: KE994039), but due to the fragmentary nature of the Adgb genes they were not included in further analyses.
Phylogeny of Vertebrate Globins
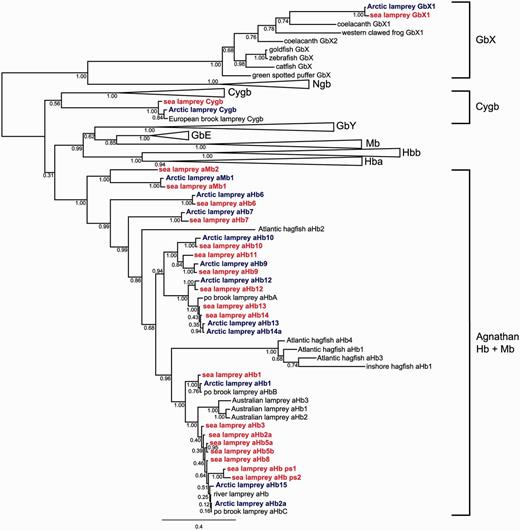
To assess orthologous and paralogous relationships of lamprey globins, a phylogenetic analysis was conducted based on an alignment of 136 vertebrate globins (supplementary table S3 and Supplementary Data, Supplementary Material online). The Bayesian tree showed well-supported monophyletic clades of vertebrate Ngb, Cygb, GbY, Mb, GbE, and α- and β-Hb, respectively, which were therefore collapsed (fig. 2). The full version of the tree is given in supplementary figure S3, Supplementary Material online.
Simplified Bayesian phylogenetic tree of agnathan globins. The numbers at the nodes are posterior probabilities. The bar represents 0.4 PAM distance. Sea lamprey globins are colored in red, Arctic lamprey globins are blue. The common names of the species are given. See supplementary table S1, Supplementary Material online, for details of the proteins and supplementary figure S3, Supplementary Material online, for the full version of the tree.
The lamprey globins fell into three separate clades: One sequence of each species was placed with gnathostome GbX (the GbX2 proteins were not included in the phylogenetic analyses because parts of the sequences were missing in both lamprey species [supplementary fig. S1, Supplementary Material online]), a second group included putative orthologs of Cygb from lampreys which were placed as sister to gnathostome Cygb (posterior probability [PP] = 0.56), and the third group placed the remaining lamprey globins and available hagfish globins together with high support (PP = 1.0). The clade containing the gnathostome-specific globins (GbY, Mb, GbE, α- and β-Hbs) was supported by 0.99 PP and was sister to the clade of aHbs/agnathan myoglobins (aMbs) (0.31). Within gnathostome globins, there was strong support for grouping Mb with GbE and α-Hb with β-Hb, consistent with the results of previous phylogenetic and synteny analyses (Hoffmann et al. 2011; Hoffmann, Opazo, and Storz 2012; Hoffmann, Opazo, Hoogewijs, et al. 2012; Schwarze and Burmester 2013).
Two Paralogous GbX Genes in Lampreys
Sequence comparisons and phylogenetic analyses identified two paralogous GbX genes (GbX1 and GbX2) in both lamprey species. In the sea lamprey genome, the GbX genes are on scaffolds GL477600 and GL476484, respectively; in the Arctic lamprey genome, they reside on KE993715 and KE993935. As mentioned, neither of the two GbX2 orthologs had complete sequence. Like other vertebrate GbX genes (Roesner et al. 2005; Blank, Wollberg, et al. 2011; Blank and Burmester 2012), the GbX1 genes display a five exon/four intron structure and include the 3′- and 5′-extensions typical for GbX. The available sequences of the two sea lamprey GbX proteins exhibited 54% identity and 69% similarity (Arctic lamprey: 45/62%). Sea lamprey and Arctic lamprey GbX1 proteins share 97% of the amino acids. Both GbX1 sequences display N-terminal myristoylation and palmitoylation sites at positions 2 and 3, respectively, as observed in other vertebrate GbXs (Blank, Wollberg, et al. 2011). Gene synteny analyses moreover revealed that the lamprey GbX1 genes are flanked by SRP14 and PLEKHG; these genes and their orientation are also conserved in the GbX region of Xenopus tropicalis (supplementary fig. S4, Supplementary Material online). A PLEKHG paralog is located adjacent to GbX2 of L. camtschaticum, suggesting that the GbX paralogs in lampreys originated through duplication of the genomic region.
Identification of Lamprey Hbs
The P. marinus genome harbors at least 18 intact aHb genes and two aHb pseudogenes, as identified by sequence comparisons and phylogenetic analyses. The aHb genes are distributed on nine scaffolds of the genome assembly (supplementary fig. S5, Supplementary Material online). Scaffold GL478636 includes aHb1 and aHbs11-14, which are all in the same orientation. With the exception of aHb1, these aHbs form a common clade in the phylogenetic tree with aHbs9 and 10 (fig. 2).
