-
PDF
- Split View
-
Views
-
Cite
Cite
John M. Archibald, Christopher E. Lane, Going, Going, Not Quite Gone: Nucleomorphs as a Case Study in Nuclear Genome Reduction, Journal of Heredity, Volume 100, Issue 5, September-October 2009, Pages 582–590, https://doi.org/10.1093/jhered/esp055
Close - Share Icon Share
Abstract
Nucleomorphs are the relic nuclei of algal endosymbionts that became permanent fixtures inside nonphotosynthetic eukaryotic host cells. These unusual organelles exist in only 2 lineages, the cryptophytes, which possess nucleomorphs and plastids (chloroplasts) derived from the uptake of a red algal endosymbiont, and the chlorarachniophytes, which harbor green algal derived nucleomorphs and plastids. Despite having evolved independently of one another, the nucleomorph genomes of cryptophytes and chlorarachniophytes are strikingly similar in size and basic structure. Both are <1 Mbp in size—the smallest nuclear genomes known—and are composed of only 3 chromosomes, each with its own subtelomeric rDNA repeats. Nucleomorph-containing algae thus represent an interesting system in which to study genome and chromosome evolution in eukaryotes. Here, we provide an overview of nucleomorph genome biology and focus on new information gleaned from comparisons of complete nucleomorph genome sequences, both within and between cryptophytes and chlorarachniophytes. Such comparisons provide fascinating insight into the evolution of these highly derived organelles and, more generally, the potential causes and consequences of genome reduction in eukaryotes.
The enormous variation in the size of eukaryotic nuclear genomes is a puzzle that has challenged biologists for more than half a century. Much of the debate has revolved around the lack of correlation between genome size and organismal complexity—the so-called C-value paradox. The genome of the single-celled microbe Amoeba dubia, for example, has been estimated to be >60 000 Mbp in size, more than 200 times larger than the human genome (ca. 2900 Mbp) and more than 200 000 times larger than the 2.9-Mbp genome of the microsporidian parasite Encephalitozooncuniculi (Katinka et al. 2001; Lander et al. 2001; Gregory 2005). This variation contrasts the genome size diversity seen in prokaryotes, which spans a much narrower range, between ∼0.5 and ∼10 Mbp (Moran 2002).
Comparative genomics has provided the solution to the C-value paradox: Eukaryotic genomes vary to a much greater degree in the amount of noncoding DNA they possess than in their total gene content (Lynch 2006). For example, whereas a significant fraction of many plant and animal genomes is comprised of noncoding DNA (e.g., introns, intergenic spacers) and selfish genetic elements (Bennetzen 2002; Gregory 2005), the genomes of unicellular eukaryotes can be much more compact. Prominent examples include the recently sequenced ∼12-Mbp genome of the diplomonad Giardialamblia (Morrison et al. 2007) and the apicomplexans Plasmodium falciparum and Cryptosporidium parvum, with genomes of ∼23 and ∼9 Mbp, respectively (Gardner et al. 2002; Abrahamsen et al. 2004). These genomes possess little in the way of repetitive elements and duplicate loci, packing thousands of genes into a tiny fraction of the space occupied by a typical animal genome. Together with E. cuniculi, these organisms are similar in that they have become parasites of other eukaryotes, a lifestyle known to precipitate significant changes in the cell biology, metabolism, and genetics of both parasite and host (Keeling and Slamovits 2004; Keeling and Slamovits 2005). Small and compact nuclear genomes are not invariably associated with parasitism, however, as evidenced by the filamentous ascomycete fungus Ashbya gossypii, whose genome is a mere 9.2 Mbp in size (Dietrich et al. 2004). Members of the green algal genus Ostreococcus have remarkably small, gene-rich genomes, in the range of 12–13 Mbp (Derelle et al. 2006; Palenik et al. 2007).
As small as the nuclear genomes of eukaryotic parasites can be, they are enormous compared with the “nucleomorph” genomes of cryptophyte and chlorarachniophyte algae. Nucleomorphs are the residual nuclei of photosynthetic eukaryotes that were engulfed by nonphotosynthetic eukaryotic host cells (Gilson and McFadden 2002; Archibald 2007). Although the origin of plastids (chloroplasts) can be traced to an ancient “primary” endosymbiosis between a eukaryote and a photoautotrophic cyanobacterium, plastids have also spread across the tree of eukaryotes by “secondary” endosymbiosis, that is, cellular mergers involving eukaryotic endosymbionts and hosts (McFadden 2001; Bhattacharya et al. 2003; Archibald and Keeling 2005). This process has generated a vast array of environmentally, economically, and medically important lineages, including primary producers such as dinoflagellates, haptophytes, and heterokonts (e.g., diatoms and giant kelp), as well as apicomplexans such as the malaria parasite Plasmodium (Delwiche 1999; McFadden 2001; Archibald and Keeling 2002; Bhattacharya et al. 2003; Palmer 2003; Archibald and Keeling 2005). The number of secondary endosymbioses that have given rise to the known spectrum of secondary plastid-containing organisms is unknown, with estimates ranging from as few as 2 to as many as 7 (see Delwiche and Palmer 1997; Cavalier-Smith 1999; Delwiche 1999; McFadden 2001; Bhattacharya et al. 2003; Palmer 2003; Keeling 2004; Archibald and Keeling 2005; Bodyl 2005 and references therein for review).
