Abstract
Tissue engineering is a branch of regenerative medicine that harnesses biomaterial and stem cell research to utilise the body's natural healing responses to regenerate tissue and organs. There remain many unanswered questions in tissue engineering, with optimal biomaterial designs still to be developed and a lack of adequate stem cell knowledge limiting successful application. Advances in artificial intelligence (AI), and deep learning specifically, offer the potential to improve both scientific understanding and clinical outcomes in regenerative medicine. With enhanced perception of how to integrate artificial intelligence into current research and clinical practice, AI offers an invaluable tool to improve patient outcome.
Export citation and abstract BibTeX RIS

Original content from this work may be used under the terms of the Creative Commons Attribution 4.0 licence. Any further distribution of this work must maintain attribution to the author(s) and the title of the work, journal citation and DOI.
1. Introduction
Regenerative medicine offers unparalleled opportunities to revolutionise UK healthcare in the 21st century, driven from the clinical context of an increasingly aged population. While medical advances have led to a welcome increase in life expectancy, this progress presents its own new challenges: increases in age-related diseases, and associated reductions in quality of life, at substantial socio-economic cost [1, 2]. Within the musculoskeletal area, for example, osteoarthritis affects up to 1 in 5 people and causes immense long-term pain and disability, and the risk of suffering as a consequence of osteoarthritis is on the rise across the world [3, 4].
Joint replacement offers an effective treatment for this multifactorial condition, yet outcomes can be poor and prosthesis lifespans are typically limited to fewer than 15 years due to fatigue and wear, even with limitations to sports activity and heavy lifting in recipients of knee joint replacements [5]. Research is currently focused on utilising the body's natural healing responses to regenerate bone through the use of bioactive materials—a dramatic shift from the implantation of bio-inert materials of the past [6]. Biofabricated bone capable of being scaled up for clinical use remains a research goal, due in part to complexities in resolving issues such as vascularisation and tissue necrosis, despite many decades of work on growing bone tissue for organ transplantation [7]. However, recent technological advances, specifically artificial intelligence (AI), offer new approaches and personalised patient-specific approaches for tissue engineering [8].
This review will discuss the current process of tissue regeneration (figure 1), and how integration with AI can enhance the field of regenerative medicine, how it is currently being applied, and the potential for tissue engineering-AI integration. AI is explained in later sections, with a focus on deep learning and application by non-computer scientists into their current research, reducing time and resources required for experimentation, without, critically, sacrificing validation or circumventing scientific discovery.
Figure 1. Bone repair using tissue engineering and biomaterial paradigm. Cells are isolated from a tissue biopsy and then cultured. Biomaterials are used with multiple properties for tissue generation and growth, including biochemical and biophysical cell-instructive properties, to aid in both proliferation and required differentiation. Generated tissue is implanted into the trauma site to aid regeneration.
Download figure:
Standard image High-resolution imageThe purpose of this review is to summarise AI without complexity, so as to be accessible to a wide audience of tissue engineers, materials scientists, or biologists less familiar with AI, machine learning, or modern computational statistics methods. This is significant because AI is a modern tool that has the potential to be used by this audience without needing a formal computational background.
2. Tissue regeneration
The human body has the innate ability to self-repair and regenerate organs and tissue, including bone. However, the reparative response can be limited and self-healing can fail due to a variety of reasons, such as infection, compromised blood supply, scale of defect as a consequence of significant trauma or disease. Osteoporosis and other musculoskeletal age-related conditions, exacerbated with an increasingly aging population, have resulted in bone becoming the most transplanted tissue after blood [7, 9]. The application of bone grafts and biomaterials to promote osteoinduction, osteoconduction, and osteogenesis to induce bone healing has resulted in the need to assess engineered biomaterials. These materials typically combine the strength of bone (capable of withstanding substantial load transfers and mechanical forces) together with properties of biocompatibility as well as the promotion of bone regeneration in combination with angiogenesis, and biodegradability to facilitate new bone growth to, eventually, replace the biomaterial.
