-
PDF
- Split View
-
Views
-
Cite
Cite
Aylin Colpan, Brian Johnston, Stephen Porter, Connie Clabots, Ruth Anway, Lao Thao, Michael A. Kuskowski, Veronika Tchesnokova, Evgeni V. Sokurenko, James R. Johnson, on behalf of the VICTORY (Veterans Influence of Clonal Types on Resistance: Year 2011) Investigators, Bradley L. Allen, Gio J. Baracco, Roger Bedimo, Mary Bessesen, Robert A. Bonomo, Stephen M. Brecher, Sheldon T. Brown, Laila Castellino, Arundhati S. Desai, Fletcher Fernau, Mark A. Fisher, James Fleckenstein, Carol S. Fleming, Narla J. Fries, Virginia L. Kan, Carol A. Kauffman, Stacey Klutts, Michael Ohl, Thomas Russo, Andrea Swiatlo, Edwin Swiatlo, Escherichia coli Sequence Type 131 (ST131) Subclone H30 as an Emergent Multidrug-Resistant Pathogen Among US Veterans, Clinical Infectious Diseases, Volume 57, Issue 9, 1 November 2013, Pages 1256–1265, https://doi.org/10.1093/cid/cit503
Close - Share Icon Share
Abstract
Background. Escherichia coli sequence type 131 (ST131), typically fluoroquinolone-resistant (FQ-R) and/or extended-spectrum β-lactamase (ESBL)–producing, has emerged globally. We assessed its prevalence and characteristics among US veterans.
Methods. In 2011, 595 de-identified E. coli clinical isolates were collected systematically within 3 resistance groups (FQ-susceptible [FQ-S], FQ-R, and ESBL-producing) from 24 nationally distributed Veterans Affairs Medical Centers (VAMCs). ST131 and its H30 subclone were detected by polymerase chain reaction and compared with other E. coli for molecular traits, source, and resistance profiles.
Results. ST131 accounted for 78% (184/236) of FQ-R and 64.2% (79/123) of ESBL-producing isolates, but only 7.2% (17/236) of FQ-S isolates (P < .001). The H30 subclone accounted for ≥95% of FQ-R and ESBL-producing, but only 12.5% of FQ-S, ST131 isolates (P < .001). By back-calculation, 28% of VAMC E. coli isolates nationally represented ST131. Overall, ST131 varied minimally in prevalence by specimen type, inpatient/outpatient source, or locale; was the most prevalent ST, followed distantly by ST95 and ST12 (13% each); and accounted for ≥40% (β-lactams), >50% (trimethoprim-sulfamethoxazole , multidrug), or >70% (ciprofloxacin, gentamicin) of total antimicrobial resistance. FQ-R and ESBL-producing ST131 isolates had higher virulence scores than corresponding non-ST131 isolates. ST131 pulsotypes overlapped extensively among VAMCs.
Conclusions. Among US veterans, ST131, primarily its H30 subclone, accounts for most antimicrobial-resistant E. coli and is the dominant E. coli strain overall. Possible contributors include multidrug resistance, extensive virulence gene content, and ongoing transmission. Focused attention to ST131, especially its H30 subclone, could reduce infection-related morbidity, mortality, and costs among veterans.
(See the Editorial Commentary by Lautenbach on pages 1266–9.)
Escherichia coli causes diverse extraintestinal infections, including urinary tract infection, bacteremia, and meningitis, resulting in considerable morbidity, mortality, and increased costs [1]. Management of such infections is complicated by the rising prevalence of resistance to preferred antimicrobial agents such as trimethoprim-sulfamethoxazole (TMP-SMZ), fluoroquinolones (FQs), and extended-spectrum cephalosporins [1–4].
Contributing to this problem is E. coli sequence type 131 (ST131), a newly emerged, disseminated lineage of multidrug-resistant E. coli [2, 5–9]. In recent surveys ST131 has accounted for up to 10%–27% of the total clinical E. coli population in various locales, and for up to 52%–67% of all extended-spectrum β-lactamase (ESBL)–producing or FQ-resistant (FQ-R) E. coli [2, 10–12].
The Department of Veterans Affairs (VA) operates the largest integrated healthcare system in the United States. [13]. Escherichia coli infections, especially urinary tract infections, are quite common among veterans [14]. Based on associations of ST131 with older age and health care contact [15], and the high prevalence of these characteristics among veterans, we hypothesized that ST131 may contribute importantly to antimicrobial resistance in E. coli among veterans. Accordingly, we assessed the prevalence, geographic distribution, and contribution to antimicrobial resistance of E. coli ST131 among US veterans during 2010–2011 at multiple VA medical centers (VAMCs) across the United States, and explored associations of ST131 with source and fitness-promoting bacterial traits.
MATERIALS AND METHODS
Patients and Isolates
During 2011 the clinical microbiology laboratories of 24 widely distributed VAMCs submitted 10 each of de-identified FQ-R and FQ-susceptible (FQ-S) extraintestinal clinical E. coli isolates from 2011, plus (because of their comparative rarity) up to 10 archived ESBL-producing E coli isolates from 2010–2011. For the FQ-S and FQ-R isolates, laboratories prospectively saved 10 consecutive FQ-R isolates and, in parallel, 10 arbitrarily selected FQ-S isolates. Isolates were submitted to the research laboratory accompanied by approximate collection date, specimen type, origin (inpatient vs outpatient), and susceptibility data, plus the source laboratory's current cumulative E. coli susceptibility data.
The 24 VAMCs were in the District of Columbia and 18 US states (California, Colorado, Florida, Idaho, Indiana, Iowa, Massachusetts, Michigan, Minnesota, Mississippi, Missouri, New York, Ohio, Tennessee, Texas, Utah, Washington, and Wisconsin). They were assigned to 1 of 4 main US census regions (ie, West, Midwest, South, and Northeast) based on location [16]. Local institutional review boards and research oversight committees approved the study protocol.