At the molecular level, the process of secondary endosymbiosis is poorly understood but clearly involves the elimination of nonessential genes from the endosymbiont nucleus and the transfer of essential genes to the nucleus of the host cell. In most secondary plastid-containing organisms, this elimination/transfer process has run its course, and the nucleus of the engulfed alga has completely disappeared. However, in the chlorarachniophytes and cryptophytes, the nucleus of the endosymbiont—the nucleomorph—persists in a much reduced and simplified form. Arguably among the most genetically complex cells in existence, cryptophytes and chlorarachniophytes possess elaborate internal membrane structures, a sophisticated protein-targeting apparatus and 4 genomes—2 nuclear genomes (host and endosymbiont), a mitochondrial genome, and a plastid genome (Gilson, Maier, and McFadden 1997; Gilson 2001; Gilson and McFadden 2002; Archibald 2007). Genome sequencing has revealed that nucleomorphs harbor the smallest nuclear genomes known: At <1 Mbp in size, these genomes are as small or smaller than the most highly reduced prokaryotic genomes, yet very little is known about the processes underlying their miniaturization. In this article, we provide an overview of the current state of knowledge with respect to the origins and evolution of nucleomorph genomes in cryptophytes and chlorarachniophytes, with an emphasis on the results of recent comparisons of complete nucleomorph genome sequences within and between members of both groups.
Nucleomorph Genome Biology
The term “nucleomorph” was first coined in the late 1970s by Greenwood and colleagues (Greenwood [1974] and Greenwood et al. [1977]) to describe a small membrane-bound body nested between the inner and outer pairs of membranes surrounding the cryptophyte plastid. A similar structure was later observed in the chlorarachniophyte alga Chlorarachnion reptans (Hibberd and Norris 1984), and with the goal of confirming speculation that these entities were the residual nuclei of ingested algal cells (e.g., Ludwig and Gibbs 1987, 1989), the cryptophytes and chlorarachniophytes became the focus of microscopic, cytochemical, and, eventually, molecular investigation (see McFadden 1993; McFadden and Gilson 1995; Kawach et al. 2006; Archibald 2007 for comprehensive review). As predicted, nucleomorphs were shown to contain DNA (Hansmann et al. 1985; Ludwig and Gibbs 1987; Hansmann 1988; Ludwig and Gibbs 1989) and to possess rRNA genes unrelated to those encoded in the host cell nuclear genome (Douglas et al. 1991; Hansmann and Eschbach 1991; Maier et al. 1991; McFadden, Gilson, Hofmann, et al. 1994). Preliminary karyotype analyses using pulsed-field gel electrophoresis revealed that the nucleomorph genomes of cryptophytes and chlorarachniophytes were orders of magnitude smaller than canonical nuclear genomes (Eschbach et al. 1991; Maier et al. 1991; McFadden, Gilson, and Douglas 1994; McFadden, Gilson, Hofmann, et al. 1994; Rensing et al. 1994; Gilson and McFadden 1996), and complete nucleomorph genome sequences from the cryptophyte Guillardia theta (Douglas et al. 2001) and the chlorarachniophyte Bigelowiella natans (Gilson et al. 2006) have convincingly shown that nucleomorphs are indeed remnant eukaryotic nuclei.
The G. theta genome is 551 kbp in size and is comprised of 3 similarly sized chromosomes, each with unusually large telomeric repeats ([{AG}7AAG6A]11; Table 1). The genome encodes 513 genes (465 coding for protein), many of which are predicted to have roles in typical eukaryotic “housekeeping” processes such as transcription, translation, protein folding/degradation, and splicing. The genome also encodes genes for plastid-targeted proteins, albeit a much reduced set (30 in total) compared with the nuclear genomes of free-living algae. Seventeen G. theta genes possess spliceosomal introns (42–52 bp in size) with standard GT-AG intron boundaries.
