-
PDF
- Split View
-
Views
-
Cite
Cite
Michael W. Gaunt, Michael A. Miles, An Insect Molecular Clock Dates the Origin of the Insects and Accords with Palaeontological and Biogeographic Landmarks, Molecular Biology and Evolution, Volume 19, Issue 5, May 2002, Pages 748–761, https://doi.org/10.1093/oxfordjournals.molbev.a004133
Close - Share Icon Share
Abstract
A unified understanding of >390 Myr of insect evolution requires insight into their origin. Molecular clocks are widely applied for evolutionary dating, but clocks for the class Insecta have remained elusive. We now define a robust nucleotide and amino acid mitochondrial molecular clock encompassing five insect orders, including the Blattaria (cockroaches), Orthoptera (crickets and locusts), Hemiptera (true bugs), Diptera, and Lepidoptera (butterflies and moths). Calibration of the clock using one of the earliest, most extensive fossil records for insects (the early ancestors of extant Blattaria) was congruent with all available insect fossils, with biogeographic history, with the Cambrian explosion, and with independent dating estimates from Lepidopteran families. In addition, dates obtained from both nucleotide and amino acid clocks were congruent with each other.
Of particular interest to vector biology is the early date of the emergence of triatomine bugs (99.8–93.5 MYA), coincident with the formation of the South American continent during the breakup of Gondwanaland. More generally, we reveal the insects arising from a common ancestor with the Anostraca (fairy shrimps) at around the Silurian-Ordovician boundary (434.2–421.1 MYA) coinciding with the earliest plant megafossil. We explore Tilyard's theory proposing that the terrestrial transition of the aquatic arthropod ancestor to the insects is associated with a particular plant group (early vascular plants). The major output of the study is a comprehensive series of dates for deep-branching points within insect evolution that can act as calibration points for further dating studies within insect families and genera.
Introduction
The molecular clock hypothesis is the basis of the modern molecular phylogenetic approach for dating and resolving evolutionary divergence. Major studies include estimates for the ages of mammals (Hedges et al. 1996 ) and birds (Cooper and Penny 1997 ). Nevertheless, deviations from a molecular clock, caused by rate heterogeneity between lineages, have been observed for most genetic markers (Hedges et al. 1996 ; Bromham, Rambaut, and Harvey 2000 ; Yoder and Yang 2000 ). However, recent approaches to molecular clock calculations provide the potential to obtain robust temporal reconstructions by assuming any number of local molecular clocks within a global phylogeny (Yoder and Yang 2000 ). More deterministic approaches for relaxing the molecular clock have been based on the assumption that nucleotide substitutions along the length of a branch approximate to lognormal or Poisson distributions. The assumptions underlying how these molecular clocks are relaxed differ, with one approach assuming there is a correlation of evolutionary rates among closely related lineages, usually referred to as “autocorrelation” (Sanderson 1997 ; Thorne, Kishino and Painter 1998 ) and the second approach allowing rates to change anywhere on a phylogenetic tree (Huelsenbeck, Larget, and Swofford 2000 ). An earlier approach to molecular dating involved the selective removal of lineages that deviated from a molecular clock “backbone,” identified using a relative rates test (Li and Tanimura 1987 ), also known as the two-cluster test (Takezaki, Rzhetsky, and Nei 1995 ).
Although insects demonstrate the greatest biodiversity in the animal kingdom, no molecular clock has previously been identified either for the class as a whole or for any order therein. Moreover, few robust examples of rate uniformity have been observed even at the family level. In addition, unlike studies of other animals, few molecular dating methods for insects have been calibrated using the available fossil record; calibrations have predominantly assumed population isolation following tectonic movements (Russo, Takezaki, and Nei 1995 ; Pellmyr and Leebens-Mack 1999 ). The use of geographic calibration points for molecular clocks could be strongly criticized because by taking the most prominent example, the breakup of Gondwanaland, few molecular or palaeontological studies (e.g., Hedges et al. 1996 ) substantiate that this is a major driving force for global speciation; therefore, assuming that continental breakup is the single source of diversity would appear to be open to error.
Methods
Complete amino acid and nucleotide data sets were obtained from GenBank, aligned using Clustal X, and edited by hand (Thompson et al. 1997 ). Selection of genetic loci was based on three factors: the most commonly sequenced loci throughout the Insecta, that is, 16S, 18S, cytochrome b (cob), cytochrome oxidase I (cox1), and elongation factor 1α (EF-1α) (Caterino, Cho, and Sperling 2000 ); the availability of nucleotide sequence data from the insect orders Blatteria or Odonata which both offer the earliest and most extensive fossil records for extant insects to allow for calibration of the clock; existing sequence data for the triatomine bugs; and, finally, availability of sequence data for Brachiopoda as the closest ancestor to the insects. Partial length sequence data were used in each instance, comprising 477, 350, and 133 amino acid residues for cox1, EF-1α, and cob data sets, respectively, 1,396 nucleotide base pairs (bp) for 18S, and 258 bp for 16S. All alignments are available on request. 16S and 18S alignments comprised sequence data for all available hematophagous insect vectors. For protein loci two examples of each species were included, and for cox1 data particular attention was paid to the extensive molecular dating available for the two Lepidopteran families, Proxidadae and Incurvidae, to enable a direct comparison with previous date estimates (Pellmyr and Leebens-Mack 1999 ).
cox1 Phylogeny
An initial assessment of the above loci suggested that cox1 provided the most suitable locus for a molecular clock study (see Molecular Clock section later for details of this initial assessment). The cox1 data set was then supplemented with a new cox1 sequence from triatomine bugs of the tribes Rhodniini (Rhodnius pictipes [GenBank accession number AF449136], R. robustus [AF449137], and R. prolixus [AF449138]) and Triatomini (Triatoma maculata [AF449139], Eratyrus mucronatus [AF449140], and Panstrongylus herreri [AF449141]). Amplicons were obtained using a two-round, or “nested,” PCR, initially using primers 5′ TGT ATT TTM TRT TCG GGG CYT GA 3′ and 5′ TCG TGG AAG AAG ATA AGT TGT T 3′ and then using primers 5′ GAG CTG GAA TAA TAG GAA CAT C 3′ and 5′ TGC ATT AAT CTG CCA YAT TA 3′. The second-round primers had the standard M13 promoter site sequences linked to their 5′ terminus (5′ TGT AAA ACG ACG GCC AGT 3′ and 5′ CAG GAA ACA GCT ATG ACC 3′). The second-round amplicons were custom sequenced using M13 primers to ensure high-quality sequence data (MWG-Biotech, U.K.). All phylogenies were outgrouped using all available sequences from the Crustacea.
The robustness of the data sets was examined for deviations in nucleotide or amino acid base composition between taxa because nucleotide or amino acid base homogeneity is a prerequisite for the maximum likelihood (ML) mutation models used throughout this study. We felt this criterion to be particularly important to observe for molecular dating purposes. Mitochondrial Apis mellifera ligustica sequence (NC_001566) was not included in the analysis because of a low level of amino acid homogeneity (P = 0.1 and >0.99 in all other cases) using the ML quartet puzzling software Puzzle (Strimmer and von Haeseler 1996 ). For nucleotide data sets, first- and second-nucleotide codon positions were used for evolutionary reconstructions because of the significant deviation of the third base from nucleotide base homogeneity.