Functioning bone tissue is extremely important to human health as, alongside helping protect various organs in the body, it produces both red and white blood cells, stores minerals, and enables mobility and support for the body [10]. This multi-functional tissue therefore contains complex internal and external structures, and is made up of multiple types of bone cells, such as osteoblasts, osteocytes (important for formation and mineralisation), and osteoclasts (important for resorption). While the mineralised tissue of cortical bone and cancellous bone makes up bone tissue, there are multiple other tissue types found in bones, such as bone marrow, cartilage, nerves, and blood vessels. Bone is actively constructed and remodelled by the previously mentioned bone cells throughout life, with the flexible bone matrix hardened by the binding of inorganic mineral salt, calcium phosphate, in a chemical arrangement known as calcium hydroxyapatite. With mechanical, synthetic, and metabolic functions, bone regeneration is a challenging area in tissue engineering, and there are multiple reviews to cover important advances in the field [11–17].
Tissue engineering is defined by the National Institute of Biomedical Imaging and Bioengineering (NIBIB) as 'an interdisciplinary and multidisciplinary field that aims at the development of biological substitutes that restore, maintain, or improve tissue function' [18]. Utilisation of biomaterials alongside natural in vivo bone repair methods produce an enhanced method for bone regeneration, via harnessing the developmental potential of stem cells, which would overcome self-healing limitations. Reliance on ex vivo culture can limit the pain and donor site morbidity associated with autografts (a common form of bone grafting using a patient's own tissue) but stem cell expansion is often a stumbling block for advancing tissue engineering into clinical practices (figure 2) [19]. This is a key and multifaceted issue within tissue engineering, as culturing a stem cell population for successful bone repair requires conditions that allow for stem cell proliferation without differentiation, and also identification and isolation of the target cells for in vivo bone regeneration. Often called mesenchymal stem cells, skeletal stem cells reside in blood vessels in bone marrow, and can therefore be found and isolated from human bone marrow stromal cells (HBMSCs), with proven differentiation potential and regenerative capabilities: HBMSCs can be expanded and transplanted in vivo to form heterotopic bone marrow organs [20–23]. However, not all HBMSC colonies are created equal. Significant heterogeneity exists within skeletal stem-progenitor cell fraction, with studies indicating variation in the differentiation potential along the osteo-chondro-adipogenic lineages, characteristic of the tri-potent primitive mesenchymal stem cell population, emphasising that stem-early osteoprogenitor cell population differences can lead to colonies with varied differentiation capacities, where there is variation in cell stem-progenitor capacity [20].
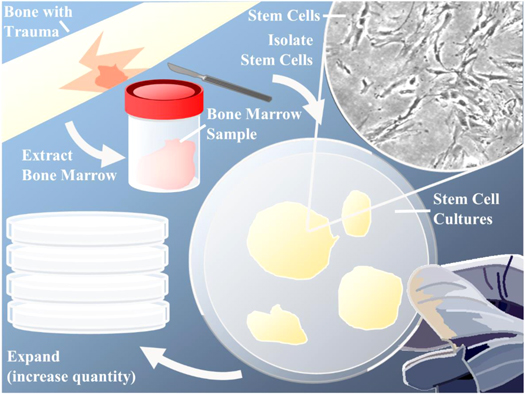
Figure 2. Acquiring stem cells for tissue engineering through donor site sampling, stem cell isolation and expansion. A sample of bone marrow is extracted from the patient. The desired cells are isolated using select stem cell markers, and then cultured into stem cell colonies. These colonies can be expanded to increase the total amount of stem cells available for seeding onto scaffolds, differentiation into desired cell lineages, or transplanting directly into the patient to aid in tissue regeneration.
Download figure:
Standard image High-resolution imageIsolation of the skeletal stem cell has been hampered, to date, by the lack of specific markers and the need to rely on current proposed surface markers for ex vivo populations with proliferative and multi-lineage potential, which may not, and typically do not, adequately reflect bone regeneration in vivo [19]. A good example of this in vivo/ex vivo mismatch is the use of Stro-1 as a stem cell marker. While a plethora of studies use the Stro-1 antibody for the enrichment of the skeletal stem cell, only a few studies have examined and demonstrated Stro-1 reactivity in the tissues from which the cells originated, such as in sub-endothelial positions near sinusoids (a type of blood vessel within bone marrow) [23, 24]. That does not detract from evidence demonstrating Stro-1 selected populations can result in fibroblasts, adipoblasts, myoblasts, and osteoblasts, highlighting that further research into stem cell markers is necessary [25].