Characteristics of the nucleomorph genome sequence of the cryptophytes Guillardia theta and Hemiselmis andersenii and the chlorarachniophyte Bigelowiella natans
| Genome characteristics | Guillardia thetaa | Hemiselmisanderseniia | Bigelowiella natansa |
| Evolutionary origin | Red algae | Red algae | Green algae |
| Genome size (bp) | 551 264 | 571 872 | 372 870 |
| Chromosome number/size | 3 (196.2, 180.9 and 174.1 kbp) | 3 (207.5, 184.7, 179.6 kbp) | 3 (140.6, 134.1 and 98.1 kbp) |
| Chromosome structure | Subtelomeric inverted repeats including rDNA genes | Subtelomeric inverted repeats, only 3 with complete rDNAs | Subtelomeric inverted repeats including rDNA genes |
| Telomeric sequence/length | ([AG]7AAG6A)11 | (G[A]17)4–7 | (TCTAGGG)25–45 |
| Genomic A + T content | |||
| Inverted repeats (including rDNA) (%) | ∼55 | ∼60 | ∼50 |
| Single-copy DNA (%) | 65–77 | ∼75 | >65 |
| Number of genes | |||
| Protein genes | 465b | 472 | 293 |
| Non-mRNA (rRNA, tRNA, snRNA, and snoRNA)g | 67 | 53 | 42 |
| Pseudogenes | 1 | 1c | 5 |
| Total | 513 | 525 | 340 |
| Gene densityd | 1.07 kb/gene | 1.09 kb/gene | 1.10 kb/gene |
| Mean intergenic distance (bp) | 70e | 97f | 113e |
| Overlapping genes | 44 (maximum 76 bp overlap) | None | Not determined (maximum 101 bp overlap) |
| Introns and size range | 17 (42–52 bp) | None | 852 (18–21 bp) |
| Plastid genes | 30 | 30 | 17 |
| Genome characteristics | Guillardia thetaa | Hemiselmisanderseniia | Bigelowiella natansa |
| Evolutionary origin | Red algae | Red algae | Green algae |
| Genome size (bp) | 551 264 | 571 872 | 372 870 |
| Chromosome number/size | 3 (196.2, 180.9 and 174.1 kbp) | 3 (207.5, 184.7, 179.6 kbp) | 3 (140.6, 134.1 and 98.1 kbp) |
| Chromosome structure | Subtelomeric inverted repeats including rDNA genes | Subtelomeric inverted repeats, only 3 with complete rDNAs | Subtelomeric inverted repeats including rDNA genes |
| Telomeric sequence/length | ([AG]7AAG6A)11 | (G[A]17)4–7 | (TCTAGGG)25–45 |
| Genomic A + T content | |||
| Inverted repeats (including rDNA) (%) | ∼55 | ∼60 | ∼50 |
| Single-copy DNA (%) | 65–77 | ∼75 | >65 |
| Number of genes | |||
| Protein genes | 465b | 472 | 293 |
| Non-mRNA (rRNA, tRNA, snRNA, and snoRNA)g | 67 | 53 | 42 |
| Pseudogenes | 1 | 1c | 5 |
| Total | 513 | 525 | 340 |
| Gene densityd | 1.07 kb/gene | 1.09 kb/gene | 1.10 kb/gene |
| Mean intergenic distance (bp) | 70e | 97f | 113e |
| Overlapping genes | 44 (maximum 76 bp overlap) | None | Not determined (maximum 101 bp overlap) |
| Introns and size range | 17 (42–52 bp) | None | 852 (18–21 bp) |
| Plastid genes | 30 | 30 | 17 |
Data taken primarily from Douglas et al. (2001), Lane et al. (2007), and Gilson et al. (2006). Numbers may vary slightly, depending on updated analyses and method of calculation.
Williams et al. (2005) identified an rpl30 gene not annotated in the original G. theta nucleomorph sequence.
The Nip7 gene possesses an as yet unidentified alternate start codon.
Calculated as genome size/total gene number.
Numbers taken from Keeling and Slamovits (2005).
Calculated from a set of 164 spacers determined to be homologous between the G. theta and H. andersenii genomes (Lane et al. 2007).
snRNA; small nuclear RNA, snoRNA; small nucleolar RNA.