Molecular Methods
Isolates were assessed for ST131 genotype by polymerase chain reaction (PCR)–based detection of ST131-specific single-nucleotide polymorphisms (SNPs) in gyrB and mdh [17], with selective confirmation by multilocus sequence typing (MLST) (http://mlst.ucc.ie/mlst/dbs/Ecoli). ST131 isolates were tested by allele-specific primers for allele 30 of fimH (encoding a variant of the type 1 fimbrial adhesin) corresponding with the main FQ resistance–associated subset within ST131, the H30 subclone [11, 17]. Primers fimH30F-21 (CCGCCAATGGTACCGCTATT) and fimH30R-20 (CAGCTTTAATCGCCACCCCA) (354 bp product) underwent PCR as follows: 8′ at 95°; 30 cycles of (20 seconds at 94° and 45 seconds at 68°); 5′ at 72°; hold at 4°. Additionally, 20 each of randomly selected FQ-S and FQ-R non-ST131 isolates underwent MLST, followed by sub–sequence type (ST) stratification using fumC-fimH (CH) typing, which utilizes a 489-nucleotide (nt) internal fragment of fimH to resolve within-ST subclones [18].
Major E. coli phylogenetic groups (A, B1, B2, and D) was determined by triplex PCR [19]. Presence of 54 extraintestinal virulence genes was assessed by multiplex PCR [17, 20, 21]. The virulence factor (VF) score was the total number of virulence genes detected, adjusted for multiple detection of the pap (P fimbriae), sfa/foc (S and F1C fimbriae), and kps (group 2 capsule) operons. Isolates were classified as extraintestinal pathogenic E. coli (ExPEC) if positive for ≥2 of the following: papAH and/or papC (P fimbriae), sfa/focDE, afa/draBC (Dr-family adhesins), iutA (aerobactin receptor), and kpsM II (group 2 capsule synthesis) [22].
XbaI pulsed-field gel electrophoresis (PFGE) analysis was used to assign isolates to pulsotypes based on 94% profile similarity to reference strains [23]. A PFGE dendrogram was inferred within BioNumerics, version 6.6 (Applied Maths, Austin, Texas) according to the unweighted pair group method based on Dice coefficients [16]. Profiles also were compared with a large private PFGE profile reference library [16].
Susceptibility Testing
Susceptibility results for 9 antimicrobial agents (ampicillin, ampicillin/sulbactam, cefazolin, ceftriaxone, ciprofloxacin, imipenem, gentamicin, nitrofurantoin, and TMP-SMZ) were as provided by participating VAMCs based on local broth microdilution or disk diffusion testing. Reported susceptibility results, if conflicting with the assigned resistance category, were reassessed by disk diffusion, using Clinical and Laboratory Standards Institute–specified methods, ATCC reference strains, and interpretive criteria [24], and isolates were reclassified accordingly. Intermediate interpretations were analyzed as resistant. The resistance score was the number of agents to which an isolate exhibited resistance. Multidrug resistance was defined using 2 thresholds, that is, resistance to ≥3 or ≥5 drug classes (counting penicillins and cephalosporins separately) [25].
Population Estimates
The overall population prevalence of ST131, other clonal groups, and resistance to individual or combined antimicrobial agents were estimated by back-calculations based on the observed prevalence of each clonal group or resistance phenotype among the FQ-R and FQ-S study isolates, respectively, and the relative sizes of the FQ-R and FQ-S populations, according to the reported prevalence of ciprofloxacin resistance in E. coli at the participating laboratories (median, 29%; range, 21%–53%; Table 1). ST131-specific resistance contributions were calculated similarly for each resistance phenotype as the product of (1) the observed prevalence of the particular phenotype among FQ-S and FQ-R ST131 isolates, respectively; (2) the proportion of FQ-S and FQ-R isolates that were ST131 (Figure 1); and (3) the relative sizes of the FQ-R and FQ-S populations (determined as above) (Table 1). The overall prevalence of each resistance phenotype among ST131, H30 ST131, and other E. coli was estimated similarly (Table 2).
Estimated Overall Contribution of ST131 to Antimicrobial Resistance in Escherichia coli Among US Veterans
| Resistance Phenotype . | Overall Prevalence in Population, %a . | Estimated Fraction due to ST131b . |
|---|---|---|
| Ampicillin | 46 | 0.46 |
| Ampicillin/sulbactam | 34 | 0.48 |
| Cefazolin | 17 | 0.47 |
| Ceftriaxone | 2 | 0.43 |
| Ciprofloxacin | 29 | 0.78 |
| Gentamicin | 11 | 0.76 |
| TMP-SMZ | 22 | 0.56 |
| Imipenem | 0.25 | 0.50 |
| Nitrofurantoin | 6 | 0.36 |
| Multidrug resistant to ≥3 classes | 24 | 0.70 |
| Multidrug resistant to ≥5 classes | 2 | 0.55 |
| Ciprofloxacin + TMP-SMZ | 15 | 0.76 |
| Ciprofloxacin + TMP-SMZ + ampicillin | 14 | 0.76 |
| Resistance Phenotype . | Overall Prevalence in Population, %a . | Estimated Fraction due to ST131b . |
|---|---|---|
| Ampicillin | 46 | 0.46 |
| Ampicillin/sulbactam | 34 | 0.48 |
| Cefazolin | 17 | 0.47 |
| Ceftriaxone | 2 | 0.43 |
| Ciprofloxacin | 29 | 0.78 |
| Gentamicin | 11 | 0.76 |
| TMP-SMZ | 22 | 0.56 |
| Imipenem | 0.25 | 0.50 |
| Nitrofurantoin | 6 | 0.36 |
| Multidrug resistant to ≥3 classes | 24 | 0.70 |
| Multidrug resistant to ≥5 classes | 2 | 0.55 |
| Ciprofloxacin + TMP-SMZ | 15 | 0.76 |
| Ciprofloxacin + TMP-SMZ + ampicillin | 14 | 0.76 |
Abbreviation: TMP-SMZ,trimethoprim-sulfamethoxazole.
a Based on the median value for cumulative prevalence of fluoroquinolone (FQ) resistance in E. coli as reported by the participating Veterans Affairs Medical Center laboratories (29%) and the observed prevalence of the listed resistance phenotypes among fluoroquinolone-resistant (FQ-R) and fluoroquinolone-susceptible (FQ-S) study isolates, respectively. The calculation was as follows: overall prevalence of phenotype in source population = .29 (prevalence of phenotype among FQ-R study isolates) + .71 (prevalence of phenotype among FQ-S study isolates).
b Based on the assumed 29% overall prevalence of FQ resistance (see above), the observed prevalence of ST131 among FQ-R and FQ-S study isolates (Figure 1), and the observed prevalence of the listed resistance phenotypes among the FQ-R and FQ-S ST131 study isolates, respectively.