Even after a population of stem cells has been isolated, or at least a population enriched for the desired mix of stem-progenitor cells for bone (re)generation, there is the need to expand the cell population. Unfortunately, HBMSC populations lose their proliferative and, more importantly, multipotential qualities upon passage. The mechanisms behind this, such as a relative loss of telomerase activity of isolated HBMSCs ex vivo, when compared to in vivo, can be difficult to overcome, as well as variations in culture protocol and techniques [19]. Given the challenges that reside in stem-progenitor cell expansion, tissue engineering has simultaneously branched into in situ approaches for tissue regeneration (figure 3) [16]. Where traditional tissue engineering utilises biomaterials such as 3D scaffolds pre-loaded with stem cells and exposed to in-vitro conditioning, in-situ techniques utilise biomaterials that are implanted directly into the body without the need for any pre-seeding of cells. Both methods require engineered bio-responsive materials, which employ biophysical and/or biochemical cues to direct cells into bone regeneration, aiding the body in self-healing. In situ biomaterials have different regulatory hurdles and barriers due to the absence of seeded cells, affording the potential for more rapid clinical translation. Nevertheless, the safety and performance of biomaterials within the in vivo environment can be challenging to predict and monitor, with issues from excess inflammation to rejection, and even a potential cancer risk with growing evidence associating textured (but not smooth) silicon-based implants with non-Hodgkins lymphoma, leading to UK and USA regulators recalling textured implants specifically due to the risk of breast implant-associated anaplastic large cell lymphoma [26–28]. A lack of complete understanding of how textures and other biophysical cues interact in complex multi-cellular environments, in both short and long term, is as important a consideration to biomaterial engineering as toxicity and therapeutic effects of biochemical cues, and eventual degradation.
Figure 3. Ex vivo and in situ tissue engineering both use the body's own regenerative ability, boosted by tissue-engineered materials. For ex vivo, cells are extracted and then cultured on biomaterial scaffolds and in bioreactor environments. The modified tissue is then implanted into the body. For in situ, there is no extraction necessary.
Download figure:
Standard image High-resolution image3. Cell-instructive biomaterials
Tissue regeneration is one of the most complex multi-cellular environments within the body, with highly evolved and tightly controlled processes of migration, proliferation, differentiation and coordination of multiple cell types, all of which change depending on the specific location and variety of trauma. Understanding how cells function is a key concept within tissue engineering [29]. There has therefore been a focus on developing understanding of natural cues and the immune response of the body, which has resulted in tissue-derived extracellular matrix (ECM) scaffolds capable of modifying the default response to injury and manipulating the immune system to stimulate tissue regeneration (figure 4) [30, 31]. Development of scalable, reproducible, and inexpensive biomaterials, which are capable of similar alteration/enhancement of the immune system necessitates an understanding of biomaterial behaviour and interaction within an immune environment. In addition, improved knowledge of biomaterial characteristics across macro-, micro -and nano-scales are necessary to understand the cues and factors involved in regeneration of tissue (e.g. bone), facilitate future development of biomaterials, and validate biomaterial optimisation, while also excluding the possibility of long-term mal-effects associated with uncontrolled bioactive implants, such as cancer [26–28, 32].
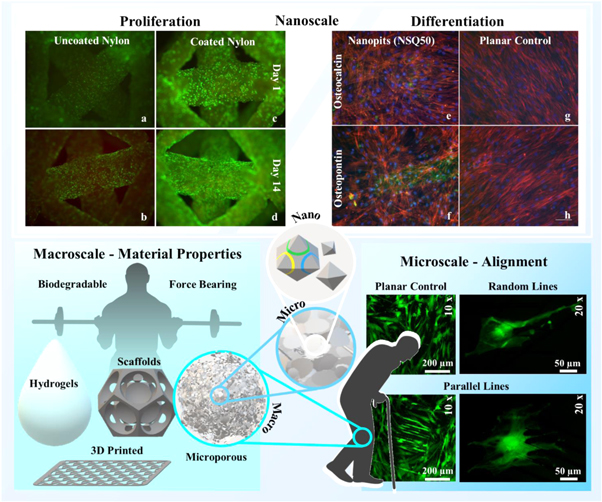
Figure 4. Cell-instructive biomaterials with a variety of characteristics at different scales of interest. At a macroscale, different types and production range from hydrogels to solid scaffolds and regimented 3D printed to more randomised microporous structures, all of which need to be degradable and capable of withstanding large forces. This is relevant for tissue regeneration, as covered in the section Tissue Regeneration. Features relevant at the microscale can include the alignment of cells, both at cellular and multi-cellular level. Alignment behaviour is important in development of angiogenesis, biological neural networks, and the growth plate (chondrocyte columns) in the growth of long bones. At the nanoscale, both chemical (a)–(d) and physical (e)–(h) characteristics overlap to promote a large range of responses, from proliferation (a)–(d) (green are live cells and red are dead or dying cells, with noticeable difference in levels of dead cells between (b) and (d)) to differentiation (e)–(h) (green is expression of osteopontin or osteocalcein, seen in (e)–(f) but not (g)–(h), while red and blue show the cell body and nucleus respectively) of HBMSCs. (e-h) reproduced with permission from [33].