Characteristics of the nucleomorph genome sequence of the cryptophytes Guillardia theta and Hemiselmis andersenii and the chlorarachniophyte Bigelowiella natans
| Genome characteristics | Guillardia thetaa | Hemiselmisanderseniia | Bigelowiella natansa |
| Evolutionary origin | Red algae | Red algae | Green algae |
| Genome size (bp) | 551 264 | 571 872 | 372 870 |
| Chromosome number/size | 3 (196.2, 180.9 and 174.1 kbp) | 3 (207.5, 184.7, 179.6 kbp) | 3 (140.6, 134.1 and 98.1 kbp) |
| Chromosome structure | Subtelomeric inverted repeats including rDNA genes | Subtelomeric inverted repeats, only 3 with complete rDNAs | Subtelomeric inverted repeats including rDNA genes |
| Telomeric sequence/length | ([AG]7AAG6A)11 | (G[A]17)4–7 | (TCTAGGG)25–45 |
| Genomic A + T content | |||
| Inverted repeats (including rDNA) (%) | ∼55 | ∼60 | ∼50 |
| Single-copy DNA (%) | 65–77 | ∼75 | >65 |
| Number of genes | |||
| Protein genes | 465b | 472 | 293 |
| Non-mRNA (rRNA, tRNA, snRNA, and snoRNA)g | 67 | 53 | 42 |
| Pseudogenes | 1 | 1c | 5 |
| Total | 513 | 525 | 340 |
| Gene densityd | 1.07 kb/gene | 1.09 kb/gene | 1.10 kb/gene |
| Mean intergenic distance (bp) | 70e | 97f | 113e |
| Overlapping genes | 44 (maximum 76 bp overlap) | None | Not determined (maximum 101 bp overlap) |
| Introns and size range | 17 (42–52 bp) | None | 852 (18–21 bp) |
| Plastid genes | 30 | 30 | 17 |
| Genome characteristics | Guillardia thetaa | Hemiselmisanderseniia | Bigelowiella natansa |
| Evolutionary origin | Red algae | Red algae | Green algae |
| Genome size (bp) | 551 264 | 571 872 | 372 870 |
| Chromosome number/size | 3 (196.2, 180.9 and 174.1 kbp) | 3 (207.5, 184.7, 179.6 kbp) | 3 (140.6, 134.1 and 98.1 kbp) |
| Chromosome structure | Subtelomeric inverted repeats including rDNA genes | Subtelomeric inverted repeats, only 3 with complete rDNAs | Subtelomeric inverted repeats including rDNA genes |
| Telomeric sequence/length | ([AG]7AAG6A)11 | (G[A]17)4–7 | (TCTAGGG)25–45 |
| Genomic A + T content | |||
| Inverted repeats (including rDNA) (%) | ∼55 | ∼60 | ∼50 |
| Single-copy DNA (%) | 65–77 | ∼75 | >65 |
| Number of genes | |||
| Protein genes | 465b | 472 | 293 |
| Non-mRNA (rRNA, tRNA, snRNA, and snoRNA)g | 67 | 53 | 42 |
| Pseudogenes | 1 | 1c | 5 |
| Total | 513 | 525 | 340 |
| Gene densityd | 1.07 kb/gene | 1.09 kb/gene | 1.10 kb/gene |
| Mean intergenic distance (bp) | 70e | 97f | 113e |
| Overlapping genes | 44 (maximum 76 bp overlap) | None | Not determined (maximum 101 bp overlap) |
| Introns and size range | 17 (42–52 bp) | None | 852 (18–21 bp) |
| Plastid genes | 30 | 30 | 17 |
Data taken primarily from Douglas et al. (2001), Lane et al. (2007), and Gilson et al. (2006). Numbers may vary slightly, depending on updated analyses and method of calculation.
Williams et al. (2005) identified an rpl30 gene not annotated in the original G. theta nucleomorph sequence.
The Nip7 gene possesses an as yet unidentified alternate start codon.
Calculated as genome size/total gene number.
Numbers taken from Keeling and Slamovits (2005).
Calculated from a set of 164 spacers determined to be homologous between the G. theta and H. andersenii genomes (Lane et al. 2007).
snRNA; small nuclear RNA, snoRNA; small nucleolar RNA.
The B. natans nucleomorph genome is also made up of 3 chromosomes and at 373 kbp is even smaller than the G. theta genome and encodes fewer genes (340 in total, 293 protein coding). However, the functional distribution of these genes is, for the most part, similar to the G. theta nucleomorph (Gilson et al. 2006). One notable exception is that the B. natans genome is “enriched” for genes involved in RNA metabolism relative to G. theta. This observation is potentially significant given that the B. natans genome contains many more introns than G. theta (Gilson et al. 2006; see below). The B. natans chromosomes are capped with classic eukaryotic telomeres comprised of a (TCTAGGG)25–45 repeat, similar to that seen in plants, algae, and many other eukaryotes.
Although the G. theta and B. natans nucleomorph genomes possess a variety of features seen in “typical” nuclear genomes, they are also quite unusual, exhibiting many of the characteristics of genomes undergoing reductive evolution (Moran 2002; Keeling and Slamovits 2005). For example, the G. theta and B. natans genomes have significantly elevated A + T contents (ca. 75%) and, as a result, encode numerous proteins with biased amino acid compositions (see below). Many nucleomorph genes/proteins are also very divergent in sequence relative to their homologues in other organisms, making them difficult to accurately place in phylogenetic analyses (Ishida et al. 1999; Keeling et al. 1999; Archibald et al. 2001; Brinkmann et al. 2005). Gene density in the 2 genomes is extremely high (1.07 kbp/gene for G. theta and 1.10 kbp/gene for B. natans, calculated as genome size/total gene number; Table 1), a feature that has a significant impact on the process of transcription. Gilson and McFadden (1996) first demonstrated cotranscription of 2 protein-coding genes in the B. natans nucleomorph and a more recent study by Williams et al. (2005) showed that a high frequency of nucleomorph mRNAs in both B. natans and G. theta encode more than one gene. These observations suggest that as nucleomorph genomes became more and more compact, cis elements regulating the initiation and termination of transcription were forced to move within or beyond adjacent genes. Multigene transcripts are also found in the reduced genome of the microsporidian E. cuniculi (Williams et al. 2005).