Estimated Overall Contribution of ST131 to Antimicrobial Resistance in Escherichia coli Among US Veterans
| Resistance Phenotype . | Overall Prevalence in Population, %a . | Estimated Fraction due to ST131b . |
|---|---|---|
| Ampicillin | 46 | 0.46 |
| Ampicillin/sulbactam | 34 | 0.48 |
| Cefazolin | 17 | 0.47 |
| Ceftriaxone | 2 | 0.43 |
| Ciprofloxacin | 29 | 0.78 |
| Gentamicin | 11 | 0.76 |
| TMP-SMZ | 22 | 0.56 |
| Imipenem | 0.25 | 0.50 |
| Nitrofurantoin | 6 | 0.36 |
| Multidrug resistant to ≥3 classes | 24 | 0.70 |
| Multidrug resistant to ≥5 classes | 2 | 0.55 |
| Ciprofloxacin + TMP-SMZ | 15 | 0.76 |
| Ciprofloxacin + TMP-SMZ + ampicillin | 14 | 0.76 |
| Resistance Phenotype . | Overall Prevalence in Population, %a . | Estimated Fraction due to ST131b . |
|---|---|---|
| Ampicillin | 46 | 0.46 |
| Ampicillin/sulbactam | 34 | 0.48 |
| Cefazolin | 17 | 0.47 |
| Ceftriaxone | 2 | 0.43 |
| Ciprofloxacin | 29 | 0.78 |
| Gentamicin | 11 | 0.76 |
| TMP-SMZ | 22 | 0.56 |
| Imipenem | 0.25 | 0.50 |
| Nitrofurantoin | 6 | 0.36 |
| Multidrug resistant to ≥3 classes | 24 | 0.70 |
| Multidrug resistant to ≥5 classes | 2 | 0.55 |
| Ciprofloxacin + TMP-SMZ | 15 | 0.76 |
| Ciprofloxacin + TMP-SMZ + ampicillin | 14 | 0.76 |
Abbreviation: TMP-SMZ,trimethoprim-sulfamethoxazole.
a Based on the median value for cumulative prevalence of fluoroquinolone (FQ) resistance in E. coli as reported by the participating Veterans Affairs Medical Center laboratories (29%) and the observed prevalence of the listed resistance phenotypes among fluoroquinolone-resistant (FQ-R) and fluoroquinolone-susceptible (FQ-S) study isolates, respectively. The calculation was as follows: overall prevalence of phenotype in source population = .29 (prevalence of phenotype among FQ-R study isolates) + .71 (prevalence of phenotype among FQ-S study isolates).
b Based on the assumed 29% overall prevalence of FQ resistance (see above), the observed prevalence of ST131 among FQ-R and FQ-S study isolates (Figure 1), and the observed prevalence of the listed resistance phenotypes among the FQ-R and FQ-S ST131 study isolates, respectively.
Overall Prevalence of Antimicrobial Resistance Among ST131 and H30 Subclone Isolates, Compared With Other Isolates, Among Escherichia coli Clinical Isolates from US Veterans
| . | Prevalence of Resistancea and Hazard Ratiob . | ||||||
|---|---|---|---|---|---|---|---|
| . | ST131 vs Others . | H30 Subclone vs Others . | |||||
| Resistance Phenotype . | Totala (%) . | ST131a (%) . | Othersa (%) . | Hazard Ratiob . | H30 ST131a (%) . | Othersa (%) . | Hazard Ratiob . |
| Ampicillin | 46 | 77 | 34 | 2.3 | 76 | 36 | 2.1 |
| Ampicillin/sulbactam | 34 | 58 | 24 | 2.4 | 57 | 27 | 2.1 |
| Cefazolin | 17 | 29 | 12 | 2.3 | 27 | 14 | 2.0 |
| Ceftriaxone | 2 | 3 | 2 | 1.9 | 4 | 2 | 2.0 |
| Ciprofloxacin | 29 | 81 | 9 | 9.2 | 90 | 10 | 9.3 |
| Gentamicin | 11 | 29 | 3 | 8.2 | 28 | 5 | 5.4 |
| TMP-SMZ | 22 | 42 | 13 | 2.4 | 47 | 14 | 3.3 |
| Imipenem | 0.25 | 0.4 | 0.2 | 1.4 | 0.5 | 0.2 | 1.4 |
| Nitrofurantoin | 6 | 8 | 5 | 3.2 | 8 | 5 | 3.4 |
| Multidrug resistant to ≥3 classes | 24 | 59 | 10 | 6.2 | 63 | 11 | 5.8 |
| Multidrug resistant to ≥5 classes | 2 | 5 | 1 | 3.2 | 6 | 1 | 3.9 |
| Ciprofloxacin + TMP-SMZ | 15 | 41 | 5 | 8.3 | 46 | 5 | 9.2 |
| Ciprofloxacin + TMP-SMZ + ampicillin | 14 | 37 | 5 | 8.1 | 42 | 5 | 9.0 |
| . | Prevalence of Resistancea and Hazard Ratiob . | ||||||
|---|---|---|---|---|---|---|---|
| . | ST131 vs Others . | H30 Subclone vs Others . | |||||
| Resistance Phenotype . | Totala (%) . | ST131a (%) . | Othersa (%) . | Hazard Ratiob . | H30 ST131a (%) . | Othersa (%) . | Hazard Ratiob . |
| Ampicillin | 46 | 77 | 34 | 2.3 | 76 | 36 | 2.1 |
| Ampicillin/sulbactam | 34 | 58 | 24 | 2.4 | 57 | 27 | 2.1 |
| Cefazolin | 17 | 29 | 12 | 2.3 | 27 | 14 | 2.0 |
| Ceftriaxone | 2 | 3 | 2 | 1.9 | 4 | 2 | 2.0 |
| Ciprofloxacin | 29 | 81 | 9 | 9.2 | 90 | 10 | 9.3 |
| Gentamicin | 11 | 29 | 3 | 8.2 | 28 | 5 | 5.4 |
| TMP-SMZ | 22 | 42 | 13 | 2.4 | 47 | 14 | 3.3 |
| Imipenem | 0.25 | 0.4 | 0.2 | 1.4 | 0.5 | 0.2 | 1.4 |
| Nitrofurantoin | 6 | 8 | 5 | 3.2 | 8 | 5 | 3.4 |
| Multidrug resistant to ≥3 classes | 24 | 59 | 10 | 6.2 | 63 | 11 | 5.8 |
| Multidrug resistant to ≥5 classes | 2 | 5 | 1 | 3.2 | 6 | 1 | 3.9 |
| Ciprofloxacin + TMP-SMZ | 15 | 41 | 5 | 8.3 | 46 | 5 | 9.2 |
| Ciprofloxacin + TMP-SMZ + ampicillin | 14 | 37 | 5 | 8.1 | 42 | 5 | 9.0 |
Abbreviation: TMP-SMZ,trimethoprim-sulfamethoxazole.