Download figure:
Standard image High-resolution imageCharacteristics of biomaterials are a combination of both the biophysical (including biomechanical) and biochemical features, resulting in a wide range of biomaterial properties, many of which have been detailed by Gaharwar et al, including improved proliferation and targeted differentiation [16]. Through manipulation of these characteristics via a wide range of manufacturing techniques, from additive manufacturing processes such as 3D printing to ablation processes such as 2D laser femtosecond laser machining, the behaviour from a single cell to a whole colony can be modulated, resulting in cell-response altering (cell-instructive) biomaterials [34, 35]. Biophysical cues can result from a wide range of features, including the stiffness, structure and, importantly, topography of the biomaterial. Surface topographies have the ability to alter cell behaviour through both intracellular and intercellular signalling, with variations to surface nanotopography affecting cell adhesion [36, 37], density and spreading [38, 39], cytokine secretion (important to cell signalling) [40], proliferation and even differentiation [41]. Biochemical characteristics range from the chemical structure of the material, to the introduction of inflammatory-suppressing agents, reprogramming factors, cytokines, growth factors, and various minerals and ions for induced differentiation and migration control. In addition, biomaterial degradation results in changes to biophysical cues, such as a reduction in stiffness (promoting chondrogenic over osteogenic lineages), and biochemical cues, such as increased release of signalling ions (altering the local microenvironment), which continue to vary throughout the process of the biomaterial breaking down [16]. These biomaterial alterations must also align with in vivo mechanical and practical applications. Can the biomaterial withstand the mechanical forces of bone and degrade at a rate that allows new bone migration to support load? Will the biomaterial remain biocompatible as it degrades by design over time? And, furthermore, prior to implantation, can the biomaterial be readily and effectively sterilised? Thus a multitude of parameters remain to be addressed in the search for the ideal material for tissue application [42].
The mechanical properties of bone vary depending on race, sex, age, and, regionally, throughout the body [43]. To tailor biomaterials on a case-by-case basis, designed specifically for the patient and the target injury, further understanding into how different characteristics interact on both a cellular and multi-cellular level is an important area of current research. Coincidently, emerging advances in controlling the bioreactor environment, alongside a new deeper understanding of how in vivo environmental cues change throughout bone formation over time, reintroduces traditional ex vivo stem cell expansion as a possibility for future tissue engineering approaches, because the in vivo environment can be more accurately replicated and optimised ex vivo [44, 45]. Therefore, research focus should not be reduced to either in-situ or ex vivo biomaterials, but rather focus on how cells respond to specific characteristics for a variety of environments, so the balance between controllable cell-instructive characteristics of a biomaterial, and relatively less controlled environmental cues, can be utilised to aid tissue regeneration.
4. A parameter problem
There have been a raft of articles reviewing how different physical characteristics alter the behaviour of cells [16, 42, 46], and several examples of how nanoscale physical cues alone can be used to instruct stem cells for control of differentiation and proliferation [33, 41, 47–50]. Surface topography has been shown to be an important parameter when designing cell-instructive biomaterials and a growing consensus has emerged that topography is as effective as biochemical cues and signals in controlling cell fate and function [46, 51, 52]. Once thought to be little more than a passive frame for cells, the ECM is now considered to play an important role in modifying and maintaining cell behaviours. Physical cues (mechanical signalling), can modulate the ECM-cell interaction to create new material-cytoskeleton links, where surface topographies instruct cells via modifications to cell adhesion and, subsequently, morphology. This, in turn, can modify cell spreading, migration, proliferation and, importantly, differentiation.