Convergent Evolution on a Genome Scale
Molecular phylogenetic analyses have shed light on the evolutionary origins of the host and endosymbiont components of cryptophytes and chlorarachniophytes and in doing so provide an important reference point for interpreting the evolution of nucleomorph genomes in the 2 groups. The cryptophyte and chlorarachniophyte host cells belong to 2 very distantly related superassemblages of eukaryotes, the Chromalveolata and Rhizaria, respectively (Keeling et al. 2005). Similarly, their nucleomorphs and secondary plastids are derived from 2 different lineages of primary plastid-containing organisms: red algae in the case of cryptophytes (Douglas et al. 1991; Van der Auwera et al. 1998; Douglas and Penny 1999; Archibald et al. 2001) and green algae in the chlorarachniophytes (McFadden, Gilson, Hofmann, et al. 1994; McFadden et al. 1995; Ishida et al. 1997, 1999; Archibald et al. 2003; Rogers et al. 2007). Attempts to accurately pinpoint the closest modern day relatives of the cryptophyte and chlorarachniophyte endosymbionts within red and green algae have been hampered by the accelerated rates of sequence evolution seen in many nucleomorph genes (Ishida et al. 1999; Keeling et al. 1999; Archibald et al. 2001; Brinkmann et al. 2005), as well as the limited taxon sampling of red and green algae for genes other than rDNA. Regardless, the cryptophyte and chlorarachniophyte nucleomorphs are clearly of independent origin—any similarities in their genomes that are not features of green and red algal nuclear genomes are very likely the result of convergent evolution.
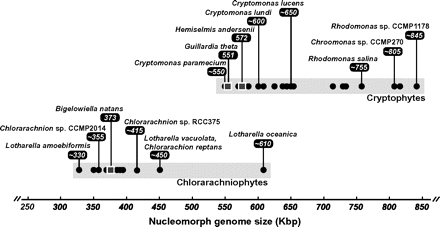
One of the most obvious similarities between the G. theta and B. natans nucleomorph genomes is the presence of 3 chromosomes. Karyotype surveys of diverse species within both groups indicate that this 3-chromosome architecture is not limited to these 2 organisms but appears to be a universal feature of cryptophyte and chlorarachniophyte nucleomorphs (Rensing et al. 1994; Gilson and McFadden 1999; Lane and Archibald 2006, 2008; Lane et al. 2006; Silver et al. 2007; Phipps et al. 2008). It would appear that early in the evolution of cryptophytes and chlorarachniophytes their nucleomorph genomes converged on the same basic karyotype from different algal endosymbionts that presumably each harbored dozens of nuclear chromosomes (if the karyotype diversity seen in modern day red and green algae is any indication). What—if anything—is the biological significance of this observation? Douglas et al. (2001) and Cavalier-Smith (2002) have suggested that it might simply be a function of striking a balance between having chromosomes that are small enough to fit inside the nucleomorph (the degree of chromatin condensation in nucleomorphs is thought to be only to the level of 30-nm fibers) and yet large enough to be stably inherited. The sizes of the B. natans and G. theta genomes are roughly similar, and thus, their genomes are divided into 3 similarly sized chromosomes. If one considers the known breadth of species diversity, nucleomorph genome sizes within chlorarachniophytes range from ∼330 to ∼610 kbp (Figure 1) with individual chromosomes between ∼95 and ∼210 kbp, whereas cryptophyte genomes range between ∼550 and ∼845 kbp, with chromosomes between ∼160 and ∼300 kbp (Rensing et al. 1994; Gilson and McFadden 1999; Lane and Archibald 2006, 2008; Lane et al. 2006; Silver et al. 2007; Phipps et al. 2008). As proposed by Gilson and McFadden (2002), it will be interesting to determine whether a relationship exists between nucleomorph DNA content, nucleomorph volume, and the volume of residual endosymbiont cytosol in the same way that in cryptophytes host nuclear DNA content appears to scale with cell size (Beaton and Cavalier-Smith 1999; Cavalier-Smith and Beaton 1999).
Summary of known or estimated nucleomorph genome sizes in cryptophyte and chlorarachniophyte algae. Genome sizes determined by complete genome sequencing (Douglas et al. 2001; Gilson et al. 2006; Lane et al. 2007) are represented as squares, whereas size estimates obtained by pulsed-field gel electrophoresis are indicated as circles (Eschbach et al. 1991; Rensing et al. 1994; Gilson and McFadden 1999; Lane et al. 2006; Silver et al. 2007; Lane and Archibald 2008; Phipps et al. 2008).