a Prevalence of resistance (overall and within each listed genotype) was calculated based on the median prevalence of fluoroquinolone (FQ) resistance at the participating Veterans Affairs Medical Centers (29%), the observed proportion of FQ-resistant and FQ-susceptible study isolates that represented ST131 or the H30 ST131 subclone (Figure 1), and the observed prevalence of each resistance phenotype within these subgroups.
b Resistance prevalence among ST131 isolates relative to all other isolates, or among H30 ST131 subclone isolates relative to all other isolates.
Overall Prevalence of Antimicrobial Resistance Among ST131 and H30 Subclone Isolates, Compared With Other Isolates, Among Escherichia coli Clinical Isolates from US Veterans
| . | Prevalence of Resistancea and Hazard Ratiob . | ||||||
|---|---|---|---|---|---|---|---|
| . | ST131 vs Others . | H30 Subclone vs Others . | |||||
| Resistance Phenotype . | Totala (%) . | ST131a (%) . | Othersa (%) . | Hazard Ratiob . | H30 ST131a (%) . | Othersa (%) . | Hazard Ratiob . |
| Ampicillin | 46 | 77 | 34 | 2.3 | 76 | 36 | 2.1 |
| Ampicillin/sulbactam | 34 | 58 | 24 | 2.4 | 57 | 27 | 2.1 |
| Cefazolin | 17 | 29 | 12 | 2.3 | 27 | 14 | 2.0 |
| Ceftriaxone | 2 | 3 | 2 | 1.9 | 4 | 2 | 2.0 |
| Ciprofloxacin | 29 | 81 | 9 | 9.2 | 90 | 10 | 9.3 |
| Gentamicin | 11 | 29 | 3 | 8.2 | 28 | 5 | 5.4 |
| TMP-SMZ | 22 | 42 | 13 | 2.4 | 47 | 14 | 3.3 |
| Imipenem | 0.25 | 0.4 | 0.2 | 1.4 | 0.5 | 0.2 | 1.4 |
| Nitrofurantoin | 6 | 8 | 5 | 3.2 | 8 | 5 | 3.4 |
| Multidrug resistant to ≥3 classes | 24 | 59 | 10 | 6.2 | 63 | 11 | 5.8 |
| Multidrug resistant to ≥5 classes | 2 | 5 | 1 | 3.2 | 6 | 1 | 3.9 |
| Ciprofloxacin + TMP-SMZ | 15 | 41 | 5 | 8.3 | 46 | 5 | 9.2 |
| Ciprofloxacin + TMP-SMZ + ampicillin | 14 | 37 | 5 | 8.1 | 42 | 5 | 9.0 |
| . | Prevalence of Resistancea and Hazard Ratiob . | ||||||
|---|---|---|---|---|---|---|---|
| . | ST131 vs Others . | H30 Subclone vs Others . | |||||
| Resistance Phenotype . | Totala (%) . | ST131a (%) . | Othersa (%) . | Hazard Ratiob . | H30 ST131a (%) . | Othersa (%) . | Hazard Ratiob . |
| Ampicillin | 46 | 77 | 34 | 2.3 | 76 | 36 | 2.1 |
| Ampicillin/sulbactam | 34 | 58 | 24 | 2.4 | 57 | 27 | 2.1 |
| Cefazolin | 17 | 29 | 12 | 2.3 | 27 | 14 | 2.0 |
| Ceftriaxone | 2 | 3 | 2 | 1.9 | 4 | 2 | 2.0 |
| Ciprofloxacin | 29 | 81 | 9 | 9.2 | 90 | 10 | 9.3 |
| Gentamicin | 11 | 29 | 3 | 8.2 | 28 | 5 | 5.4 |
| TMP-SMZ | 22 | 42 | 13 | 2.4 | 47 | 14 | 3.3 |
| Imipenem | 0.25 | 0.4 | 0.2 | 1.4 | 0.5 | 0.2 | 1.4 |
| Nitrofurantoin | 6 | 8 | 5 | 3.2 | 8 | 5 | 3.4 |
| Multidrug resistant to ≥3 classes | 24 | 59 | 10 | 6.2 | 63 | 11 | 5.8 |
| Multidrug resistant to ≥5 classes | 2 | 5 | 1 | 3.2 | 6 | 1 | 3.9 |
| Ciprofloxacin + TMP-SMZ | 15 | 41 | 5 | 8.3 | 46 | 5 | 9.2 |
| Ciprofloxacin + TMP-SMZ + ampicillin | 14 | 37 | 5 | 8.1 | 42 | 5 | 9.0 |
Abbreviation: TMP-SMZ,trimethoprim-sulfamethoxazole.
a Prevalence of resistance (overall and within each listed genotype) was calculated based on the median prevalence of fluoroquinolone (FQ) resistance at the participating Veterans Affairs Medical Centers (29%), the observed proportion of FQ-resistant and FQ-susceptible study isolates that represented ST131 or the H30 ST131 subclone (Figure 1), and the observed prevalence of each resistance phenotype within these subgroups.
b Resistance prevalence among ST131 isolates relative to all other isolates, or among H30 ST131 subclone isolates relative to all other isolates.