However, even this single parameter (topography) contains a large number of sub-parameters (for example roughness, ordering and shape), with additional further parameters (i.e. average height deviation amplitude, density of summits, texture aspect ratio, skewness, etc). Thus, if a more regulated set of topographies is selected, the potential parameter space expands exponentially. This results in significant problems with traditional investigative approaches: to comprehensively evaluate which nanotopographies are most effective (i.e. gratings, pits, randomised, or patterning not yet investigated) using traditional methods of culturing cells on virtually all patterns, with consequent analysis, the time taken would be unfeasible (figure 5). When including microtopographies, let alone other biomaterial characteristics, this problem only amplifies. To elaborate, if only nanoscale gratings and pits were used, that is two parameters. However, the variation in grating could include line separation, line thickness, grating length, increased disorder, different angles, etc., and the pits could be circular, triangular, rectangular, square, or hexagonal (or any other shape). That is now 10 parameters within a non-exhaustive list. Investigating a range of sizes from 100 μm to 100 nm with 10 nm precision would increase the parameter number to a thousand (1 × 103). When two variations are combined, for example line thickness and line separation, the parameter count is at least a million (1 × 106). If all the variations previously listed are combined, the parameter count becomes larger than 1 × 1030. Just considering the imaging of topographies, the fastest part in the cell culture- imaging-analysis chain, if an automated fluorescent microscope can take a bright field-fluorescent image pair on a single topography every millisecond, imaging alone would take 30 quintillion years without any repetition (substantially longer than the current approximation of the age of the universe of around 13.7 billion years [53]).
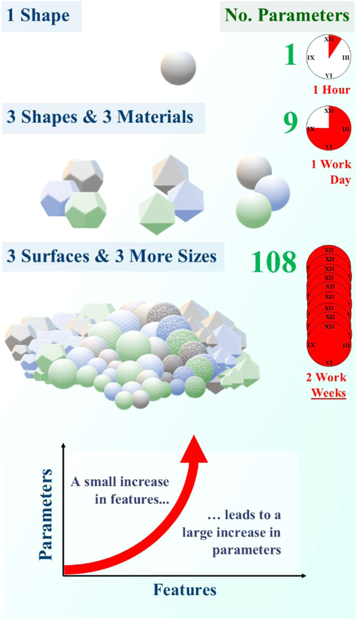
Figure 5. The number of parameters rises exponentially as the number of features is increased. With 4 characteristics, in this case shape, material, size, and surface type, the parameter count increases from 1 to 108 with only 13 features (3 shapes + 3 materials + 3 surfaces + 4 sizes). Assuming one parameter takes an hour to investigate, 2 characteristics could take a whole day to investigate while 4 characteristics two weeks. There are hundreds of different biomaterial characteristics, with thousands of different features, leading to millions of experimental research hours.
Download figure:
Standard image High-resolution imageThere are, therefore, too many parameters to traditionally investigate all interactions for the optimal cell-instructive biomaterial. However, advances over the last decade have shown that, when AI is trained on sufficiently varied data, a complete data set can be unnecessary to successfully determine underlying pattern recognition (figure 6). Humans do not need to see every breed of dog to recognise a dog that has not been seen before. Similarly, a person can describe what makes a dog, the formula or rules of determining a dog, from seeing a few dogs, without the need for advanced DNA and biological/zoological arguments. This was originally utilised outside the medical field: AI did not need to see every laser machined structure possible to determine the 3D depth profile of an unseen laser beam profile [54]. Only from the varied training set, the AI was also able to determine further useful laser machining parameters without any underlying physical processes encoded (no physics was taught to the AI before exposure to training data). To stick with the previous comparison, it could state what defines a dog without needing DNA or learning zoology. Once adapted for stem cell biology, AI was able to predict the statistically likely skeletal stem cell response to an unseen surface topography, using only 203 fluorescent images of live stained cells [35]. More importantly, using the AI predictions led to an experimentally validated topographical parameter for inducing cell alignment, without a single piece of cellular, chemical or physical biology encoded.
Figure 6. AI is capable of accurate predictions of unseen scenarios with a limited training dataset, as long as data is sufficiently varied. For an AI to predict whether an unseen animal is a dog or not, a dataset needs to include a variety of dogs and animals which are not dogs, without becoming biased or limiting to particular breeds of dog or species of alternate animals. A smaller quantity dataset (blue) is a higher quality than the larger quality dataset (yellow), even though there is less data for the AI to work with. (Both datasets must be expanded by several magnitudes to train a neural network adequately.)