The ability of the cox1 locus in comparison with the other mitochondrial protein-encoding genes to demonstrate a robust molecular clock was reanalyzed for representative insects for which a complete mitochondrial genome had previously been deposited in GenBank. Each mitochondrial protein-encoding gene (adenosine triphosphate synthase 6 and 8 [atp6 and atp8] combined, cox1, cox2, cox3, cob, nicotinamide adenine dinucleotide dehydrogenases 1–3, 5, 6 [nad1, nad2, nad3, nad5, nad6], with nad4 and nad4L combined) was separately aligned and assessed for a molecular clock using amino acid ML quartet puzzling (Puzzle) and second-codon–position nucleotide ML, using a seven-parameter model. The insects used represented the Diptera (Anopheles quadrimaculatus [NC_000875]; A. gambiae [NC_002084]; Cochliomyia hominivorax [AF260826]; Ceratitis capitata [NC_000857]; Drosophila yakuba [NC_001322]; D. melanogaster [NC_001709]), the Lepidoptera (Bombyx mori [NC_002355]), and the Hemiptera (T. dimidiata [AF301594]), the Orthoptera (Locusta migratoria [X80245]), and the phylogeny was rooted using the Brachiopoda (Daphnia pulex [NC_000844]; Artemia franciscana [X69067]).
A prerequisite of the study was to reconstruct constrained molecular clock ML phylogenies using nucleotide and amino acid data and compare them with the unconstrained models. The basic model of nucleotide evolution was determined using MODELTEST (Posada and Crandall 1998 ). Seven ML model parameters, including a four-category discrete gamma distribution (one parameter, Γ), the general time-reversible model of nucleotide substitution (six parameters, GTR) and invariant rate parameter (one parameter, PINVAR), were estimated during a heuristic tree search for a reconstruction limit of 1 using PAUP*4.0b (Swofford 2000 ). The ML amino acid phylogeny (described later) was then used as a backbone to resolve the nucleotide ML phylogeny because nucleotide data could not adequately resolve the interorder topology, and the seven parameters were reestimated. All parameter estimates were incorporated into a full ML heuristic search for nucleotide data (PAUP*4.0b). All parameter estimates are available on request.
ML amino acid phylogenies were constructed by a heuristic search using the JTT (Jones, Taylor, and Thornton 1992 ) and mtREV24 (Adachi and Hasegawa 1996a ) models of amino acid substitution where appropriate by PROTML in the MOLPHY package (Adachi and Hasegawa 1996b ). Nucleotide and amino acid parsimony trees were calculated using a heuristic search with Tree bisection-reconnection (TBR) branch swapping for 1,000 random sequence additions (PAUP*4.0b). Minimum distance phylogenies were built using an ML model for nucleotide data with the parameters defined by MODELTEST and using p-distances for amino acid data (PAUP* 4.0). The cox1 topology was initially assessed using parsimony and minimum distance nonparametric bootstrapping. Further topological differences between the cox1 ML, parsimony, and minimum distance nucleotide and amino acid phylogenies were assessed by the Shimodaira-Hasegawa test (Shimodaira and Hasegawa 1999 ), using the full optimization (100 replications) or the RELL approximation (1,000 replications) to the ML bootstrap with the seven-parameter ML model for nucleotide data previously described (Γ, GTR and PINVAR).
Tenebrio molitorcox1 sequence (X88966) was omitted because of a topological discrepancy with insect morphologically-based insect phylogenies. For morphologically-based insect phylogenies, Tenebrio sp. (Coleoptera) is an out-group to Diptera and Lepidoptera (Whiting et al. 1997 ; Wheeler et al. 2001 ), whereas in the amino acid phylogeny of cox1, Diptera is an out-group to Coleoptera and Lepidoptera; therefore, the phylogenetic reconstructions were repeated. The suitability of EF-1α analysis for dating within Insecta was questionable when a gene duplication event was observed from the quartet puzzling ML phylogeny, resulting in EF-1α F1 and EF-1α F2 of Drosophilia melanogaster disrupting the Brachycera suborder and possibly causing splits in the dipteran monophyly (data not shown). This observation is supported by previous studies on EF-1α (Danforth and Ji 1998 ).
Molecular Clock
Molecular clock tests were conducted both before any loci were sequenced and after the sequence data were incorporated to ensure that a molecular clock was still observed. Initial screening of the 16S, 18S, cob, cox1, and EF-1α loci for mutation rate consistency between different lineages was performed for nucleotide and for amino acid data using the likelihood ratio test 2ΔL = n − 2, where n is the number of taxa and L the negative log likelihood score; if a constant rate of evolution occurs between lineages, then sequence data represented in an ML additive tree, where branch length differences represent sequence divergence, should not be significantly different from an ML ultrametric tree, where the tips of the tree are equidistant from the root. All molecular clocks for 16S, 18S, cob, and EF-1α were significantly rejected, even for small numbers of taxa, using amino acid ML quartet puzzling (Puzzle; Strimmer and von Haeseler 1996 ) with JTT (Jones, Taylor, and Thornton 1992 ) or mtREV24 (Adachi and Hasegawa 1996) mutation models and for second-codon positions for nucleotide data, using a seven-parameter model (discussed elsewhere; PAUP*4.0b). However, cox1 locus robust molecular clocks were obtained for some, but not all, taxa using ML quartet puzzling for amino acid data (not shown) and, in particular, for second-codon positions for nucleotide data using a seven-parameter ML model (discussed later in Methods), indicating the suitability of this locus for dating studies.
The calibration point for the cox1 nucleotide and amino acid molecular clock phylogenies was set at the Blattaria (cockroaches)-Orthoptera (crickets and locusts) divergence. Early ancestors of extant Blattaria, initially called Pinnule Insects (Henning 1981 ), have more recently been reclassified as members of this order (Jarzembowski 1994 ) which represents one of the earliest, most extensive fossil records of all extant insects (Labandeira and Sepkoski 1993 ; Ross and Jarzembowski 1993 ). Furthermore, the onset of their radiation coincided with the radiation seen in the extensive fossil record of an extinct member of the neopteran clade, the Protorthoptera, which are more closely related to Orthoptera than to Blattaria. It is noteworthy that Blattaria are one of three closely related orders that include Mantoidea (mantids) and Isoptera (termites), and at least Isoptera are believed to have evolved from wood-eating Blattaria (Lo et al. 2000 ).
The cox1 phylogeny of nucleotide sequence, including the triatomine bug sequences, was resubjected to a likelihood ratio test for a global molecular clock, i.e., assuming the same rate for all lineages, using second-codon–position nucleotide data under the seven-parameter model using a four-category discrete Γ distribution, GTR, PINVAR (PAUP 4.0*b). Model parameters for the molecular clock test were estimated for the unconstrained phylogeny and used for the global clock.