Nine aHb genes could be assigned to one of the four previously sequenced adult aHb chains of the sea lamprey (Li and Riggs 1970; Hombrados et al. 1983, 1987; Qiu et al. 2000). The genes ENSPMAG00000007266, ENSPMAG00000007259, and ENSPMAG00000007276, which reside on scaffold GL477137, translate into proteins with identical amino acid sequences that correspond to the major aHb component of this species, PM II (AF248645; (Qiu et al. 2000) and were thus named aHbs2a–c (supplementary fig. S5, Supplementary Material online). The nucleotide sequences of genes ENSPMAG00000007266 (aHb5a) and ENSPMAG00000007259 (aHb5b) on scaffold GL477423 were identical, reflecting either a recent duplication event or an assembly artifact. They correspond to the Hb component PM V, which is the best studied Hb subunit of the sea lamprey (Li and Riggs 1970; Hendrickson et al. 1973; Hombrados et al. 1983; Honzatko et al. 1985). A gene fragment represented by a 3′-exon on scaffold GL480013 (30,606-30,701; aHb5c) is also identical to PM V. Gene ENSPMAG00000005317 on scaffold GL477423 closely resembles PM V (99% identity) and was named aHb5d. Genes ENSPMAG00000001587 (aHb1; incorrectly annotated on scaffold GL478636) and ENSPMAG00000005328 (aHb3 on scaffold GL477423) match the protein sequences of PM I (P09967) and PM III (P09968), respectively (Hombrados et al. 1987).
Nine newly identified genes represent previously uncharacterized aHbs of the sea lamprey. They were named according to the topology of the phylogenetic tree (fig. 2). aHb6 (scaffold GL479302: 428,488–432,549) was not annotated by ENSEMBL, but has an ortholog in the Arctic lamprey (see below). aHb6 is highly represented in ESTs of embryonic P. marinus. aHb7 is also found among the embryonic ESTs (full sequence in EE278870), though only exon 3 is present in the genome assembly (GL487383: 3,904-4,026). aHb8 (ENSPMAG00000005367) is another embryonic Hb and shares 98% amino acid identity with Hb1 mRNA of L. camtschaticum (see below). aHb9 (ENSPMAG00000008540) resides on scaffold GL476782, whereas aHbs11–14 are on GL478636. aHb11 is coded by ENSPMAG00000001592 whereas aHb12–14 correspond to a misannotated gene, which is covered by ENSPMAG00000001587. aHb10 is represented by an EST (FD718926) though only the 5′-exon is located on GL478504. Transcripts of all six genes, aHb9–14, were found in the ESTs from P. marinus embryos or larvae, suggesting a specific function in early life stages (supplementary table S4, Supplementary Material online).
The genome of the Arctic lamprey L. camtschaticum also revealed 18 aHb genes (supplementary table S2, Supplementary Material online), of which 14 full-length coding sequences could be deduced. Four additional aHb genes with partial sequences were identified. In cases where the orthology of lamprey globins could be inferred, the L. camtschaticum aHb genes were named according to the putative P. marinus ortholog. Sequence comparisons and phylogenetic analyses revealed seven aHb genes in the Arctic lamprey that appeared to have 1:1 orthologs in P. marinus: aHb1, aHb6, aHb7, aHb9, aHb10, aHb11, and aHb12. This approach did not allow a reliable assignment of orthology of aHb13 and aHb14, which was thus deduced from the positions of the genes in the genome. aHb14 appears to have been duplicated in the Arctic lamprey. Three genes on scaffold KE993857 and a gene on contig APJL01123255 differ in only 2–6 bp and translate into identical amino acid sequences. Three aHb genes on scaffold KE993857 resemble the aHb2 genes of the sea lamprey, which reside on scaffold GL477137 of that species. In addition, the two scaffolds share conserved synteny of the genes AZIN1 and KLHL10 (supplementary figs. S5–S7, Supplementary Material online); the Arctic lamprey genes were therefore named aHb2-c according to their positions in the genome. An additional gene on APJL01123255 which closely resembles the aHb2-c genes was named aHb2d. BLAST searches showed that L. camtschaticum aHb2c corresponds to the previously identified Hb1 mRNA of this species. The aHb2 proteins differ in five to six amino acids from the major components of the adult Hb of P. marinus, aHb2, whereas aHb3 and 5 are apparently not represented in the L. camtschaticum genome. Arctic lamprey aHb15 closely resembles aHb2, but no clear ortholog could be assigned and we continued the numbering of the aHb genes. Because L. camtschaticum aHb16–18 are only represented by one or two exons, no clear ortholog could be assigned.
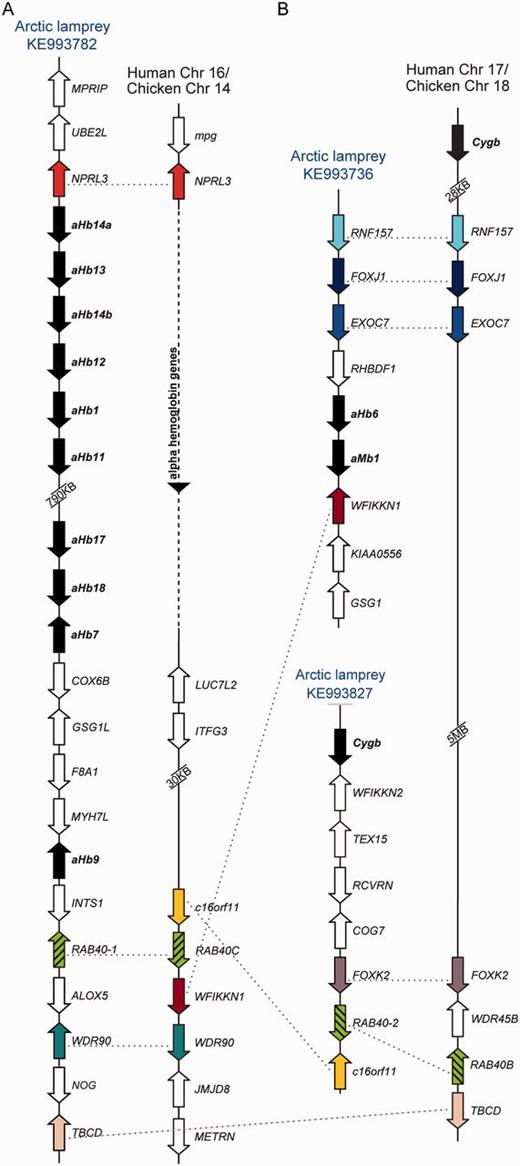
The globin genes of L. camtschaticum were found on four scaffolds of the current genome assembly (supplementary fig. S6, Supplementary Material online). In addition, three contigs include aHb genes. Scaffold KE993782 is orthologous with scaffolds GL478636 and GL476782 of P. marinus (supplementary fig. S7, Supplementary Material online). This large scaffold includes the region corresponding to scaffold GL478636 of P. marinus where aHb1, aHbs11–13, and aHb14a+b are located, and the region corresponding to P. marinus GL476782 that harbors aHb9. Scaffold KE993782 also includes aHb7 and the fragmentary genes aHb17 and aHb18. The gene NPRL3 was found adjacent to this aHb cluster of both lampreys. Synteny analyses moreover show the conservation of NPRL3 5′ to the α-Hb cluster of gnathostome vertebrates in a tail-to-tail orientation (fig. 3A). Moreover, scaffolds GL478636 of P. marinus and KE993782 of L. camtschaticum also share WDR90 and RAB40 genes, which both reside downstream of the gnathostome α-Hb cluster (fig. 3A).