Download figure:
Standard image High-resolution imageAI is not a magic wand to solve problems in science. Rather, AI should be viewed as a powerful statistical tool to aid understanding in parameter-heavy or time extensive fields. Tissue engineering is both, which makes AI an important addition. By using AI in crucial areas where current understanding is lacking, the process of designing a patient-specific and wound-specific biomaterial can be accelerated. Timescales of more than 30 quintillion years can be safely reduced to more grant funding friendly levels.
5. Understanding AI
Rather than go into the mathematics of AI, a more generalised overview allows for using the tool without a mathematical or computer science background. For a more mathematical or in-depth approach for deeper understanding, there are many well-referenced books and reviews available [55, 56]. Both the AI capable of determining the depth of laser machined surfaces and the AI able to predict the skeletal stem cell response to topography use a subsection of artificial neural networks called deep learning. Deep learning is an increasingly popular choice of machine learning architectures, with advanced computational methods and nonlinear mathematics, but relatively easy implementation. In part, this is how AI gained the moniker of a 'black box'. With this magical black box used in common smartphone apps, through the use of large image databases such as ImageNet and Open Images, a picture of a cat can go in and a correct tag label can come out [57, 58]. A photo of a face can go in and a younger, beauty-enhanced image is returned, allowing everyone to alter their virtual appearance in real-time, with all the good and bad that entails [59, 60]. However, this is not magic and, while reverse-engineering the massive computational architecture of a deep neural network is difficult, a basic understanding of how data is transformed (how the input is altered to become the output), is not.
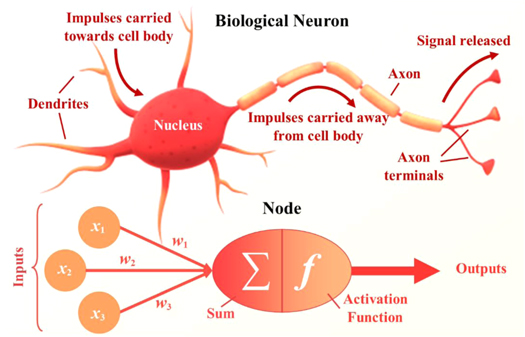
Artificial neural networks are computing algorithms for data transformation, which require no understanding of the underlying properties or science behind the transformation. They are universal approximators, capable of offering statistically likely solutions to problems that are too complex or time extensive to solve directly from fundamental principles and current scientific understanding [61, 62]. Conceptually, artificial neural networks have multiple similarities to biological neural networks, specifically the animal visual cortex. Both are made up of an assembly of nodes (neurons), interconnected through synapses (figure 7). What makes neural networks so effective is how data is transformed as it is propagated through the network. Every node sums the output of previous connected nodes, which are all given individual weightings, to form a new output, which is then weighted and summed with outputs of others in a new node. This can be compared to how dendrites propagate the electrochemical stimulation received from other neurons, before the impulse is carried away from the cell body and passed on through axon terminals. In both biological and artificial neural networks, the weights of the synapses can change as the network learns. For artificial neural networks, this process of altering the weights so the data transformation better fits training examples is called backpropagation, and the error is automatically minimised as the network 'learns' [63]. As with animals, connections are strengthened through larger weightings if they produce the correct outputs, simultaneously weakening those that do not, analogous to brain plasticity.
Figure 7. Biological neurons which make up the visual cortex and artificial nodes are conceptually analogous. Dendrites carry impulses towards the cell body, where the nucleus (the brain of the cell) can process this information. A new impulse is generated and carried away from the cell body through the axon to the axon terminals, where the signal is released and becomes an input signal to another neuron. Weighted inputs from several nodes are summed within a new node, which then processes the information through an activation function. This new output is then weighted and input to another node in a latter layer. Both process multiple inputs and transmit as a new output for further processing.
Download figure:
Standard image High-resolution imageDeep neural networks (DNNs), or deep learning, is an artificial neural network that has layers of 'hidden' nodes between the input and output layer (figure 8). The function of these central hidden layers changes at each level. The first will process the data directly from the input, and yet the second will only process data transformed by the first, which means it is capable of operating at a comparatively more abstract level. Further hidden layers would operate at a higher level still. With enough layers, and with computing power to run those layers, and enough varied input data to finely tune the synapses, deep learning architectures need no representation design or interference from human computer engineers. Via backpropagation through the hidden layers, the network learns what features of the input data are relevant at each layer with no concept of science or prior knowledge required. Using a relatively small number of hidden layers, less than 20, a deep learning system can implement highly intricate functions to its inputs, which are sensitive to minute details (such as distinguishing between different types of canine) and insensitive to large irrelevant variations such as the background, pose, lighting and surrounding objects [55].