The cox1 phylogeny of amino acid sequence was tested for a molecular clock with a 12-category discrete gamma distribution using PAML, written by Z. Yang (Yoder and Yang 2000 ). This statistical method allows for variable evolutionary rates among lineages to address complex evolutionary models, where some branches have the same rate, a “local molecular clock,” whereas different parts of the tree have different rates. The approach is attractive because it allows the user to specify any given evolutionary model and, in addition, allows a likelihood ratio test to assess the robustness of the clock. One feature of this approach is that by assigning discrete rate categories to given lineages, it often results in abrupt changes in evolutionary rate between species boundaries.
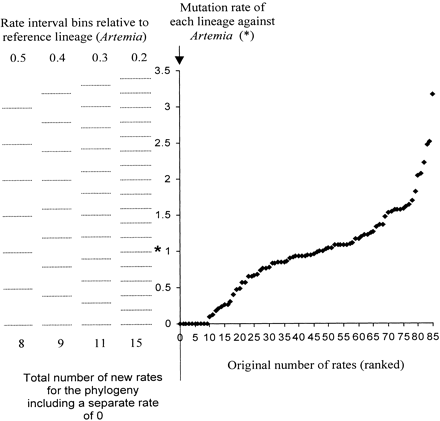
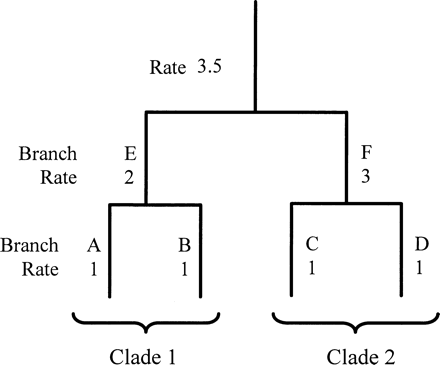
Amino acid ML models for the cox1 phylogeny assuming a global molecular clock or a local molecular clock for each insect order and different rates for lineages between each order were significantly rejected using a likelihood ratio test 2ΔL = n − k − 2 parameters, where k is the number of evolutionary rates in the model, and n and L are as previously described (PAML; Yoder and Yang 2000 ). These models and all subsequent models discarded ambiguous amino acid sites. The local molecular clock method (PAML) gives the user the potential to estimate the local rate of molecular evolution for each branch in the tree. Given this potential, we adopted a philosophy similar to that of Sanderson (1997) by calculating the local rate of molecular evolution for separate branches, then minimized the rate difference between branches, and incorporated a method of smoothing the differences between rates. However, we sought to avoid reliance on any one assumption of mutation rate heterogeneity. To simplify the calculation, we assumed that autocorrelation of rates occurred within each insect genus, i.e., a local molecular clock was assigned to each genus, but separate mutation rates could occur between genera, with the exception of the separate rates that were assigned for R. pictipes and T. vitticeps because of their very short branch lengths; in addition, mutation rates were calculated for all intergenera branches. This resulted in a molecular clock phylogeny with 85 ML rates, having almost as many mutation rates as branches, with a maximum and minimum mutation rate of 0.093 and a 3.59-fold difference in substitutions per site relative to A. franciscana, the reference lineage (rate = 1; fig. 1 ). To reduce the number of rate parameters, we defined a series of bin ranges, or “rate categories,” for the frequency distribution of ranked mutation rates of the 85-ML parameter model, effectively pooling lineages that showed similar mutation rates regardless of where they occurred on the phylogeny. There was no objective way of defining any given bin range, and categorizing what appeared to be continuous data (fig. 1 ) could result in problems; therefore, in order to smooth the differences between mutation rates a large number of different bin ranges was defined, resulting in a series of mutation rate histograms for the cox1 phylogeny. All histogram bin ranges were arbitrarily based on two different boundary values, 0+ to 0.1 and 0.05+ to 0.15 relative to the mutation rate of A. franciscana, where + is an incremental increase on the stated value. Four bin ranges comprising 0.2, 0.3, 0.4, and 0.5 intervals relative to the A. franciscana mutation rate were constructed (fig. 1 ), in addition to a 0-rate category, defining 8–16 different mutation rates for the phylogeny. To further minimize rate interval “edge effects,” each combination of 0.1-unit boundary values within each rate interval was described; for example, a 0.5-unit interval size has five possible 0.1-unit boundary value combinations and therefore five different bin ranges, whereas a 0.2 interval size had two different bin ranges. In total, 28 different models describing the evolutionary rates of the cox1 phylogeny were calculated (PAML); 14 rate models were derived from different bin intervals (5 + 4 + 3 + 2) based on two different boundary values. Models that did not support the unconstrained model, by a collapse in the branch length of the ancestral lineage to the Branchipoda and Insecta presumably reflecting instability in the malacostracan root, were discarded (17 models were retained). In summary, 17 date estimates were obtained for each node based on the hypothesis that identical rates occur at any part of the phylogeny, although this hypothesis may not represent the true behavior of local clocks within the phylogeny. We therefore employed a second assumption that autocorrelation of mutation rates occurs between insect genera, families, or suborders (fig. 2 ). Two approaches to autocorrelation were adopted using the 17 models of rate estimates derived from the previous hypothesis; when this hypothesis described two or more clades at different parts of the tree as having identical mutation rates, the assumption of rate identity between unrelated clades was removed, and the model was recalculated (approach 1; 17 models), and in addition, when two “sister” branches showed identical mutation rates, then the mutation rate of the “parental” branch was also assumed to be identical regardless of the value previously assigned to it, and the model was recalculated (approach 2; 17 models).
A weakness of the approach described here, in common with that of Sanderson (1997) , is that no external criteria are used for defining which lineages are autocorrelated, such as the comparative reproductive rate between species. Moreover, the rate hypothesis is derived from the data and is also tested against the same data, which is effectively the situation with the “tree pruning” methods and is only relevant if it is compared with an approach where this assumption is removed or minimized, i.e., compared with the nucleotide clock. Ideally, the nucleotide data set should be bootstrapped to obtain a better estimate of the confidence limits, but in conjunction with the models described so far this would result in an unrealistically large number of models.
Root Stability
Concerns for the stability of the malacostracan root were further investigated for the autocorrelation-based approaches (fig. 2 ), because of a further collapse in the branch length of the ancestral lineage to the Branchipoda and Insecta in 10 of the 34 models. The collapsed branch length occurred in models where each of the five branches in the malacostracan root was assigned an independent mutation rate. The remaining models showed identical rates between the parental branch to the Penaeus monodon and P. notialis sister groups and the parental branch to the Pagurus longicarpus and Penaeus branches. To test root stability, a final group of models was made assigning independent rates for the Penaeus and Pagurus-Penaeus parental branches (nine models for approach 1 and nine models for approach 2; fig. 2 ). A direct comparison of the difference in date estimates could then be made for each model, with and without the extra rate category in the root, and the stability of the root assessed. The maximum difference between date estimates from models with the extra rate category in the root compared with those without it for all 18 models was less than 5% for all insect nodes (branching points), but the nodes within the Brachycera suborder (i.e., the Drosophila-Ceratitis split) showed a difference of up to 17%. The maximum difference in date estimates for basal lineages was 7% for the nodes of Branchiopoda, 10.5% for the Malacostraca-Branchiopoda ancestral node, and over 70% variation for the nodes within Malacostraca. Although the root was unstable, and the date estimates for Malacostraca were not considered further, this did not greatly affect the divergence of most lineages, except for the Brachycera suborder which was unusually unstable.