Synteny analyses of selected lamprey globin genes. Orthologous genes are shown in the same color. (A) The genes NPRL3, RAB40, and WDR90 link the aHb cluster on scaffold KE993782 with the gnathostome αHb cluster. (B) FOXK2 and RAB40 paralogs (hatched) link the Arctic lamprey Cygb-scaffold (KE993827) with the gnathostome Cygb locus, whereas RNF157, FOXJ1 and EXOC7 link this latter scaffold to aMb1–aHb6 cluster (scaffold KE993736).
Two Functional Mbs in Lampreys
Romero-Herrera et al. (1979) reported the tryptic pattern and the amino acid composition of a putative Mb from the cardiac muscle of P. marinus. The translated amino acid sequence of ENSPMAG00000006056 provided an identical match to this protein and was thus designated as agnathan myoglobin 1 (aMb1). The phylogenetic analysis revealed a close affinity between aMb1 and the translated product of ENSPMAG00000008310 (fig. 2), which was therefore named aMb2. Sequence comparison further identified a putative ortholog of P. marinus aMb1 on KE993736 of the Arctic lamprey, plus a partial sequence that corresponds to exon 2 on contigs APJL01135086 and APJL01176948.
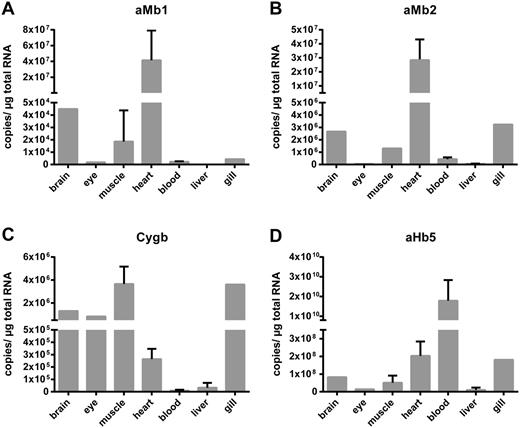
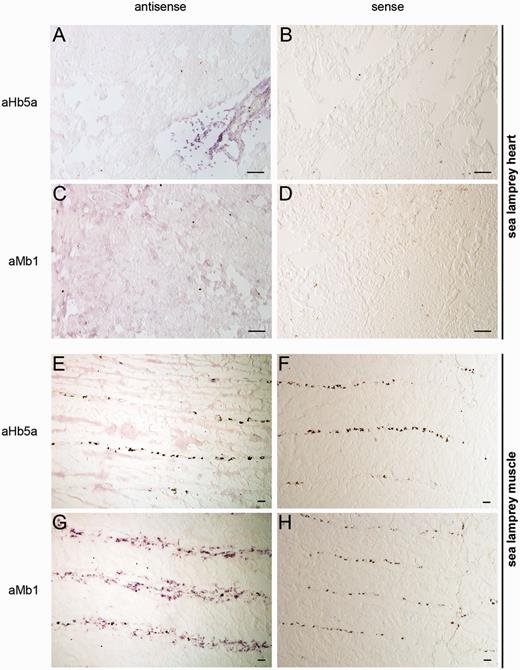
Quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR) experiments revealed the presence of mRNAs of both aMb1 and aMb2 in the heart of the sea lamprey, whereas expression levels in most other tissues, including blood, were much lower (fig. 4A and B). Moderate levels of aMb1 and aMb2 mRNA were also found in skeletal muscle. This suggests that both aMb1 and aMb2 act as functional Mbs in the sea lamprey. aHb5a, which was used as control, showed the highest mRNA level in the blood (fig. 4D). mRNA in situ hybridization (ISH) studies showed strong aMb1 antisense signals in the myonucleus of the myofibers and a more diffuse staining in the remaining tissue (fig. 5G). Hybridization with sense probes, which served as negative controls, gave no signal (fig. 4B, D, F, and H). In heart tissue, aMb1 antisense probe showed a diffuse expression pattern similar to the aHb5a mRNA distribution in heart and skeletal muscle (fig. 5). By contrast, antisense probes of aHb5a gave strong ISH signals in the erythrocytes in blood vessels (fig. 5A).