Figure 8. A concept image of a deep neural network. The input layer, consisting of several different nodes (circles), is connected to the first hidden layer, where the data from the input layer is weighted, summed and transformed without human interaction before progression into the next layer. The second hidden layer is only input data from the first hidden layer, allowing for more abstract feature extraction. Data processed from this hidden layer is then propagated to the output layer. The number of hidden layers, the number of nodes and the type of data transformation between each layer is completely customisable between different networks.
Download figure:
Standard image High-resolution imageAll the AI examples so far have used a type of deep neural network that has excelled in imaging tasks for several decades, referred to as convolutional neural networks (ConvNets or CNNs) [64–73], especially in the medical field [74–80]. Going back to the biological comparisons, the general architecture is similar to the hierarchy in the visual cortex ventral pathway, necessary for visual perception [81, 82]. The intrinsic parts to a ConvNet, the convolutional and pooling layers, are similar to (and partially inspired by) archetypal concepts of simple and complex cells in visual neuroscience [55, 83–85]. The most important part of processing an image is feature extraction. However, spatial features are only noticeable through the positional arrangement of the pixels. It is not possible to determine whether a dog has rounded or pointed ears, important for assessing breeds, when looking at a highly zoomed in area of the ear, which would only give you information on the colour (a pixel value) and not the shape (spatial pixel arrangement). The convolutional layer processes an image by transforming the pixel data through a specialised weighted matrix, an array of numbers, so specific features are extracted without losing spatial information (figure 9). A weight combination may extract a variety of features, with one focused on colour, another on edges, and another blurring unimportant noise from the image, for example, where the features are learned by the ConvNet (weights altered by the ConvNet) to minimise loss. The discovery of what should be considered features, and which features to focus on, are done without human intervention, besides a single target to aim for and against which the error is calculated. Next, the pooling matrix reduces the size of the image and therefore the number of parameters, in this case pixels, for the network to consider. This merges semantically similar features and creates invariance to small shifts and distortions (a brown dog is still a brown dog, no matter where it sits in an image or how good or bad the lighting is).
Figure 9. Feature extraction is simple for ConvNets, as the relatively large data in an image is easily reduced (convolved) from large pixel arrangements to smaller pixel distributions without losing spatial data, which is often necessary (such as determining whether there is a dog ear, which cannot be done through pixel value alone). Generating data is harder, as it is a one-to-many problem. While one DNN can determine which features are essential, placement and detail require a realistic rather than simply statistically averaged pixel arrangement.
Download figure:
Standard image High-resolution imageGenerating data is not as simple as processing data. During processing, small shifts in data (the dog is on the left or the dog is on the right) become obsolete (there is a dog either way). However, generating an image from important information (a picture of a dog is required) requires a level of choice (where to put the dog), and therefore a level of probability (50% chance it should be on the left and 50% chance it should be on the right) of matching the target image used to train the ConvNet, which is not required in relatively simpler feature extraction. When simple ConvNets are used to generate images, the result over time will average all statistically likely variations rather than produce a single highly likely outcome (the dog is on both sides and neither side simultaneously—a blurry mess—rather than a clear picture of a dog on a single side). This output minimises loss, but it is not very useful because the generated data is not realistic. Therefore, another ConvNet is added to the architecture to modify the loss function with a very important question—does the output from the generator ConvNet look realistic? Blurry images are no longer viable outputs as they are identifiably fake. Together, this generative ConvNet (also called U-Nets due to the architectural design) and discriminator ConvNet pair forms a conditional generative adversarial network (cGAN), so called because the different ConvNets are trained adversarially. The discriminator is responsible for determining whether an image is real or generated, while the generator is responsible for fooling the discriminator.
Deep learning can encompass a large variety of network architectures, of which the cGAN is only one of many. However, popular architectures share the same staple features: a number of hidden layers, with increasingly abstract and difficult-to-visualise data transformations at each layer; ability to learn and improve without human interference; and a loss function. Whether it is supervised learning, where the network learns from training on examples, reinforcement learning, where the network interacts directly with the data environment, or another type of machine learning, deep learning is highly adaptive and becoming more easily accessible as improvements in computation drive down the cost for GPUs and other basic hardware requirements.