To examine the robustness of date estimates to model assumptions, we obtained the 95% confidence interval (CI) using median bootstrapping for 1,000 replications with the STATA package (http://www.stata.com). We did not attempt to determine which is the correct model but simply bootstrapped all dates for each node obtained from the nonautocorrelated- and autocorrelated-based approaches (3 × 17 models) and the additional models obtained from analyzing the stability of the malacostran root (2 × 9 models), giving 69 models in total. Median bootstrapping was used to minimize outlier effects and a priori assumptions of the CI estimation. In addition, this approach sought to minimize a priori assumptions of rate heterogeneity between insect lineages and rate changes only at speciation boundaries (Huelsenbeck, Larget, and Swofford 2000 ).
Stratigraphic Dating
Permian stratigraphic ranges were taken from a single source (Jin et al. 1997 ); all other stratigraphic ranges used were taken from Gradstein and Ogg (1996) . This geological time scale incorporates several improvements over previous studies (Harland et al. 1989, p. 265) which includes more accurate Mesozoic chronology (Triassic-Cretaceous) using radiometric dating rather than interpolations from marine magnetic anomaly profiles (Gradstein et al. 1995 ) and relatively precise isotope datings for Paleozoic chronology (pre-Triassic) (Roberts, Claoue-Long, and Jones 1995 ; Tucker and McKerrow 1995 ).
Data deposition
The new nucleotide sequence data described here are available at GenBank under accession numbers AF449136–AF449141. All amino acid and nucleotide alignments and ML parameter estimates are available on request.
Results
As a route to estimating the evolutionary dates for deep divergences within insect evolution, we used near full-length cox1 nucleotide and amino acid data using ML global and local molecular clocks, respectively. Several lines of evidence suggested that cox1 provided the most suitable locus for this study. Firstly, it was the only locus that recovered global molecular clocks for some, but not all, of the taxa examined particularly for nucleotide data (fig. 3 ). Secondly, in comparison with all other protein-encoding mitochondrial genes, using representatives of four of the five insect orders (described in Methods) for which there was a complete mitochondrial genome sequence available and including the out-groups of Brachipoda, cox1 provided the smallest difference between the global molecular clock and the unconstrained models for amino acid data without Branchiopoda as the root (2ΔL = 15.0 at df = 7; P = 0.036) and nucleotide data with Branchiopoda as the root (2ΔL = 11.1 at df = 9; P = 0.27). In contrast, all other protein-encoding mitochondrial genes produced a substantial deviation between the unconstrained and clock models (P < 0.003 amino acid and P < 0.05 nucleotide data), with the exception of cox3 nucleotide data (2ΔL = 15.2; P = 0.084 at df = 9). Furthermore, cox1, cox2, cox3, nad1, and nad4 were the only loci that did not show significant deviation of the amino acid composition between lineages using the mtREV24 mutation model, i.e., the model derived from the empirical mitochondrial sequence.
Robustness of cox1 Phylogeny
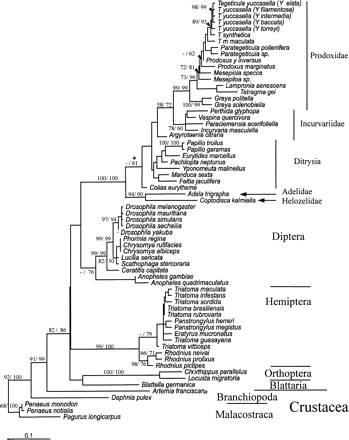
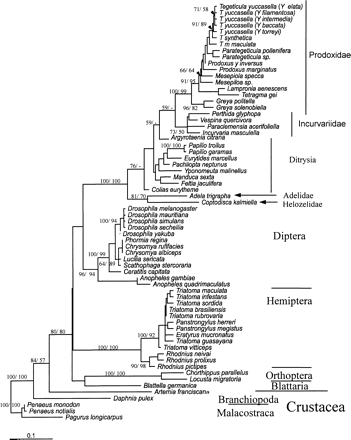
Unequivocal congruence in branching order was obtained within each order for amino acid phylogenies by both parsimony and ML methods, with the exception of the placement of C. capitata and small differences within Triatoma spp., and provided the basic phylogeny. Moreover, regardless of the method used, either ML or parsimony using amino acids or nucleotides, very similar topologies were recovered within each order. Nucleotide data using first- and second-codon–position nucleotides, however, did not recover the interorder topology (Methods). All family and superfamily relationships within each order were strongly supported using neighbor-joining bootstrap analysis for nucleotide data sets, with the exception of taxonomic relationships within Lepidoptera (Appendix 1 ). Bootstrap analysis of cox1 amino acid data sets generally resulted in lower bootstrap values; however, unlike nucleotide data, amino acid data recovered the correct interorder topology (Appendix 2 ). The topology describing Proxididae, Incurvivdae, Ditrysia, Adelidae, and Helozelidae within Lepidoptera in figures 3 and 4 was recovered for distance, parsimony, and ML tree-building methods using both amino acid and nucleotide data sets. In addition, the robustness of the Adelidae and Helozelidae outgroup within the Lepidoptera was supported by the Shimodair-Hasegawa test using full optimization to the ML bootstrap for nucleotide data, against the alternative topology of Ditryia forming an out-group to this order (ΔL = 11.3; P = 0.04; Appendix 1 ). Alternative branching orders were obtained by examining topological differences between amino acid and nucleotide data sets among the ML, parsimony, and distance methods and involved the placement of C. capitata within the dipteran suborder Brachycera and lineages within Triatoma spp. (although topological comparisons were not exhaustive in this instance) and Drosophila spp. using the Shimodaira-Hasegawa test with the RELL approximation to the ML bootstrap for nucleotide data as a quicker alternative to the full-optimization test. The phylogeny was rooted using Malacostraca as previously described (Friedrich and Tautz 1995 ; Regier and Shultz 1997, 1998 ; Boore, Lavrov, and Brown 1998 ).