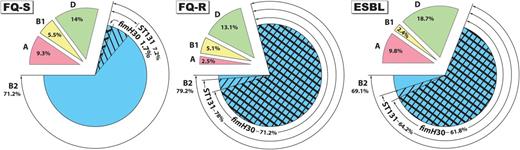
Distribution by resistance group of major Escherichia coli phylogenetic groups, ST131, and the fimH30 subclone among 595 E. coli isolates from veterans. Major phylogenetic groups: A (pink), B1 (yellow), B2 (blue), and D (green). ST131, fine cross-hatching; fimH30 subclone, bold cross-hatching. For prevalence of ST131 and the fimH30 ST131 subclone in the fluoroquinolone-susceptible group vs the fluoroquinolone-resistant or extended-spectrum β-lactamase group, P < .001. Abbreviations: ESBL, extended-spectrum β-lactamase; FQ-R, fluoroquinolone-resistant; FQ-S, fluoroquinolone-susceptible.
Statistical Analysis
Comparisons of proportions and continuous variables were tested by using Fisher exact test and the Mann-Whitney U test, respectively (both 2-tailed). The significance criterion was P < .05.
RESULTS
Prevalence of ST131 and the H30 ST131 Subclone
The 595 E. coli study isolates were from 24 widely distributed VAMCs and constituted 3 susceptibility groups: FQ-S (n = 236), FQ-R (n = 236), and ESBL (n = 123). Although the overall distribution of phylogenetic groups A, B1, B2, and D was fairly similar across the 3 susceptibility groups, with group B2 consistently predominating, ST131 accounted for 78% of the FQ-R isolates and 64.2% of the ESBL isolates, but only 7.2% of the FQ-S isolates (P < .001, vs FQ-R or ESBL; Figure 1). Moreover, the H30 ST131 subclone—a recently emerged, FQ resistance-associated lineage within ST131 [17, 21]—accounted for 95%–97.8% of ST131 isolates within the FQ-R and ESBL groups, but only 12.5% of those within the FQ-S group (P < .001, vs FQ-R or ESBL; Figure 1).
ST131 was broadly distributed geographically and exhibited consistent associations with FQ resistance and ESBL production (Supplementary Table 1). Among FQ-S isolates, ST131 was identified at only 13 VAMCs, and accounted for only 10%–20% of FQ-S isolates per VAMC. In contrast, among FQ-R isolates, ST131 was identified at all 24 VAMCs, and accounted for 50%–100% of FQ-R isolates per VAMC. Similarly, among ESBL isolates ST131 was encountered at each VAMC that provided ≥3 ESBL isolates, and accounted for 33%–100% of ESBL isolates per VAMC.
ST131 was similarly prevalent across the 4 major US census regions among the FQ-S and FQ-R isolates (Supplementary Table 2). In contrast, among ESBL isolates its prevalence was significantly lower in the Midwest region, at 37.2%, than in other census regions, which had ST131 prevalence values of 74%–84% (Supplementary Table 2).
Specimen type was documented for 545 (92%) isolates and included urine (85%), bloodstream (7%), and miscellaneous (8%: 1.8% respiratory, 1.7% wound, <0.8% each for 12 others). Inpatient vs outpatient source was documented for 414 (70%) isolates, with 304 (73.4%) being from outpatients. ST131 did not vary significantly in prevalence by either variable (not shown).
Prevalence of STs
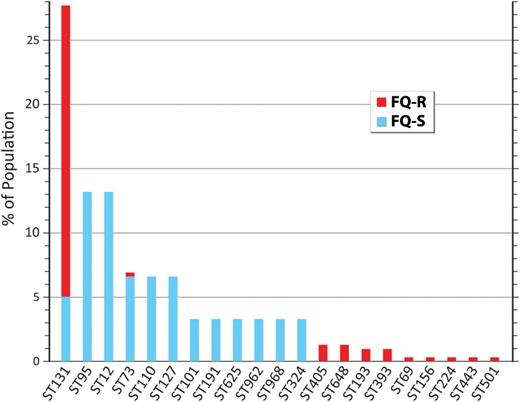
Based on ST131's proportional contribution to the FQ-R and FQ-S subgroups, plus the respective sizes of these subgroups, ST131 was estimated to account for 27.7% of all VAMC E. coli isolates nationwide. Seven-locus MLST of a randomly selected subset of 20 each FQ-S and FQ-R non-ST131 isolates identified a diversity of STs within each resistance group, with minimal overlap across groups. According to back-calculations for total population prevalence, in descending order the most prevalent non-ST131 STs contributing FQ-S isolates were ST95, ST12, ST73, ST10, and ST127 (6.6%–13.2% prevalence each), whereas the most prevalent contributing FQ-R isolates were ST405, ST1193, ST648, and ST393 (0.9%–1.3% prevalence each). Therefore, ST131 was by far the most prevalent ST overall (27.7% total prevalence), far outnumbering the next most prevalent STs, ST95, and ST12 (13.2% each; Figure 2). Based on similar calculations, the overall prevalence of the H30 ST131 subclone was estimated at 22.8%, >3-fold greater than the next most prevalent fimH-based CH subclones, 38–18 and 38–41 (from ST95: 6.6% each).
Overall population prevalence of ST131 and other sequence types (STs) among Escherichia coli clinical isolates from veterans. The 19 most prevalent STs are shown. Estimated overall prevalence was calculated based on subsamples. Nearly all fluoroquinolone-resistant ST131 isolates represented the fimH30 ST131 subclone. Abbreviations: FQ-R, fluoroquinolone-resistant; FQ-S, fluoroquinolone-susceptible.
PFGE Analysis
XbaI PFGE analysis of 85 randomly selected ST131 isolates (Figure 3) showed a predominance of pulsotypes 968 (26%), 800 (12%), and 812 (4%), as in a recent global survey of ST131 isolates [16]. Pulsotypes were distributed broadly across VAMCs and census regions. Of the 7 profile clusters with ≥98% similarity (2 isolates each), 4 comprised isolates from widely separated VAMCs.