Quantification of mRNA levels of selected sea lamprey globins in different tissues. Using qRT-PCR the mRNA copy numbers of the aMb1 (A) and Mb2 (B), the putative Cygb (C), and aHb5a (D) were obtained. aMb1 and aMb2 were detected in heart, brain, gill, and skeletal muscle, aHb5a was most highly expressed in blood, whereas Cygb showed a widespread distribution.
ISH of sea lamprey aHb5a (A, E) and aMb1 (C, G) antisense RNA probes in heart (A, C) and muscle (E, G) cryosections. aHb5a mRNA was detected in erythrocytes (A), which reside in the blood vessels, but not in the muscle tissue (E). Expression of aMb1 mRNA was detected as diffuse staining in heart sections (C) and in myonucleus of myofibers in muscle (G). Sense probes, which were used as negative controls, showed no signals (B, D, F, H). Scale bar = 100 µm.
A Putative Cygb in Lampreys
The agnathan Cygbs (from the sea lamprey, the Arctic lamprey, and the European brook lamprey) were identified as putative orthologs of gnathostome Cygb on the basis of sequence comparisons and expression patterns. In the Bayesian tree, these proteins grouped with the gnathostome Cygbs, albeit with low support (0.56 PP) (fig. 2). qRT-PCR analyses showed low to moderate expression levels in a variety of sea lamprey tissues, with brain, eyes, gills, and muscles having the highest Cygb mRNA levels (fig. 4C).
Exon 1 of the sea lamprey Cygb resides on the genomic scaffold GL478089, whereas exons 2 and 3 reside on GL477469 (supplementary fig. S5, Supplementary Material online). In the Arctic lamprey, the full length Cygb is on scaffold KE993827 (supplementary fig. S6, Supplementary Material online). Synteny analyses showed that WFIKKN2 is present on the 3′-side of the Cygb genes (supplementary fig. S7, Supplementary Material online). Notably, FOXK2 and RAB40 genes reside downstream of both agnathan and gnathostome Cygb (fig. 3B). In addition, genes RNF157, FOXJ1, and EXOC7 are located upstream of sea lamprey aHb6–aMb1, whereas homologous genes are positioned downstream of the gnathostome Cygb (fig. 3B).
Discussion
The Diversity of Lamprey Hbs
We identified 18 functional aHb and two aHb pseudogenes in the genome of the sea lamprey P. marinus. Only four Hb chains had been identified previously in protein biochemical studies (named here aHb1, aHb2, aHb3, and aHb5) and represent subunit components of the adult aHb (Li and Riggs 1970; Hombrados et al. 1983, 1987; Qiu et al. 2000) (supplementary table S1, Supplementary Material online). Another five aHb genes closely resemble one of these chains (>98% identity), and most likely represent recent duplicates whose products were not distinguishable from one another in the original protein studies. This interpretation is supported by tandemly linked chromosomal arrangements of aHb2a, b, and c, and aHb5a and d, respectively.
Products of the other nine intact aHb genes were not previously identified as subunits in studies on the native adult Hb proteins. These studies did not examine earlier life stages and it is likely that these loci are predominantly expressed prior to metamorphosis. This interpretation is supported by the expression pattern, which was derived from the transcriptomes and analyzed at Biosample (http://www.ncbi.nlm.nih.gov/biosample/, last accessed July 22, 2014). These data show preferential expression of aHb6, aHb7, and aHb12 in the eggs, aHb9, aHb10, aHb11, and aHb14 in the embryos, and aHb7, aHb9, aHb11, aHb12, aHb13, and aHb14 in the larvae (supplementary table S4, Supplementary Material online). This observation confirms a differential expression of aHbs in adults and earlier life stages (Lanfranchi et al. 1994), which may reflect functional differentiation of the aHb isoforms that have distinct O2-binding properties (Bird et al. 1976). Of note, the phylogenetic tree shows that two of the aHbs expressed in eggs (aHb6 and aHb7) represent the earliest branching lineages (fig. 2).
The aHb repertoire of the Arctic lamprey L. camtschaticum includes at least 18 distinct genes, but the aHb protein of this species has not been functionally characterized. Nevertheless, three aHb mRNA sequences are available at EMBL/GenBank, which correspond to aHb1, aHb2, and aHb7 of this study. It is unknown whether these chains code for components of the adult aHb. However, such an interpretation is supported by the fact that the orthologs of aHb1 and aHb2 are also present in the adult Hb of the sea lamprey. Notably, both sea lamprey aHb5 and aHb2, and Arctic lamprey aHb2 genes have multiple copies in the genomes, suggesting a high level of expression and that their encoded products are incorporated as major subunit isoforms of adult aHb.
Gene Duplication, Genome Duplication, and the Origins of Vertebrate-Specific Globins
Phylogenetic analyses indicate that Adgb, GbX, and Ngb are ancient globins that originated prior to the radiation of Protostomia and Deuterostomia (Roesner et al. 2005; Blank and Burmester 2012; Hoffmann, Opazo, Hoogewijs, et al. 2012; Hoogewijs et al. 2012; Storz et al. 2013) (fig. 6). We identified two globins that correspond to GbX, confirming the early divergence of this globin type. Putative Adgb genes were found in the genomes, but were not further analyzed because of their fragmentary nature. Notably, the assemblies of the P. marinus and L. camtschaticum genomes do not contain an Ngb ortholog, and no Ngb-like transcripts were found in the ESTs of the agnathans. This suggests that Ngb has been deleted in the Agnatha—a surprising finding given that this ancient, highly conserved globin protein is present in every gnathostome taxon that has been examined to date (with the possible exception of sharks; see Venkatesh et al. 2007, 2014).