Nucleotide Molecular Clock for cox1
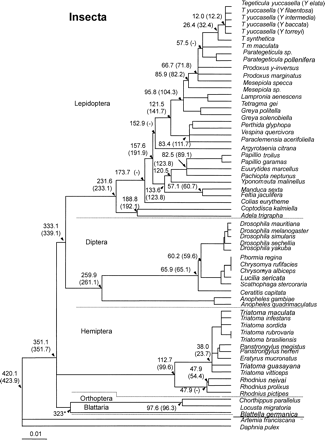
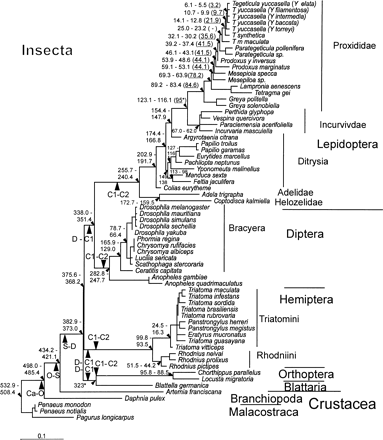
Figure 3 describes the molecular clock for cox1 nucleotide data for second-codon positions for all insect taxa and including Branchiopoda. Although the mutation rate heterogeneity between lineages is significant using a likelihood ratio test, the vast majority of lineages do obey a molecular clock. For instance, if the lineages that demonstrate the fastest or slowest evolutionary rates in Hemiptera and Lepidoptera are excluded under a seven-parameter ML nucleotide model (Γ, GTR and PINVAR; Methods), namely, R. pictipes, P. megistus, Colias eurytheme, and Argyrotaenia citrana, along with the fastest or slowest evolutionary rates in Incurvidae, Incurvaria masculella and Proxidadae, Prodoxus y-inversus, a robust molecular clock is observed, although there is a collapse in the root (2ΔL = 65.9 at df = 55; P = 0.15). The purpose of this analysis was not to perform a “relative rates” test (e.g., the two-cluster test; Russo, Takezaki, and Nei 1995 ) but primarily to demonstrate that most lineages conform to a molecular clock, and this provides an indication that the locus is suitable for further analysis to account for rate heterogeneity between lineages. Moreover, not even first- and second-codon positions were capable of recovering the phylogeny and raised doubts about an exclusive reliance on second-codon positions for evolutionary dating. Figure 4 presents an amino acid molecular clock phylogeny, accounting for rate heterogeneity between lineages. The date estimates from this analysis arise from 69 models resulting from the assumptions of insect amino acid mutation, including the models that examined root stability, using near–full-length cox1 sequence for five orders of insect with all the available crustacean cox1 sequences (figs. 1 and 2 ; see Methods).
Amino Acid Molecular Clock for cox1
Each of the 69 local clock models was compared with the unconstrained model. All amino acid clock models demonstrated a robust molecular clock using a likelihood ratio test described previously (P > 0.05 for all models [minimum 2ΔL = 16.4 at df = 9], P > 0.225 for 96% models [2ΔL = 12.89 at df = 10], and P > 0.7 for 62% models [2ΔL = 15.32 at df = 19], where L = ML log likelihood value).
The nucleotide data were primarily used to assess the ability of a locus to form a molecular clock. However, a striking congruence in date estimation was observed between the cox1 nucleotide and amino acid data sets for all deep nodes in the five orders of Insecta. Thus, the Daphnia pulex and Artemia franciscana divergence is 498.0–485.4 and 434.2–421.1 MYA, respectively, for the amino acid clock and 420.1/423.9 MYA for the nucleotide clock, where 420.1 MYA is the date from the global nucleotide clock, and 423.9 MYA is the global clock with lineages removed; the nucleotide clock gives single dates because of a collapse in the root. The divergence between the Blattaria/Orthoptera lineage and Hemiptera is 382.9–373.0 and 375.6–368.2 MYA for the amino acid clock, respectively, and 351.1/351.7 MYA for the nucleotide clock, because of a collapse in the bifurcating topology. Furthermore, congruence is also observed for the divergence between the dipteran suborders Nematocera containing the Anopheles lineages and Brachycera containing the Drosophila lineages (282.8–247.7 MYA amino acid clock and 259.9/261.1 MYA nucleotide clock), Triatomini and Rhodniini divergence within the Hemiptera (99.8–93.5 MYA amino acid clock and 112.7/96.3 MYA nucleotide clock), Chorthippus parallelus and L. migratoria within the Orthoptera (95.8–88.5 MYA amino acid clock and 97.6/99.4 MYA nucleotide clock) as well as for the deepest split in Lepidoptera (255.7–240.4 MYA amino acid clock and 231.6/233.1 MYA nucleotide clock). Date estimates for other nodes within the Lepidoptera were also generally congruent between the amino acid and nucleotide clocks, although for the family Proxididae the nucleotide clock gave dates no more than 10% older than did the amino acid clock. The major exception was the deepest nodes within the Brachycera suborder that produced substantially different dates between the amino acid clock (165.9–129.0 MYA) and the nucleotide clock (65.9/65.1 MYA); the dates of the amino acid clock are congruent with the fossil record (table 1 ). It is worth noting that the dates of the nodes within the Brachycera suborder were affected by instability in the malacostran root (Methods). However, the second deepest node within the Brachycera root, the Scahophaga stercoraria-Drosophila split, is more congruent between the amino acid (78.7–66.4 MYA) and nucleotide clocks (60.2/59.6 MYA).
Robustness of Insect Dating Estimates
The evolutionary dates obtained in this study provide robust date estimates when compared with the earliest dates for the stratigraphic ranges of all available insect fossils known to us and for the latest date of the Cambrian explosion (table 1 ). In addition to the fossil record, relevant divergence dates are also robust against proposed biogeographic events affecting speciation in the phylogeny (table 1 ). In particular, the continental breakup of Gondwanaland (105–95 MYA) provides the earliest date for the isolation of triatomines in South America (95.5–93.6 MYA) and the latest date for the divergence between the globally distributed Lepidoptera Prodoxidae and Incurvariidae (123.1–116.1 MYA). Moreover, the breakup of Gondwanaland was used in a previous study to provide a minimum calibration point for the divergence between moths of the families Prodoxidae and Incurvariidae (Pellmyr and Leebens-Mack 1999 ), which enabled the minimum dates of nodes within Prodoxidae to be estimated using the two-cluster test (fig. 4 , numbers in brackets). Excluding a single date obtained by the two-cluster test because it was incongruent with the chronology for all other phylogenies (Pellmyr and Leebens-Mack 1999 ), no significant difference using the chi-square test (χ2) is observed between the dates for Prodoxidae derived using either the two-cluster test calibrated from the breakup of Gondwanaland (expected result) or the minimum bootstrap values obtained from the ML insect local clocks calibrated from the divergence of Blattaria (observed results) (χ2: P = 0.09 at 10 df values given in fig. 4 ). This result is strengthened by removing the two-cluster–test calibration point (χ2: P = 0.23 at df = 9). However, although the dates in this study fit with those of Pellmyr and Leebens-Mack (1999) , it must be noted that generally biogeographic calibration points for molecular clocks are subject to error. In summary, virtually all the evidence of the date estimates obtained here converges to suggest a robust basis for reconstructing over 400 Myr of insect evolution. The temporal congruence verifies the rationale behind using Blattaria fossils as the calibration point of the clock.