Another curious similarity in the structure of the B. natans and G. theta nucleomorph genomes is the presence of rDNA repeats next to the telomeres. In G. theta, each chromosome end has a 5S rDNA locus on one strand and an 18S-5.8S–28S rDNA operon on the other, the latter transcribed toward the telomere (Douglas et al. 2001). In B. natans, a 5S gene is absent, and the 18S-5.8S–28S rDNA repeat is in the opposite orientation (Gilson et al. 2006). Again, this arrangement evolved independently in the chlorarachniophyte and cryptophyte nucleomorphs, and with the exception of a single cryptophyte genus (see below), appears to be a universal feature of nucleomorphs in both lineages. Interestingly, subtelomeric repeats are also seen in the reduced genomes of 2 unrelated eukaryotic parasites mentioned previously, E. cuniculi (Katinka et al. 2001) and G. lamblia (Upcroft et al. 2005; Morrison et al. 2007). Although subtelomeric regions are typically highly recombinagenic and thus an obvious home for a gene family known to evolve in a concerted fashion, they have also been shown to be areas of repressed gene activity in a variety of organisms (the so-called “telomere position effect”; Ottaviani et al. 2007). Clearly, there is much to be learned about the relationship between chromosome position and gene expression in nucleomorphs and other reduced nuclear genomes.
A Third Nucleomorph Genome
Toward the goal of better understanding the process of genome evolution in cryptophyte and chlorarachniophyte nucleomorphs, we recently sequenced the 572-kbp nucleomorph genome of another cryptophyte alga, Hemiselmis andersenii (Lane et al. 2007). This sequence has made it possible to obtain a first glimpse into the similarities and differences between distantly related nucleomorphs that are nevertheless the product of the same secondary endosymbiosis. The genus Hemiselmis first caught our attention when initial karyotypic surveys revealed the presence of fragmented rDNA repeats on the second nucleomorph chromosome of H. andersenii and its closest relatives (Lane and Archibald 2006), and the complete genome has shown that only the 5S rDNA locus remains on both ends of chromosome II and one end of chromosome III (Lane et al. 2007). These interchromosomal variations in subtelomeric repeat structure are presumably the result of ongoing recombination between the rDNA loci at each chromosome end. With respect to overall genome structure, a high degree of gene order conservation is observed between the H. andersenii and G. theta genomes, with numerous stretches of synteny >20 kbp in size. This was unexpected given that the 2 organisms are not closely related (Lane and Archibald 2006; Lane et al. 2007; Lane and Archibald 2008). We postulate that the retention of large syntenic blocks over significant evolutionary time scales is the result of the high gene density seen in the cryptophyte nucleomorph, which presumably diminishes the frequency with which nonhomologous recombination can scramble the genome without disrupting coding sequences (Lane et al. 2007). This hypothesis has also been used to explain the high degree of synteny seen in the reduced genomes of microsporidian parasites (Slamovits et al. 2004).
Perhaps, the most surprising feature of the H. andersenii nucleomorph genome is that it has lost all spliceosomal introns (Lane et al. 2007). Even the most highly reduced nuclear genomes retain at least a few introns (e.g., Katinka et al. 2001; Morrison et al. 2007), and, as noted above, the G. theta nucleomorph genome possesses 17 small introns and encodes a U6 small nuclear RNA and 13 protein factors with known or predicted roles in splicing (Douglas et al. 2001). These include 2 subunits of the U5 snRNP complex and the highly conserved spliceosome-specific protein prp8. Homologues of 15 of the 17 intron-containing genes in G. theta are found in the H. andersenii genome, but none have introns and no introns are present in any of the other H. andersenii open reading frames (ORFs). Significantly, whereas the G. theta and H. andersenii genomes both encode a variety of genes with predicted functions in ribosome biogenesis (e.g., cbf5, nop56), H. andersenii is missing all 5 small nuclear RNAs, prp8, U5 snRNP proteins, and several other important spliceosome components (Lane et al. 2007). The H. andersenii nucleomorph genome thus represents the first described instance of complete intron loss in a nuclear genome.
Elimination of introns would seem to be an obvious consequence of genome size reduction in nucleomorphs. However, it is worth noting that this has not taken place in the chlorarachniophyte B. natans, whose nucleomorph genome is inundated with introns—852 in total, with an average of ∼3.1 introns per gene. Instead, the B. natans introns have shrunk to a mere 18–21 bp in size, the smallest known (Gilson and McFadden 1996; Gilson et al. 2006). In terms of overall abundance, it is also necessary to consider that intron densities in cryptophyte and chlorarachniophyte nucleomorph genomes may be correlated with the intron densities of red and green algal nuclear genomes, which are believed to be intron-poor and intron-rich, respectively (Gilson et al. 2006). Overall, the pattern and process of intron evolution in cryptophyte and chlorarachniophyte nucleomorph genomes is very poorly understood, and it will be important to determine the presence/absence, size, and abundance of nucleomorph introns in the diverse species shown in Figure 1, in particular, those with genomes that are significantly larger and smaller than the 3 “reference” genomes that have been sequenced.
Nucleomorph-Specific Genes: Where Did They Come From and What Are They Doing?