Xbal pulsed-field gel electrophoresis–based dendrogram for 85 ST131 Escherichia coli isolates from veterans. The 85 isolates were selected randomly from the total ST131 population. Region denotes the 4 main US census regions. Horizontal lines bound the 5 most prevalent pulsotypes. Vertical line separates isolates with ≥98% overall profile similarity from less similar isolates. Abbreviations: ESBL, extended-spectrum β-lactamase; FQ, fluoroquinolone phenotype (R, resistant; S, susceptible); PFGE, pulsed-field gel electrophoresis.
Virulence Genes
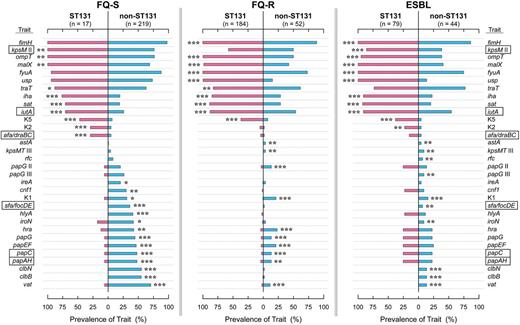
Virulence traits were assessed as a possible contributor to ST131's high prevalence. Of the studied virulence genes, 57% (31/54) varied significantly in prevalence with ST131 genotype in 1 or more resistance groups (Figure 4). ST131-associated virulence genes included certain adhesins (afa/dra, iha, fimH), a toxin (sat), siderophore receptors (iutA, fyuA), capsule variants (kpsMT II, K2, K5), and miscellaneous traits (usp, ompT, traT, and malX). Non-ST131–associated genes included other adhesins (papAHCEFG, papG alleles I and II, sfa/focDE), toxins (hlyA, cnf1, hlyF, pic, vat, astA), siderophore receptors (iroN, ireA), protectins (K1 capsule, O4 lipopolysaccharide [rfc]), and microcins/colibactins (clbB, clbN, cvaC).
Virulence genotypes of 595 Escherichia coli isolates in relation to ST131 genotype, by antimicrobial resistance group. Traits shown are those (among 54 total) that yielded P < .05 for comparisons of ST131 (pink bars) vs non-ST131 (blue bars) isolates in at least 1 resistance group. Traits are arranged, from top to bottom, in order of descending prevalence among the fluoroquinolone-susceptible (FQ-S) ST131 isolates (if positively associated with ST131), then ascending prevalence among the FQ-S non-ST131 isolates (if negatively associated with ST131). P value symbols are shown adjacent to the higher-prevalence group when P < .05, and are as follows: *P < .05, **P < .01, ***P < .001. Rectangles enclose traits contributing to molecular definition of extraintestinal pathogenic E. coli. Trait definitions: afa/draBC, Dr-family adhesins; clbB and clbN, colibactin synthesis; cnf1, cytotoxic necrotizing factor; fimH, type 1 fimbriae; fyuA, yersiniabactin receptor; hlyA, α hemolysin; hra, heat-resistant agglutinin; iha, adhesin-siderophore; ireA, siderophore receptor; iroN, salmochelin receptor; iutA, aerobactin receptor; kpsM II, group 2 capsule; K1, K2, and K5, group 2 capsule variants; malX, pathogenicity island marker; ompT, outer membrane protease T; papA, papC, papEF, and papG, P fimbrial structural subunit, assembly, tip pilins, and adhesin, respectively; papG allele II, P adhesin variant; sat, secreted autotransporter toxin; sfa/foc, S or F1C fimbriae; traT, serum resistance-associated; usp, uropathogenic-specific protein; vat, vacuolating toxin. Abbreviations: ESBL, extended-spectrum β-lactamase; FQ-R, fluoroquinolone-resistant; FQ-S, fluoroquinolone-susceptible.
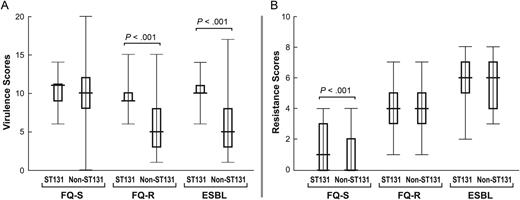
Virulence profiles among ST131 isolates were fairly consistent across resistance groups, but among non-ST131 isolates varied greatly by resistance group, being much sparser among FQ-R and ESBL isolates than FQ-S isolates (Figure 4). Within each resistance group a significantly greater proportion of ST131 than non-ST131 isolates qualified molecularly as ExPEC (FQ-S, 83% vs 57%: P = .04; FQ-R, 54% vs 35%: P = .012; ESBL, 85% vs 27%: P < .001). Among FQ-S isolates, VF scores were similarly high regardless of ST131 genotype (Figure 5). In contrast, among FQ-R and ESBL isolates, VF scores were much higher among ST131 isolates than non-ST131 isolates.
Virulence and resistance scores among ST131 and non-ST131 Escherichia coli isolates within 3 resistance groups. Box-and-whisker plots show group medians (heavy horizontal bar), 25th and 75th percentiles (bottom and top of boxes, respectively), and maximum and minimum values (light horizontal bars). P values, as determined by the Mann-Whitney U test (2-tailed), are shown for ST131 vs non-ST131 comparisons when P < .05. A, Virulence scores (number of distinct virulence genes) among ST131 vs non-ST131 isolates within each resistance group. B, Resistance scores (number of resistance markers detected) among ST131 isolates vs non-ST131 isolates within each resistance group. Abbreviations: ESBL, extended-spectrum β-lactamase; FQ-R, fluoroquinolone-resistant; FQ-S, fluoroquinolone-susceptible.
Antimicrobial Resistance
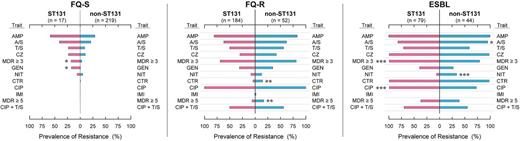
Resistance to the studied antimicrobial agents, both individually and combined, varied greatly in prevalence by agent and resistance group, but minimally by ST131 genotype (Figure 6). Paralleling these trends, aggregate resistance scores increased progressively by resistance group, from FQ-S, through FQ-R, to ESBL isolates (Figure 5). Within a given resistance group the ST131 isolates had similarly high (FQ-R and ESBL group) or slightly but significantly higher (FQ-S group) scores compared with non-ST131 isolates.