The molecular clock phylogenies describe the aquatic-terrestrial transition of the insect-Artemia (Anostraca, fairy shrimp) ancestor during the early/late Silurian (428.0–419.0 MYA). The date coincides with the earliest plant megafossils, notably Cooksonia cambrensis (423.0–419.0 MYA) (Edwards, Feehan, and Smith 1983 ). The radiation of the deepest insect order lineages under investigation, notably the Blattaria/Orthoptera lineage, occurred from the middle to late Devonian (approx. Eifelian-Famennian; 380.0–354.0 MYA), or perhaps very early Carboniferous, and provides the latest date for the evolution of insect flight. The date obtained here for the divergence of the Hemiptera lineage also occurred in the middle to late Devonian and denotes the divergence between hemimetabolous and holometabolous insects (Whiting et al. 1997 ).
Discussion
There are significant weaknesses in the insect fossil record for Lepidoptera and for fossil insects in general, particularly before 323 MYA (i.e., for the first 70–60 Myr of insect evolution). Insect palaeontology depends on assigning fossils to extant taxa usually on the basis of wing characters (Shear and Kukalova-Peck 1990 ). Consequently, no fossil has been identified that could represent a “missing link” between insect orders, although more recent ancestors of extant insects are seldom classified with certainty. In addition, insect fossilization occurs almost exclusively in water-deposited sediments (Shear and Kukalova-Peck 1990 ).
Molecular clock approaches enable a temporal reconstruction of ancestral nodes regardless of palaeontological difficulties (Hedges et al. 1996 ; Cooper and Penny 1997 ), and this study provides a vital tool to complement the insect fossil record. Most previous molecular dating studies in insects have almost exclusively focused on one gene or two adjacent mitochondrial loci (Brown et al. 1994 ; Takezaki, Rzhetsky, and Nei 1995 ; Esseghir et al. 1997 ; Foley et al. 1998 ; Pellmyr and Leebens-Mack 1999 ). The cox1 locus used in this study provided the closest example to a global molecular clock gene against all the other candidate loci examined both at the nucleotide and the amino acid level for the Insecta by providing the most consistent rates of evolution between different lineages. The cox1 locus also showed similar amino acid base compositions between most orders, a prerequisite for the amino acid mutation models used in this study. EF-1α amino acid data were also constructed as a series of local molecular clocks as described for cox1 amino acid data but resulted in unrealistically large values for the divergence between Decapoda and Brachiopoda (>1,000 MYA). It is noteworthy that many studies have identified the ability of cox1 to form a robust molecular clock in insects, including the butterfly Heliconius sp. (Brower 1994 ), chewing lice (Phthiraptera; Hafner et al. 1994 ), Tetraopes sp. (Coleoptera; Farrell 2001 ), and Alpheus sp. (Decapoda; Knowlton 1993 ).
The cox1 phylogenies obtained were congruent with the insect molecular phylogenies of previous studies, although we do not attempt to resolve the phylogeny with certainty (Russo, Takezaki, and Nei 1995 ; Caterino and Sperling 1999 ; Lyman et al. 1999 ; Pellmyr and Leebens-Mack 1999 ). In addition, the cox1 phylogeny of the insect orders was identical with the 18S and 28S phylogeny and a morphologically based insect phylogeny (Whiting et al. 1997 ; Wheeler et al. 2001 ). Furthermore, our phylogeny is supported by previous phylogenetic analyses that described Malacostraca and Branchiopoda (including Anostraca) forming a sister group to Insecta, using combined EF-1α and RNA polymerase II (POL II) amino acid sequence data (Regier and Shultz 1997, 1998 ; Shultz and Regier 2000 ), combined 18S and 28S data (Friedrich and Tautz 1995 ), and mitochondrial gene translocations (Boore, Lavrov, and Brown 1998 ). Finally, our use of the Malacostraca as an out-group to the Anostraca-Insecta clade is also supported by the EF-1α and POL II analyses (Regier and Shultz 1997 ).
Molecular clock methods are particularly susceptible to the problems of mutation rate heterogeneity between lineages. We addressed this issue for amino acid data using a large number of local molecular clocks to represent the large range possible of temporal estimates each model could generate and to compare this method with a global molecular clock using second-codon–position nucleotides. The congruence in date estimates between these methods was surprising and particularly strong for deep nodes, despite the nucleotide data containing less phylogenetic information. Moreover, the temporal estimates from both the amino acid local clock models and the global nucleotide clocks were compatible with all known insect fossils, with the breakup of Gondwanaland and the Cambrian explosion. In addition, the use of a clock method that does not rely on a global molecular clock, the use of ML techniques, and the use of models that do not assume any one mechanism of rate variation should minimize or remove dating errors caused by incomplete taxon sampling. We recognize that the sample size defined in this study is far from complete, representing 5 of the 26 orders of extant insects, although the nodal dates described provide a series of potential calibration points for other insect molecular clocks where their fossil record is sparse (most insects) and would provide a more suitable method of calibration than using biogeographic assumptions. The nodal date for the earliest extant hexapod, Collembola, is desirable because it would provide a direct comparison with the fossil record, but the nodal dates from the surrounding lineages were compatible with a date of around 400 MYA.
The calibration point is crucial in all molecular clock calculations. One piece of data validating the calibration point used in this study is the lack of significant difference between the temporal estimates observed for the lepidopteran families Proxodidae and Incurvariidae despite two contrasting calibration points being used, the fossil record of the early ancestors of extant Blattaria used in this study for calibrating amino acid local clock models and the breakup of Gondwanaland used previously for calibrating first- and second-codon–position nucleotide data using the relative rates test (Pellmyr and Leebens-Mack 1999 ), although in this instance the second-codon–position nucleotide clock described here gives older dates for both these families. Although fossils of the earliest extant family of Blattaria, Blaberidae, are found in the Triassic period, five families of ancestral Blattaria (Pinnule Insects), namely, Adeloblattidae, Cobaloblattidae, Mylacridae, Necymylacridae, and Phyloblattidae, have been unequivocally assigned to the late Carboniferous (Bashkirian epoch, 323 MYA; reviewed by Ross and Jarzembowski 1993 ; Labandeira 1994 ). Both the ancestors of extant Blattaria and the Blattaria fossils are particularly well preserved, perhaps because they favor damp habitats that favor fossilization, and in addition, entire specimens fossilize, reducing the reliance on wing fragments for identification (Wooton 1981). Apart from morphological analysis (Jarzembowski 1994 ), support for the classification of the extinct ancestors of Blattaria as members of this order is observed in one phylogenetic study where extant Blattaria are ancestral to Isoptera, their closest relative (Lo et al. 2000 ). In addition, it is possible that predatory behavior in the closest relative to the Blattaria/Isoptera lineage, the Mantoidea (mantids), was recently derived because it is a rare characteristic within Neoptera, and this would be consistent with the much later appearance of their fossils in Cretaceous and late Triassic strata (Labandeira 1994 ; Grimaldi 1997 ). Other members of this clade include Orthoptera and a paraphyletic clade of diverse Protothoptera now extinct. The early radiation of Protorthoptera also coincides with the earliest Blattaria fossils (Labadeira 1994) and suggests that this date was close to the Blattaria/Orthoptera divergence.