Another interesting observation in the comparison of the H. andersenii and G. theta nucleomorph genomes is the unexpectedly small number of identifiable genes the 2 genomes share. Out of 472 protein-coding genes in the H. andersenii genome (Table 1), only 314 have an obvious counterpart in G. theta on the basis of sequence similarity alone (Lane et al. 2007). Furthermore, 140 of the remaining predicted ORFs are apparently H. andersenii specific, having no counterpart in any other sequenced genome, including those of red algae, green algae, and plants. However, 30 of these genes with no obvious sequence similarity are nested within syntenic blocks in the same position as an unidentified ORF of similar size in G. theta. Pairwise comparisons reveal that the majority of these ORF pairs encode proteins with a similar number of membrane-spanning helices (when present) and similar isoelectric points. We thus proposed that these “syntenic” ORFs, many of which encode predicted proteins >300 amino acids in length, are derived from a common ancestral locus but have diverged from one another beyond the point of recognition (Lane et al. 2007). Why do these ORFs persist? What could be the function of their gene products if their sequences are so unconstrained?
As noted above, nucleomorph-encoded proteins are often biased in amino acid sequence and the G. theta– and H. andersenii–specific ORFs described above encode proteins that are truly striking in this regard. In some cases, >50% of their primary amino acid sequence is comprised of so-called “FINKY” residues (phenylalanine, isoleucine, asparagine, lysine, and tyrosine), a set of amino acids encoded by A + T–rich codons (Singer and Hickey 2000). Such a combination of basic, hydrophobic, and polar residues raises the possibility that these proteins interact with membranes, and indeed, a significant fraction of the G. theta– and H. andersenii–specific proteins possess 2 or more predicted transmembrane spanning domains (Lane et al. 2007). Experiments are currently underway to explore this possibility further.
The Smaller the Genome, the Smaller the Genes
In addition to providing a first glimpse at differences in the structure and coding capacity of 2 evolutionarily distant cryptophyte nucleomorph genomes, comparison of the G. theta and H. andersenii sequences has yielded fascinating insight into the consequences of genome reduction and compaction in nucleomorphs. Most notably, cryptophyte nucleomorph genes/proteins are significantly smaller than their homologues in canonical nuclear genomes (Lane et al. 2007). Specifically, we showed that 92% of a set of 198 proteins encoded in both the G. theta and H. andersenii nucleomorphs were shorter than their counterparts in the red alga Cyanidioschyzon merolae and the land plant Arabidopsis thaliana. At the protein level, this shortening manifests itself as truncations at the amino and carboxyl termini, short internal deletions as small as a few amino acids, as well as the removal of entire domains, in some cases with important functional implications such as the removal of the evolutionarily conserved C-terminal domain on the largest subunit of RNA polymerase II (RPB1; Lane et al. 2007). Surprisingly, protein sizes within nucleomorphs also differ from one another: Eighty-one percent of 290 orthologs were smaller in the 551-kbp genome of G. theta than in the 572-kbp H. andersenii nucleomorph genome (Lane et al. 2007). Statistical analysis of these pairwise protein comparisons revealed that the differences were significant using a variety of methods.
A link between genome compaction and gene/protein size has also been reported in the 2.9-Mbp genome of the intracellular parasite E. cuniculi (Katinka et al. 2001). Eighty-five percent of E. cuniculi genes are smaller than their orthologs in the larger genome of the yeast Saccharomycescerevisiae. Katinka et al. (2001) proposed that this difference was due to the fact that E. cuniculi has a smaller proteome, a simplified interaction network, and, consequently, fewer interaction domains in its proteins. In the case of cryptophyte nucleomorphs, given that the coding capacity of the H. andersenii and G. theta genomes is very similar (Table 1) and the 2 organisms presumably import roughly the same number of proteins from the host cytosol into their respective endosymbiont compartments, the complexity of their proteomes should also be similar. However, we found a statistically significant size difference between the genes in the G. theta and H. andersenii nucleomorph genomes, which are 551 and 572 kbp, respectively, and have significantly different mean intergenic distances (Lane et al. 2007, Table 1). This argues against the idea that it is a simplified interactome that precipitates a reduction in protein size. We speculate that it is a deletion bias that has resulted in smaller genes in the nucleomorph genome of G. theta (albeit with some that have retained introns), as well as its smaller and more compact genome. It will be interesting to see whether a correlation between genome size and gene size holds when a larger set of cryptophyte species are considered and whether the same pattern is observed when the chlorarachniophyte nucleomorph genomes are compared with those of their green algal relatives. If our preliminary comparisons of G. theta and H. andersenii are any indication, the process of genome reduction and compaction appears to have had—and continues to have—a profound impact on nucleomorph genomes and the genes they encode.