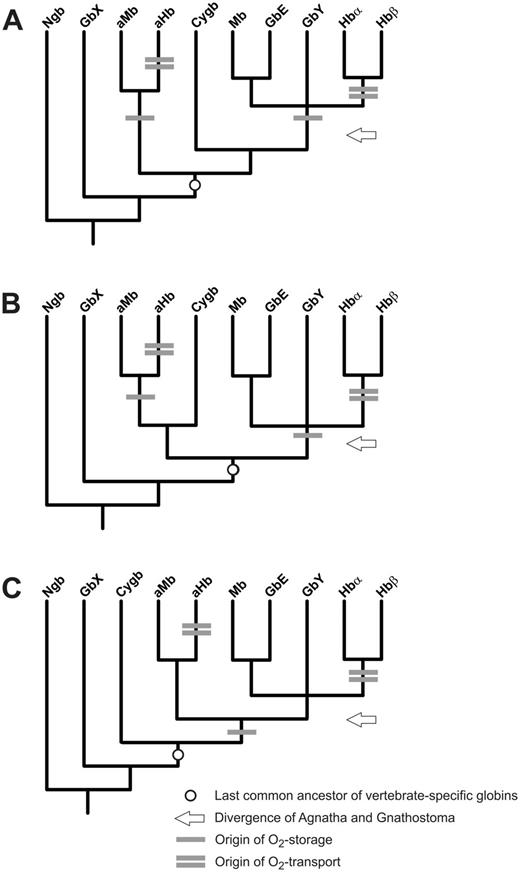
Hypothesized evolution of respiratory function in vertebrate globins. The three possible positions of Cygb are depicted in simplified models illustrating alternative relationships among the eight primary vertebrate globin types (A–C). One bar indicates the origin of O2-storage function (and, possibly, pentacoordination), whereas two bars indicate the origin of blood O2-tansport function. The circle indicates the last common ancestor of the vertebrate-specific globins and the arrow the time of divergence of Agnatha and Gnathostomata. Note that if last common ancestor of the vertebrate-specific globins already had an O2-storage function, this function may have also been lost in Cygb.
The last common ancestor of Gnathostomata and Agnatha had at least six (fig. 6A and B) or five (fig. 6C) distinct globin types. In the most parsimonious scenario (fig. 6C), the last common ancestor of Gnathostomata and Agnatha possessed Adgb, GbX, Ngb, Cygb, and a globin locus that eventually gave rise to agnathan aHbs and aMbs, gnathostome Hbs and Mbs, as well as gnathostome GbE and GbY. Thus, remarkably, Cygb is the only orthologous vertebrate-specific globin lineage that has been retained in both gnathostomes and agnathans. Invertebrate globins, including those from tunicates, hemichordates and cephalochordates, may well group with Adgb, Ngb and GbX, but they are not nested within the set of vertebrate-specific globin discussed here (Storz et al. 2011, 2013; Blank and Burmester 2012; Hoffmann, Opazo, Hoogewijs, et al. 2012; Hoogewijs et al. 2012).
Gene synteny may provide important clues regarding the origins of vertebrate-specific globins. Notably, the aHb locus in the lamprey genomes that includes aHb1, aHb7, aHbs11–14, aHb17, and aHb18 is flanked by the gene NPRL3 upstream and by genes RAB40 and WDR90 downstream; copies of these same genes are located in the same positions in the α-Hb gene cluster of amniote vertebrates (corresponding to the P-terminus of human Chromosome 16) (fig. 3A). This pattern of conserved synteny reflects a paralogous relationship between the agnathan aHb genes and the gnathostome α-Hb genes that likely stems from one or two rounds of whole-genome duplication (WGD) in the vertebrate common ancestor. The weight of available evidence suggests that two rounds of WGD occurred prior to the split between agnathans and gnathostomes (Kuraku and Kuratani 2006; Kuraku 2008, 2010; Kuraku et al. 2009; Smith et al. 2013), although the genomic organization of Hox clusters in the lamprey L. camtschaticum has been interpreted as evidence that WGDs may have occurred independently in the lampreys and gnathostomes (Mehta et al. 2013). Conserved synteny between the agnathan gene cluster that contains aHb1, aHb7, aHbs11–14, aHb17 and aHb18 and the gnathostome α-Hb gene cluster, and the 3:1 pattern of conserved synteny between the agnathan aHb6/aMb1 cluster, agnathan Cygb, and gnathostome Cygb (fig. 3), are both consistent with the view that at least one round of WGD occurred prior to the divergence of agnathans and gnathostomes. In combination with the phylogenetic reconstruction (fig. 2), patterns of conserved synteny suggest a possible orthologous relationship between the Cygb genes of agnathans and gnathostomes, as Cygb is flanked by FOXK2 and RAB40 genes in the genomes of both taxa (fig. 3B).