Finally, the reliability of a molecular clock is also dependent on branch lengths of the root lineages (Yoder and Yang 2000 ). Although the stability of the amino acid clock was affected by the instability of the root lineages of the cox1 phylogeny, i.e., small fluctuations in the root lineages affected the temporal estimates of all other nodes, this was only a major problem for dates within the root. The nucleotide clock also showed an unstable root, although there was no means of correcting for this using a global molecular clock.
Applications
The primary focus of this study is the resolution of deep lineages within the Insecta. The dating of the deepest nodes within the Insecta is particularly relevant for providing calibration points for dating studies within relevant families and genera and is particularly noteworthy given the congruence between the date estimates obtained and the fossil record. The haematophagous triatomine bugs are a case in question because their poor fossil record has thwarted an accurate understanding of their age. The bugs are the sole vectors of Trypanosoma cruzi (the agent of Chagas disease), and the Rhodniini (palm dwelling) and Triatomini (terrestrial and arboreal dwelling) genera describe the deepest evolutionary divergence within the family (Lyman et al. 1999 ; Gaunt and Miles 2000 ; Marcilla et al. 2001 ). The vast majority of triatomine bugs are restricted to Central and South America, and the date of divergence between the tribes Rhodniini and Triatomini (99.7–93.6 MYA) coincides with the breakup of Gondwanaland (≥95 MYA). The date most importantly provides a calibration point for current studies on the Rhodnius genus, linking the evolutionary age of bugs with their habitat. For example, the deepest split within triatomine bugs is close to the date for the earliest palm megafossils (83.5–71.3 MYA), opening up the possibility of either Rhodnius-palm coevolution or the Rhodniini-Triatomini split resulting from adaptation of one genera to a specific habitat.
A more fundamental date for insects is the origin of the insect-anostracan common ancestor. The emergence of the Insecta-Anostraca ancestor obtained here, 434.0–421.0 MYA, coincides with the earliest megafossil of vascular plants, C. cambrensis, found in the Wenlock epoch of the early Silurian 428–423 MYA (Edwards, Feehan, and Smith 1983 ). Cladistic studies of C. caledonica and C. pertonii besides several other early tracheophytes (vascular plants) suggest that numerous plant species were present in this epoch (Kenrick and Crane 1997, pp. 226–270 ). The correlation is compatible with the hypothesis that the true insects evolved from an aquatic arthropod that formed an ecological association with a specific group of terrestrial plants and subsequently coevolved with that plant group, a hypothesis that dates back almost three quarters of a century (Tilyard 1928 ). Insect-pteridophyte coemergence from an aquatic-terrestrial anostracan ancestor was also compatible with the palaeontological flora from the three aquatic Devonian localities yielding insect or hexapod fossils, at Rhynie in Scotland (Whalley and Jarzembowski 1981 ; Ross and Jarzembowski 1993 ), and at Gaspé (Labandeira, Beall, and Hueber 1988 ) and Gilboa (Shear et al. 1984 ) in northeastern North America. For example, an early vascular plant Zosterophyllopsida is found at Gaspé, and one of the earliest vascular plants Rhynopsida is found at Rhynie. The Rhynie plants show three types of surface lesions inflicted while the plant was alive, of which two types could be attributed to penetration by an arthropod (Kevan, Chaloner, and Savile 1975 ). Reexamination of the Rhynie hexapods showed that one specimen was associated with an early Rhynie plant (Whalley and Jarzembowski 1981 ).
The date obtained for the origin of the neopteran clade comprising Blattaria, Orthoptera, and Hemiptera at 382.9–373.0 MYA for amino acid data and at 350.1 for nucleotide data to some extent coincides with the early pteriodophytes (mostly ferns and horsetails) during the late Devonian at around 380 MYA (Kenrick and Crane 1997 ). For example, early macrofossils of the two largest groups of pteriodophytes, the Lycopsida and Filicopsida, occur in the early Devonian (417 MYA) and early Carboniferous (354 MYA), respectively. It is also interesting to note that this period immediately precedes the early date obtained for divergence of the Diptera and Lepidoptera lineages that occurred during the early Carboniferous and provides calibration points for further studies to estimate the number of insect orders that were established during the Carboniferous.
The origin of the neopteran insects investigated in this study also provides the latest date for the evolution of insect flight. Kevan, Chaloner, and Savile (1975) hypothesized that insect flight arose as an adaptation to the increasing height of vascular plants during the early evolution of trees. The date obtained here for the ancestor to the neopteran clade, which mostly coincides with the early pteriodophytes, i.e., plants with stems tall enough to prevent herbivory with ground-borne insects, is compatible with this hypothesis.
Richard Thomas, Reviewing Editor
Abbreviations: atp6 and atp8, adenosine triphosphate synthases 6 and 8; bp, base pair; CI, confidence interval; cob, cytochrome b;cox1, cox2, and cox3, cytochrome oxidases I, II, and III; EF-1α, elongation factor-1α; ML, maximum likelihood; nad1, nad2, nad3, nad4, nad4L, nad5, and nad6, nicotinamide adenine dinucleotide dehydrogenases 1–6; POLL II, polymerase II.
Keywords: insecta insect evolution molecular clock maximum likelihood triatomine bugs
Address for correspondence and reprints: Michael W. Gaunt, Pathogen Molecular Biology and Biochemistry Unit, Department of Infectious and Tropical Diseases, London School of Hygiene and Tropical Medicine, Keppel Street, London WC1E 7HT. michael.gaunt@lshtm.ac.uk .