Antimicrobial resistance prevalence among 595 E. coli isolates according to ST131 status and resistance group. P value symbols (from the Mann-Whitney U test) for comparison of ST131 (pink bars) vs non-ST131 (blue bars) isolates within each resistance group, which are shown next to the higher prevalence group if P < .05, are as follows: *P < .05, **P ≤ .01, ***P ≤ .001. Abbreviations: AMP, ampicillin; A/S, ampicillin-sulbactam; CIP, ciprofloxacin; CTR, ceftriaxone; CZ, cefazolin; ESBL, extended-spectrum β-lactamase; FQ-R, fluoroquinolone-resistant; FQ-S, fluoroquinolone-susceptible; GEN, gentamicin; IMI, imipenem; MDR, multidrug resistance (to ≥3 or ≥5 drug classes); NIT, nitrofurantoin; T/S, trimethoprim-sulfamethoxazole.
Back-calculations suggested that ST131's overall contribution to antimicrobial resistance within the source E. coli population was ≥40% for each β-lactam agent, >50% for TMP-SMZ resistance and multidrug resistance, and >70% for ciprofloxacin, gentamicin, and combined ciprofloxacin plus TMP-SMZ (or combined ciprofloxacin, TMP-SMZ, and ampicillin) resistance (Table 1).
The estimated overall prevalence of each resistance phenotype (Table 2) was consistently greater among the ST131 and H30 subclone isolates than other isolates (median hazard ratios, 3.2–3.4; range, 1.3–9.3). The stratified resistance prevalence values for ST131 or H30 subclone isolates vs other isolates often straddled a prevalence threshold (eg, 10%, 15%, 20%) commonly used for selecting empirical antimicrobial therapy (Table 2).
DISCUSSION
We screened for the ST131 clonal group and its H30 subclone among 595 E. coli clinical isolates, collected systematically in 2011 from 24 VAMCs distributed widely across the United States. We found that ST131 was ubiquitous and highly prevalent, especially among antimicrobial-resistant isolates, and differed from other E. coli according to its phylogenetic group B2 background, high prevalence of recognized virulence trait genes, and extensive antimicrobial resistance capabilities. These findings newly identify ST131 and its H30 subclone as extremely important pathogens among veterans, which has significant implications for the prevention, diagnosis, and management of E. coli infections in the VA population.
ST131 accounted for only 7.2% of FQ-S isolates, but for a striking 78% of FQ-R isolates and 64.2% of ESBL isolates. Moreover, as the median prevalence of FQ resistance in E. coli at the participating VAMCs was 29%, ST131 presumably accounted for approximately 28% of all clinical E. coli isolates at these VAMCs. These high prevalence values for ST131 exceed those from the most recent general surveys, which have been as high as 17%, 22%, 23%, and 27% for overall prevalence, and 24.8%, 52%, and 69% for prevalence among FQ-R isolates [2, 8, 10, 11, 15, 26]. Possible explanations for this finding include further emergence of ST131 since the previous studies, or geographical or host population differences.
Evidence against further emergence and geographical differences is provided by analysis of national surveillance isolates from the SENTRY program, showing that ST131's prevalence in the general US population was similar in 2011 (unpublished data, J. R. J.) compared to 2007 [2]. Evidence favoring host population differences is that veterans receiving care at VAMCs tend to be elderly men, often with multiple comorbidities and extensive antimicrobial use [13, 27]. Older age, antimicrobial use, and healthcare contact have been identified as risk factors for ST131 infection [15]. Therefore, the VAMC patient population may be especially susceptible to ST131.
In contrast to the ST131 isolates, the non-ST131 isolates were divided among multiple STs, none of which contributed more than 13.2% to the total population. Therefore, ST131 was by far the most prevalent clonal group among veterans, with an estimated 28% overall prevalence, exceeding the next most prevalent STs by >2-fold. Although several recent studies identified ST131 as the first or second most prevalent clonal group within collections of all E. coli clinical isolates from specific regions [2, 8, 10, 11, 15, 26], none documented such a great gap between ST131 and traditional high-prevalence ExPEC clonal groups such as ST95, ST73, ST12, and ST127.
Notably, most of the present ST131 isolates, including nearly all within the FQ-R and ESBL groups, represented the H30 ST131 subclone, which was recently shown to have a single-strain origin and to account for most FQ-R ST131 isolates, regardless of source and locale [11]. Therefore, this single, remarkably successful subclone within ST131 has achieved dominance within the veteran-associated E. coli population, especially the antimicrobial-resistant subset.
ST131 was distributed fairly uniformly across the 24 VAMCs, geographical regions, specimen types, and inpatient vs outpatient settings. The ubiquity of ST131 among US veterans indicates that the study's findings likely are applicable throughout the VA healthcare system, and in similar nonveteran populations. In this regard, the occurrence across VAMCs of ST131-associated pulsotypes that are common also in the general population suggests ongoing transmission of ST131 among VAMCs and between veterans and nonveterans, and that similar risk factors and transmission pathways for ST131 may apply in veteran and nonveteran populations.
As possible explanations for ST131's emergence and predominance, compared with other E. coli, the ST131 isolates more frequently represented phylogenetic group B2, had more extensive virulence genotypes and/or antimicrobial resistance profiles, and more commonly qualified molecularly as ExPEC. This implies that, in ST131, antimicrobial resistance has combined with extraintestinal virulence to an extent not observed previously in E. coli, thereby creating a proverbial “superbug.” Although in vivo evidence for hypervirulence in ST131 from animal models is lacking [7, 28, 29], such models may not reflect the human situation. Indeed, recent epidemiological data document a prevalence gradient of ST131 in relation to clinical severity, from fecal isolates (low), through cystitis isolates (intermediate), to pyelonephritis isolates (high), implying enhanced clinical virulence for ST131 [2, 30].