Convergent Evolution of Agnathan and Gnathostome Hbs and Mbs
Vertebrate Hb and Mb are famous for their respiratory functions: Working in tandem, they jointly ensure an adequate cellular O2 supply for aerobic energy production (Dickerson and Geis 1983; Weber and Vinogradov 2001; Wittenberg and Wittenberg 2003). Although the functional properties of agnathan aHbs have been well documented, we have conclusively demonstrated that lampreys also possess two distinct aMbs—proteins highly expressed in cardiac muscle that may have an O2-storage function analogous to that of gnathostome Mb. In fact, we could assign sea lamprey aMb1 to a protein previously isolated from the heart of this species (Romero-Herrera et al. 1979), and further documented that this gene and a second gene (aMb2) are expressed in cardiac muscle, and to lesser degrees in brain, gills, and skeletal muscle (figs. 4 and 5). Putative orthologs of both proteins were identified in the Arctic lamprey.
The lamprey aMbs are clearly not orthologous to gnathostome Mb (fig. 2); rather it appears that the aHb and aMb gene clusters represent products of repeated rounds of tandem duplication that were specific to the agnathan lineage. Thus, ancestral agnathan and gnathostome globins each independently evolved functions related to erythrocyte-based O2 transport, referred to as Hb-function, and muscle-specific O2 supply, referred to as Mb-function. This conclusion does not depend on the phylogenetic position of Cygb, which remains unresolved (fig. 2). There are good reasons to suppose that the O2-storage function more closely approximates the ancestral state of the Mb/Hb progenitor proteins, as an authentic O2-transport function requires the prior existence of a circulatory system. In principle, the evolution of a circulatory O2-transport function from an ancestral O2-storage function would involve several key steps: 1) Switching the site of expression from tissue to blood cells, 2) a reduction in O2-binding affinity, and 3) the evolution of cooperative O2-binding by means of oxygenation-linked changes in the quaternary structure of a multimeric subunit assembly (as in the tetrameric Hb of gnathostomes) or oxygenation-linked changes in polymerization state (as in the Hbs of agnathans), which are typically monomeric in oxy-state protein and self-associate into dimers or higher-level polymers upon deoxygenation (Wald and Riggs 1951, 1998; Fago et al. 2001).
Evidence for the convergent evolution of O2-transport Hbs in agnathans and gnathostomes has been documented previously (Hoffmann, Opazo, et al. 2010). The original phylogenetic analyses indicated that agnathan Hbs are more closely related to Cygb than to the progenitors of the α- and β-chain Hbs of gnathostomes. In this study, phylogenetic analysis of a far more extensive set of globin sequences (including an agnathan ortholog of Cygb) confirmed the independent origins of O2-transport Hbs in agnathans and gnathostomes and also documented that agnathan aHbs are not many-to-one orthologs of gnathostome Cygb (figs. 2 and 6). We also document evidence that suggests the possibility of convergence between muscle-specific Mbs in the two vertebrate lineages, although an O2-storage function for the last common ancestor cannot be excluded (fig. 6). This dual convergence of O2-transport Hbs and O2-storage Mbs involved the convergent co-option of different precursor proteins in the ancestral globin repertoire of vertebrates.
Notably, functional Hbs and Mbs have also been observed in a variety of invertebrates (Weber and Vinogradov 2001). From the phylogenetic trees, it appears likely that these proteins emerged several times convergently from a globin ancestor as well (see, e.g., Roesner et al. 2005; Blank and Burmester 2012; Hoffmann, Opazo, Hoogewijs, et al. 2012). For example, the emergence of a functional Hb from a muscle-based Mb analog has been demonstrated in snails (Lieb et al. 2006). During the evolution of eukaryotes, the functional versatility of the globin-based heme structure and its potential for reversible O2-binding appears to have been repeatedly recruited for respiratory functions involving O2-storage and O2-transport.
Conclusion
The Emergence of Vertebrate Globin Diversity
Once the ancestors of contemporary vertebrates reached a certain threshold of body size and internal complexity, the passive diffusion of O2 became insufficient to meet metabolic demands, and this presumably favored the evolution of specific respiratory specializations to sustain sufficient O2 supply to internal tissues. These include respiratory surfaces such as gills, a circulatory system, and proteins that reversibly bind O2 for transport and storage. Vertebrates as well as many invertebrates have recruited globin proteins to serve respiratory functions. It is uncertain whether the last common ancestor of all current metazoan globins already had a function in O2 supply. In fact, globin proteins could have evolved reversible O2-binding from an acylated, membrane-bound, hexacoordinate GbX-like ancestor with a distinct, membrane-related function in lipid protection or signaling (Blank and Burmester 2012).
Gnathostome Cygbs do not exhibit membrane binding, but are able to reversibly bind both lipids and O2 (Reeder et al. 2011). The actual position of Cygb in the vertebrate globin tree is not well resolved and three possible scenarios are conceivable (fig. 6). Because respiratory functions have been conclusively demonstrated for aHbs, Hbs, Mbs, and GbE, it is tempting to assume a similar function in the ancestral proto-globin that gave rise to these vertebrate globin types (fig. 6C). It remains uncertain whether this also applies to Cygb. Regardless, we have demonstrated that the physiological division of labor between Mb-like and Hb-like respiratory proteins evolved convergently in Agnatha and Gnathostomata, most likely with the advent of larger body size, along with the evolution of active muscles and a closed circulatory system. Finally, given the ancient origin of Ngb in Metazoa and its high sequence conservation among vertebrates—which suggests a functionally important role—the apparent loss of this gene in lampreys is surprising. It will be interesting to see whether this nerve-specific globin is similarly lost in hagfish and/or other vertebrate lineages, such as sharks (Venkatesh et al. 2007, 2014).