Table 1 Temporal Robustness of the cox1 Local Molecular Clock Against the Earliest and Latest Dates of Relevant Stratigraphic, Biogeographic, and Fossil Records
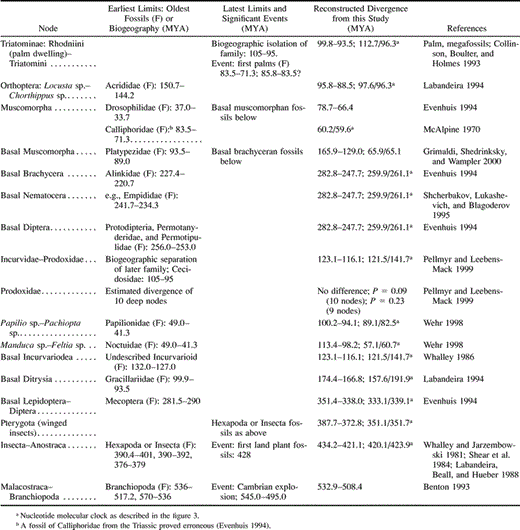
Table 1 Temporal Robustness of the cox1 Local Molecular Clock Against the Earliest and Latest Dates of Relevant Stratigraphic, Biogeographic, and Fossil Records

Fig. 1.—The ranked distribution of mutation rates from the 85–rate category model of the amino acid cox1 phylogeny described in Methods, which assumes local molecular clocks for most insect genera to define, and examples of using rate 0.2, 0.3, 0.4, and 0.5 interval bins to reduce the total number of rates in the model. In this instance, the interval bins are based on boundary values of 0+ to 0.1 as defined in Methods. * denotes Artemia reference lineage (rate = 1)
Fig. 2.—Hypothetical example of a nonautocorrelation model (fig. 1 ) where identical mutation rates occur at different parts of the phylogeny (rate in branches A and B = rate in branches C and D) and the construction of two different groups of models assuming autocorrelation where the mutation rate at different parts of the phylogeny is not assumed to be identical (rate in clade 1 ≠ rate in clade 2). Approach 1: mutation rate in branches A and B ≠ that in branches C and D, and the model is recalculated. Approach 2: mutation rate in branches A, B, and E is assumed to be identical, as is the mutation rate in branches C, D, and F, but the rate in branches A, B, E ≠ rate in branches C, D, E, and the model is recalculated
Fig. 3.—ML global molecular clock phylogeny using second-codon–position cox1 nucleotides of five insect orders and two classes of Crustacea, calibrated using the Blattaria/Orthoptera divergence (0.022% nucleotide substitutions per million year). Numbers within brackets denote dates obtained when the fastest and slowest lineages within Hemiptera and Lepidoptera were removed
Fig. 4.—ML local molecular clock phylogeny using near–full-length cox1 amino acids of five insect orders and two classes of Crustacea. The model here represents the median value of the average date per node for each of the 69 models (104.5 Myr; average rate = 0.048% amino acid subsitutions per million year) and is presented for diagrammatic purposes only. CI (95%) of date estimates were obtained by median bootstrapping (small triangles). Numbers in brackets denote minimum date estimates from a previous study obtained by the two-cluster test using nucleotide data (Pellmyr and Leebens-Mack 1999 ). Large triangles denote stratigraphic boundaries (Gradstein and Ogg 1996 ) and are drawn to scale, where Ca, Cambrian; O, Ordovician; S, Silurian; D, Devonian; C1, Early Carboniferous; C2, Late Carboniferous
Appendix 1.—ML amino acid phylogeny of cox1 showing neighbor-joining and parsimony bootstrap analysis for nucleotide data after 1,000 replications. Key: * Prodoxidae/Incurvariidae, Ditrysia, Adelidae/Helozelidae topology supported by the Shimodaira-Hasegawa test using the full optimization ML bootstrap analysis, with alternative branching order of Ditrysia forming an out-group to Prodoxidae/Incurvariidae and Adelidae/Helozelidae
Appendix 2.—ML amino acid cox1 phylogeny showing neighbor-joining and parsimony bootstrap analysis for amino acid data after 1,000 replications
We gratefully acknowledge the financial support from the Wellcome Trust. We thank the two anonymous referees and the subeditor, as well as Z. Yang (University College London), G. Clark, I. Mauricio, S. Solis Mena (LSHTM, U.K.), and the staff at the Instituto Oswaldo Cruz and the Instituto Evando Chagas (Brazil). We are extremely grateful to the relevant authorities who kindly provided current information and detailed advice on the paleontological science described here.
References
Adachi J., M. Hasegawa,
Boore J. L., D. V. Lavrov, W. M. Brown,
Bromham L. D., A. E. Rambaut, P. H. Harvey,
Brower A. V. Z.,
Brown J. M., O. Pellmyr, J. N. Thompson, R. G. Harrison,
Caterino M. S., S. Cho, F. A. H. Sperling,
Caterino M. S., F. A. H. Sperling,
Collinson M. E., A. Boulter, P. L. Holmes,
Cooper A., D. Penny,
Danforth B. N., S. Q. Ji,
Edwards D., J. Feehan, D. G. Smith,
Esseghir S., P. D. Ready, R. Killick-Kendrick, R. Ben-Ismail,
Evenhuis N. L.,
Farrell B. D.,
Foley D. H., J. H. Bryan, D. Yeates, A. Saul,
Friedrich M., D. Tautz,
Gaunt M., M. A. Miles,
Gradstein F. M., F. P. Agterberg, J. G. Ogg, J. Hardenbol, P. van Veen, J. Thierry, Z. Huang,
Grimaldi D.,
Grimaldi D., A. Shedrinksky, T. P. Wampler,
Hafner M. S., P. D. Sudman, F. X. Villablanca, T. A. Spradling, J. W. Demastes, S. A. Nalder,
Harland W. B., R. L. Armstrong, A. V. Cox, L. E. Criag, A. G. Smith, D. G. Smith,
Hedges S. B., P. H. Parker, C. G. Sibley, S. Kumar,
Huelsenbeck J. P., B. Larget, D. Swofford,
Jin Y. G., B. R. Wardlaw, B. F. Glenister, G. V. Kotlyar,
Jones D. T., W. R. Taylor, J. M. Thornton,
Kenrick P., P. R. Crane,
Kevan P. G., W. G. Chaloner, D. B. O. Savile,
Knowlton N., L. A. Weigt, L. A. Solorzano, D. K. Mills, E. Bermingham,
Labandeira C. C.,
Labandeira C. C., B. S. Beall, F. M. Hueber,
Li W.-H., M. Tanimura,
Lo N., G. Tokuda, H. Watanabe, H. Rose, M. Slaytor, K. Maekawa, C. Bandi, H. Noda,
Lyman D. E., F. A. Monteiro, A. A. Escalante, C. Cordon-Rosales, D. M. Wesson, J. P. Dujardin, C. B. Beard,
Marcilla A., M. D. Bargues, J. M. Ramsey, E. Magallon-Gastelum, P. M. Salazar-Schettino, F. Abad-Franch, J. P. Dujardin, C. J. Schofield, S. Mas-Coma,
McAlpine J. F.,
Pellmyr O., J. Leebens-Mack,
Posada D., K. A. Crandall,
Regier J. C., J. W. Shultz,
———.
Roberts J., J. Claoue-Long, P. J. Jones,
Ross A. J., E. A. Jarzembowski,
Russo C. A. M., N. Takezaki, M. Nei,
Sanderson M. J.,
Shcherbakov D. E., E. D. Lukashevich, V. A. Blagoderov,
Shear W. A., P. M. Bonamo, J. D. Grierson, W. D. I. Rolfe, E. L. Smith, R. A. Norton,
Shear W. A., J. Kukalova-Peck,
Shimodaira H., M. Hasegawa,
Shultz J. W., J. C. Regier,
Strimmer K., A. von Haeseler,
Swofford D. L.,
Takezaki N., A. Rzhetsky, M. Nei,
Thompson J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, D. G. Higgins,
Thorne J. L., H. Kishino, I. S. Painter,
Tilyard R. J.,
Tucker R. D., W. S. McKerrow,
Wehr W. C.,
Whalley P.,
Whalley P., E. A. Jarzembowski,
Wheeler W. C., M. Whiting, Q. D. Wheeler, J. M. Carpenter,
Whiting M. F., J. C. Carpenter, Q. D. Wheeler, W. C. Wheeler,