Going, Going, Not Quite Gone
No review of nucleomorphs and their unusual genomes would be complete without considering the question of why they exist. Given that all other secondary plastid-containing algae have, through the combined effects of gene loss and intracellular gene transfer, completely eliminated their endosymbiont nuclear genomes, why have the nucleomorphs of cryptophytes and chlorarachniophytes been retained? There are 2 obvious possibilities. First, it is possible that the process of gene loss and nucleomorph-to-host-nucleus gene transfer has yet to run its course, and, given enough time, the cryptophyte and chlorarachniophyte nucleomorphs will eventually disappear. Alternatively, it is possible that nucleomorph genomes have approached (or are approaching) an endpoint beyond which further genome reduction is extremely difficult or impossible. This latter scenario is analogous to the situation in plastids and mitochondria, where, with the exception of mitosomes and most hydrogenosomes (Embley et al. 2003), genomes are always present, if only to encode a handful of genes. Ideas about why mitochondria and plastids retain genomes abound (e.g., Allen 1993, 2003; Martin et al. 1998; Race et al. 1999), with perhaps the most robust hypothesis centered on the advantages of regulating the expression of important redox proteins by redox potential in the compartment in which they function (Allen 1993, 2003). However, this idea cannot be invoked to explain the persistence of nucleomorph genomes because redox reactions do not take place in the nucleomorph.
Does the modest set of genes that remain in the nucleomorph genomes of cryptophytes and chlorarachniophytes provide any clues as to why they persist? Gilson et al. (2006) showed that although the general functional distribution of B. natans and G. theta nucleomorph genes is similar, gene-for-gene the overlap between the 2 genomes is seemingly random. Most notably, only 2 of the 17 genes for plastid proteins in the B. natans genome are among the 30 plastid-protein genes in G. theta. Gilson et al. (2006) thus conclude that nucleomorphs are “evolutionary intermediates” and will eventually disappear. Nevertheless, it is possible that the cryptophyte and chlorarachniophyte nucleomorph genomes have been retained for different reasons, and in order to tackle this question it will be necessary to understand the diversity of genes in the nucleomorph genomes of closely and distantly related species within both groups. In the case of cryptophytes, whereas it is interesting that the H. andersenii and G. theta nucleomorph genomes share the exact same suite of 30 plastid protein genes (Lane et al. 2007), it is at present impossible to distinguish between shared gene content due to common ancestry versus the existence of functional/mechanistic barriers to successful nucleomorph-to-host-nucleus gene transfers. Together with multiple complete genome sequences, a fully resolved phylogeny of cryptophytes and chlorarachniophytes should make it possible to assess the degree of randomness in the pattern of nucleomorph gene loss and, by extension, predict the ultimate fate of nucleomorphs.
Future Directions
As is abundantly clear from the above discussion, nucleomorph genome research is still very much in a “data collection” phase. Indeed, with only 3 genomes in hand, it is likely that some of the most interesting and fundamental questions about the evolution of nucleomorph genomes have yet to even be formulated. This will soon change as current and future genome sequencing projects in our laboratory target the biggest and smallest known nucleomorph genomes within cryptophytes and chlorarachniophytes (Figure 1) as well as multiple genomes from closely related species within both groups. Among other things, we hope to use these sequences to 1) elucidate the extent of gene content variation in the nucleomorph genomes of diverse members within both groups, 2) determine the tempo and mode of nucleomorph spliceosomal intron loss in the cryptophyte genus Hemiselmis and its closest relatives, 3) further explore the relationship between nucleomorph genome size/density and gene/protein size, 4) address the question of whether a deletion bias is at the heart of nucleomorph genome shrinkage, and 5) better understand the process of nucleomorph-to-host-nucleus gene transfer. In addition, and related to this last point, the extremely limited coding capacity of cryptophyte and chlorarachniophyte nucleomorph genomes demands that the vast majority of proteins required for proper function of the plastid and nucleomorph are nucleus encoded and imported posttranslationally. Whereas expressed sequence tag surveys have provided a preliminary glimpse at the compliment of endosymbiont-derived, nucleus-encoded, plastid-targeted proteins in chlorarachniophytes and cryptophytes (e.g., Archibald et al. 2003; Patron et al. 2006), for the most part very little is known about the nuclear genomes of these organisms. Fortunately, the Joint Genome Institute's Community Sequencing Program is sequencing the nuclear genomes of G. theta and B. natans (http://www.jgi.doe.gov/sequencing/why/50026.html). These genome sequences should make it possible to assemble the complete “parts list” for the molecular and biochemical processes taking place in the plastid, endosymbiont cytosol, and the nucleomorph itself. Combined with further study of the “shrunken” nucleomorph-encoded proteins, such analyses should provide fundamental insight into the minimal functional units necessary for core eukaryotic cellular processes.
We thank S. Bearne and J. Rainey for stimulating discussion on the possible functions of compositionally biased nucleomorph proteins. J.M.A. is associate director and scholar of the Canadian Institute for Advanced Research Program in Integrated Microbial Biodiversity.
References
Author notes
Corresponding Editor: Michael Lynch