Materials and Methods
Data Collection and Sequence Analyses
Using the BLAST algorithm, putative globin genes were identified in the genomic sequences and ESTs of the sea lamprey that are archived in ENSEMBL (http://www.ensembl.org, last accessed July 22, 2014) and GenBank (http://www.ncbi.nlm.nih.gov, last accessed July 22, 2014). The genomic sequences of the Arctic lamprey were accessed at http://jlampreygenome.imcb.a-star.edu.sg/ (last accessed July 22, 2014) (Mehta et al. 2013). Gene models were built by hand and with the help of GenScan (http://genes.mit.edu/GENSCAN.html, last accessed July 22, 2014). These tools were also used to annotate flanking genes. Intron–exon boundaries were identified with Spidey (http://www.ncbi.nlm.nih.gov/spidey, last accessed July 22, 2014). Preliminary analyses and translation into amino acids were performed with GeneDoc 2.7 (Nicholas et al. 1997). Myristoylation and palmitoylation sites were predicted by Myristoylator (http://web.expasy.org/myristoylator/, last accessed July 22, 2014) (Bologna et al. 2004) and CSS-Palm 2.0 (http://csspalm.biocuckoo.org/, last accessed July 22, 2014) (Ren et al. 2008), respectively.
Multiple Sequence Alignment and Phylogenetic Reconstruction
Sequences of 136 vertebrate globins were collected from the lamprey genomes and from EMBL/GenBank (supplementary table S3, Supplementary Material online). The data set covered 20 globins from P. marinus and 14 globins from L. camtschaticum. Incomplete globin sequences were excluded. Moreover, some closely related globin genes translate into identical proteins (supplementary tables S1 and Supplementary Data, Supplementary Material online) and are represented only by a single sequence in phylogenetic analyses. We further collected all available globin sequences of other agnathans from the databases; the other vertebrate globins were selected to represent each of the distinct globin types and to cover a broad range of taxa. Alternative multiple alignments of the amino acid sequences were generated by MAFFT with the FFT-NS-i, L-INS-i and G-INS-i strategies (Katoh and Toh 2008; Katoh et al. 2009), MUSCLE (Edgar 2004), PROMALS3D (Pei et al. 2008), and T-coffee (Notredame et al. 2000). The quality of each alignment was evaluated with MUMSA (http://msa.sbc.su.se/, last accessed July 22, 2014) (Lassmann and Sonnhammer 2005). The alignment generated by MAFFT L-INS-i received the highest MUMSA score and was used for phylogenetic analyses. Tree reconstructions were carried out with MrBayes 3.2.1 (Huelsenbeck and Ronquist 2001; Ayres et al. 2012). ProtTest (Abascal et al. 2005) was used to select the most appropriate model of amino acid evolution (LG; Le and Gascuel 2008) applying the Akaike Information Criterion. The LG model was coded with general time reversible as fixed prior with the prset command by specifying the aarevmatpr and statefreqpr options. A gamma distribution of substitution rates was assumed, and Bayesian trees were constructed. Two independent runs with one cold and three heated chains were performed for 5,000,000 generations. Starting trees were random and the trees were sampled every 1,000th generation. Posterior probabilities were estimated on the final 3,000 trees. The Ngb and GbX proteins were defined as outgroups because they diverged from the other globins prior to the separation of Protostomia and Deuterostomia (Roesner et al. 2005; Blank and Burmester 2012).
Gene Synteny Analyses
Gene orders and sequences were obtained from the genome assemblies of Homo sapiens (Annotation Release 104), Gallus gallus (build 3.1), and X. tropicalis (build 1.1), which are available at NCBI (http://www.ncbi.nlm.nih.gov/projects/mapview/, last accessed July 22, 2014). Syntenic regions were identified by comparison with the gene orders in the globin-containing contigs from the P. marinus and L. camtschaticum genomes.
In Silico Analysis of Globin Expression Pattern
The ESTs of P. marinus, as available at GenBank, were searched with the identified globin sequences employing tBLASTn and BLASTn searches. Information regarding the stage-specific expression pattern of each hit was obtained from Biosample (http://www.ncbi.nlm.nih.gov/biosample/).
RNA Extraction and cDNA Cloning
Two adult sea lampreys (63 cm, 731.7 g and 58 cm, 535.3 g) were collected from the Elbe estuary in June 2013. Tissues samples were harvested, immediately placed on dry ice, and stored at −80 °C. Subsamples of skeletal muscle, brain, eye, liver, heart, and blood for subsequent RNA extraction were placed in RNAlater (Qiagen, Hilden, Germany). Total RNA was extracted separately from each of these tissues using the Crystal RNA Mini Kit (Biolab Products, Gödenstorf, Germany). Briefly, about 1 cm3 of tissue was placed in liquid nitrogen and ground to a fine powder with a mortar and pestle, homogenized in 1 ml peqGOLD Trifast (PEQLAB, Erlangen, Germany) and 200 µl of chloroform added. The aqueous phase was then purified using the filter and silica column method following the manufacturer’s instructions. Samples were treated with DNase (RNase-free DNase, Qiagen) and the quality of the RNA was assessed by gel electrophoresis. The RevertAid H Minus First Strand cDNA Synthesis Kit (Thermo Scientific, Bonn, Germany) was used for reverse transcription of 1 µg total RNA with oligo-(dT)18 primer in a final volume of 20 µl. For amplification of the selected sea lamprey globin cDNAs, specific oligonucleotides spanning the full-length coding sequences were designed (supplementary table S5, Supplementary Material online). The PCR products were cloned into standard cloning vectors (pGEM-T, Promega, or pJET 1.2, Thermo Scientific) and sequenced by a commercial service (GATC, Konstanz, Germany).