Finally, according to back-calculations, ST131 accounted for a majority of antimicrobial resistance among clinical isolates, particularly for certain individual agents (FQs, 78%; TMP-SMZ, 56%; gentamicin, 76%), combined TMP-SMZ plus FQ resistance (52%), and multidrug resistance (≥3 classes, 70%; ≥5 classes, 55%). Therefore, problematic antimicrobial-resistant E. coli infections among veterans are caused predominantly by ST131 and, specifically, its H30 subclone, indicating that clonal spread dominates over both horizontal transfer of resistance elements and de novo mutation to resistance in driving the current E. coli resistance epidemic.
These findings have important practical implications. First, given ST131's high overall prevalence and major contribution to antimicrobial-resistant E. coli infections, focused attention to ST131 conceivably could yield substantial reductions in morbidity and costs within the VA healthcare system. Secondly, given the ubiquity of ST131, such measures should be applicable broadly across VAMCs. They could include preventive interventions (eg, vaccines or probiotics), infection control strategies (analogous to the current VA-wide screening for methicillin-resistant Staphylococcus aureus colonization [31]), and rapid detection, especially of the H30 subclone. Rapid detection could be particularly useful in selecting empirical therapy, as for many agents the ST131 and H30 subclone isolates exhibited resistance prevalence values exceeding typical empirical therapy thresholds of 10%, 15%, or 20%, with other isolates falling below these thresholds. Third, a fuller elucidation of why ST131 rose to such striking prominence could provide novel insights into the emergence of new resistant and virulent pathogens generally, thereby enabling more effective responses to future epidemic “superbugs.” Ongoing surveillance for such emergent pathogens is needed, to provide an early warning when a new successful lineage begins to expand.
Study limitations include the absence of clinical data, the reliance on locally determined susceptibility results, the less systematic sampling of ESBL isolates than of FQ-S and FQ-R isolates, and the large number of unadjusted comparisons. Study strengths include the large, recent, and broadly distributed study population, the systematic sampling of concurrent FQ-S and FQ-R isolates, and the attention to multiple ecological variables and bacterial characteristics.
In summary, we documented an impressively high prevalence of ST131 and its fimH30 subclone among clinical E. coli isolates from US veterans in 2011. ST131 accounted for more antimicrobial resistance (especially to FQs, TMP-SMZ, gentamicin, and multiple drug classes), and exhibited greater molecularly inferred virulence, than did other E. coli. Focused attention to ST131 and its H30 subclone could help reduce infection-related morbidity, mortality, and healthcare costs among veterans.
Notes
Disclaimer. The views expressed are solely those of the authors and do not reflect the views and policies of the Department of Veterans Affairs.
The VICTORY Investigators. Bradley L. Allen, MD, PhD (Roudebush VAMC and Indiana University School of Medicine, Indianapolis); Gio J. Baracco, MD (Miami VA Healthcare System [HCS] and University of Miami Miller School of Medicine, Miami, FL); Roger Bedimo, MD, MS, FACP (VA North Texas HCS and UT Southwestern Medical Center, Dallas); Mary Bessesen, MD (Department of Veterans Affairs Eastern Colorado HCS and University of Colorado–Denver School of Medicine, Denver); Robert A. Bonomo, MD (Cleveland VAMC, Cleveland, OH); Stephen M. Brecher, PhD (VA Boston HCS and BU School of Medicine, Boston, MA); Sheldon T. Brown, MD (James J. Peters VAMC, Bronx, NY, and Mt Sinai School of Medicine, NY, NY); Laila Castellino, MD (VAMC and Wright State University Boonshoft School of Medicine, Dayton, OH); Arundhati S. Desai, MD (Kansas City VAMC, Kansas City, MO); Fletcher Fernau, BA (James J. Peters VAMC, Bronx, NY); Mark A. Fisher, PhD (University of Utah Department of Pathology, and ARUP Laboratories, Salt Lake City); James Fleckenstein, MD (Washington University, St Louis, MO); Carol S. Fleming, BA, MT(ASCP), CLS (VA NCHCS Sacramento, Mather, CA); Narla J. Fries, CLS, MT(ASCP) (James A. Haley VAMC, Tampa, FL); Virginia L. Kan, MD, FACP (VA Medical Center and George Washington University, Washington, DC); Carol A. Kauffman, MD (VA Ann Arbor HCS and University of Michigan Medical School, Ann Arbor); Stacey Klutts, PhD, and Michael Ohl, MD, MSPH (Iowa City VAMC and University of Iowa, Iowa City); Thomas Russo, MD (Buffalo VAMC and SUNY at Buffalo, NY); Andrea Swiatlo MS, MT(ASCP) (VAMC, Jackson, MS); and Edwin Swiatlo, MD, PhD (VAMC and University of Mississippi Medical Center, Jackson).
Acknowledgments. This material is based on work supported by the Office of Research and Development, Medical Research Service, Department of Veterans Affairs (grant number 1 I01 CX000192 01 to J. R. J.), and National Institutes of Health (grant number RC4-AI092828 to E. V. S. and J. R. J.). Dave Prentiss (Minneapolis VAMC) helped prepare the figures. Mark Bielke, MD (Milwaukee VAMC, Milwaukee, WI), Jill E. Clarridge III, PhD (Puget Sound HCS VAMC and University of Washington, Seattle), Christopher Graber, MD (GLAHS-WLA campus, Los Angeles, CA), Dennis Stevens, MD (Boise VAMC, Boise, ID), and Michael Tyndale (PVAHCS, San Diego, CA) provided isolates and associated source data.
Potential conflicts of interest. J. R. J. has research grants and/or contracts from Merck, Rochester Medical, ICET, and Syntiron, and has patent applications relating to diagnostic tests for ST131 and other E. coli lineages. E. V. S. has patent applications relating to diagnostic tests for ST131 and other E. coli lineages. All other authors report no potential conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
Author notes
Presented in part: IDWeek 2012, San Diego, California, 17–21 October 2012. Poster 828.
- gentamicin sulfate (usp)
- ciprofloxacin
- drug resistance, microbial
- drug resistance, multiple
- fluoroquinolones
- genes
- gentamicins
- hospitals, veterans
- inpatients
- outpatients
- trimethoprim-sulfamethoxazole combination
- veterans
- infections
- morbidity
- virulence
- pathogenic organism
- antimicrobials
- escherichia coli
- extended-spectrum beta lactamases






Comments