Quantitative Real-Time Reverse Transcription PCR
Globin mRNA expression levels were estimated by qRT-PCR on an ABI 7500 real-time PCR system using the ABI Power SYBR Green master mix (Applied Biosystems, Darmstadt, Germany). RNA samples from muscle, brain, eye, liver, heart, and blood tissue were examined. qRT-PCR amplification was performed (40 amplification cycles; 95 °C for 15 s, 60 °C for 15 s, 72 °C for 30 s) with a final cDNA amount equivalent to 50 ng total RNA, 200 nM of each oligonucleotide, and water to a final volume of 20 µl. Fluorescence was measured at the end of each amplification cycle. To avoid amplification of genomic DNA, oligonucleotide primers that included intron-spanning positions were employed (supplementary table S5, Supplementary Material online). Each experiment was performed in triplicate. Negative controls (without cDNA) were run as a single experiment. The specificity of the amplification reaction was analyzed by dissociation curve analyses. Analysis of qRT-PCR results was performed with the ABI 7500 Sequence Detection software 2.0.6 (Applied Biosystems). Absolute mRNA copy numbers were calculated by means of the standard curve method with dilutions 107–102 of the recombinant plasmid. The samples were normalized according to 1 µg total RNA.
In Situ Hybridization
Digoxigenin-labeled antisense and sense riboprobes from the annotated lamprey aMb1 and aHb5a genes were constructed using the DIG RNA Labeling Kit (Roche Diagnostics, Mannheim, Germany). The plasmids containing the globin cDNAs were linearized with NcoI (antisense probe) and NotI (sense probe), and used as templates. The labeled probes were purified by lithium chloride precipitation and their integrity was checked by gel electrophoresis. The efficiency of digoxigenin labeling was determined by dot blots.
Frozen heart and muscle samples were equilibrated for 20 min at −20 °C, and cryosectioned at 16 μm thickness. The sections were mounted on poly-l-lysine cover slides (Fisher Scientific, Schwerte, Germany), fixed for 20 min on ice in 4% paraformaldehyde in phosphate-buffered saline (PBS) (140 mM NaCl, 2.7 mM KCl, 8.1 mM Na2HPO4, 1.5 mM KH2PO4, pH 6.9), and rinsed twice in PBS at room temperature (RT). The sections were acetylated in 0.5% acetic anhydride in 0.1 M triethanolamine (pH 8.0) for 10 min, washed with PBS, dehydrated in a graded ethanol series (70%, 90%, 95%, 100%), and dried. For hybridization, the probe mix (1,000 ng/µl probe, 2.5 mg/ml tRNA, 50 mM DTT) was denatured for 10 min at 65 °C and mixed at a ratio 1:5 with hybridization buffer (50% deionized formamide, 10% dextran sulfate, 1× Denhardt’s solution, 300 mM NaCl, 10 mM Tris–HCl pH 8.0, 1 mM ethylenediaminetetraacetic acid [EDTA] pH 8.0). Hybridization was carried out at 58 °C for 16 h. The slides were rinsed twice in 4× SSC (20× SSC: 3 M NaCl, 0.3 M sodium citrate, pH 7.0) for 10 min at RT, treated for 30 min at 37 °C with RNase A (0.18 Kunitz unit/ml; Roth, Karlsruhe, Germany) in 10 mM Tris pH 8.0, 0.5 M NaCl, 0.5 mM EDTA, followed by additional washing steps (2 × 5 min at RT in 2× SSC, 1 mM DTT for, 10 min in 1× SSC, 1 mM DTT at RT, 10 min in 0.5× SSC, 1 mM DTT at RT and 30 min in 0.1× SSC, 1 mM DTT at 60 °C).
After equilibration for 5 min in PBS/0.1% Tween-20 and 5 min in Buffer B (100 mM Tris–HCl, 150 mM NaCl, pH 7.5, 0.5% blocking reagent; Roche Diagnostics, Mannheim, Germany), the slides were incubated for 2 h at 37 °C with alkaline-phosphatase-coupled antidigoxigenin antibody (Roche Diagnostics) diluted 1:5,000 in Buffer B. Unbound antibodies were removed by two 15-min washes in 100 mM Tris–HCl, 150 mM NaCl, pH 7.5, followed by an 15-min incubation in 100 mM Tris–HCl, 100 mM NaCl, 50 mM MgCl2, pH 9.5. The visualization of the probes was carried out with the nitro-blue tetrazolium/5-bromo-4-chloro-3'-indolyphosphate substrate system. After 16 h the color reaction was stopped by washing in 100 mM Tris–HCl, 1 mM EDTA, pH 7.4, for 15 min. Slides were rinsed for 30 s in 95% ethanol, air dried, embedded in 1× PBS/glycerin (1:9), covered by a coverslip, fixed by nail polish, and analyzed with an Olympus BX51 research microscope.
Acknowledgments
The authors thank Miriam Götting, Walter Zeeck and Claus Zeeck for their help with the collection of lampreys, and Katharina Kruszewski and Anthony Signore for their help with sequence data. This work is supported by a grant of the Deutsche Forschungsgemeinschaft to T.B. (BU 956/18). K.S. was supported by a PhD fellowship from the University of Hamburg. J.F.S. acknowledges support from NIH grant HL087216. F.G.H. acknowledges support from NSF grant EPS. T.H. acknowledges funding by the Johannes Gutenberg University Centre for Computational Sciences Mainz (SRFN).
References
Author notes
Associate editor: John True