AI, even deep learning specifically, is a vast field and this review has barely touched the surface. As well as CNNs and cGANs, there are numerous architecture choices, in medicine and other disciplines, which have had significant impact in both research and subsequent applications. There have been many prominent reviews throughout the last decades to help narrow down the most impactful advances, in both deep learning and artificial intelligence as a whole. The review paper 'Deep learning' by LeCun et al summarises the key concepts of deep learning, including supervised learning, backpropagation, and unsupervised learning, with no mathematical background required [55]. For an in-depth understanding into the mathematics of deep learning, one of the most well respected and comprehensive sources is the book 'Deep learning' by Goodfellow et al [56]. This will advance the reader from beginner through to intermediate level, although a background in pre-degree level mathematics is recommended.
'A guide to deep learning in healthcare' by Estava et al is a medicine (and allied fields) specific review that covers successful applications, such as medical imaging and genomics, and modern concepts, such as reinforcement learning and deep learning architectures with multiple data inputs [78]. Additionally, 'Deep learning in biomedicine' by Wainberg et al describes the rise in data-driven biomedicine and explains key mathematical concepts with accessible visuals [86]. It also reviews the current successes and pit-falls of deep learning in biomedicine, and how applications can be improved in the future through transparency and performance guarantees.
6. Current AI integration
Deep learning in healthcare is a growing field, especially with areas of big data that can be difficult for human comprehension and non-AI computational pattern recognition, such as genomics (often at the forefront of computer science integration), or gene expression data from a single patient, or public health information across a nation, transformed into specific biomedical applications [78]. Applications of deep learning in biology, healthcare and drug discovery have highlighted accomplishments that would have been virtually infeasible without deep learning (figure 10) [80, 87]. Clinical imaging, electronic health records, genomics, smart health-tracking wearable devices, even use of sensor-equipped smartphones, the reach of deep learning is both improving and generating new ways of interfacing data, patients, and medical professionals [88]. With relatively little data processing required, and the ability to extract features and characteristics without prior knowledge of biology, chemistry or medicine, advances can be made in areas where physicians currently struggle, such as determining if novel variants in a patient's genome are medically relevant, whether abnormalities in plasma could be early-onset cancerous mutations, or how cell features are linked to tumour pathology [89, 90]. Medical imaging is often at the forefront in integrating AI, as it is a field of complex and ambiguous data, which often requires lengthy manual processing, where an expert will medically interpret and analyse, and then annotate, large amounts of data [91, 92]. While ConvNets and deep learning have traditionally been utilised effectively in segmentation of tissues and organs, increasingly more complex tasks are being advanced through deep learning, such as detection of abnormalities or non-invasive cell counting, morphological identification and behavioural prediction, including with stem cells [35, 87, 93–99].
Figure 10. AI is behind many advances in clinical-related fields, including discoveries in microbiology, health interfacing through smart wearable technology, rapid medical imaging analysis enhancement, and novel potential drug discoveries.
Download figure:
Standard image High-resolution imageBiomaterial science has also been improved through use of machine learning, via design of three-dimensional scaffolds for cell control from geometrical fibre-based material matrixes [100], with optimization of piezoelectric drop-on-demand printing, vital for future experimental bioprinting research [101], and generating blueprints for 3D bioprinting of tissue-specific microenvironments and patient-specific organs [102, 103]. Through the integration of deep learning, there is the possibility of patient-specific and trauma-specific parameters for tailored production of biomaterials with optimal characteristics. One of the most important biophysical parameters for cell-instructive biomaterials is topographical characteristics, both at the microscale (associated with cell migration, elongation and proliferation) and the nanoscale (associated with adhesion, differentiation and osteogenic gene expression) and overlap between the two. Deep learning has recently been applied to microtopographies to accurately determine cell positions in response to different (unseen) surface patterning, outputting statistically significant predictions for adult skeletal stem cells from donors not used during training data acquisition [35]. Similar work has been conducted on HBMSCs response to micro-environmental cues, with morphology analysed by simpler machine learning models. However, published results focused on training accuracy, which does not allow for analysis and validation of possible real-world usage [104]. If validation results are able to match training accuracies, both studies represent promising steps in applying AI to tissue engineering problems as, in regards to the microtopography study, an accurate deep learning-based model for stem cell behaviours can dramatically reduce the amount of time and resources required for lengthy laboratory-centred experimentation.
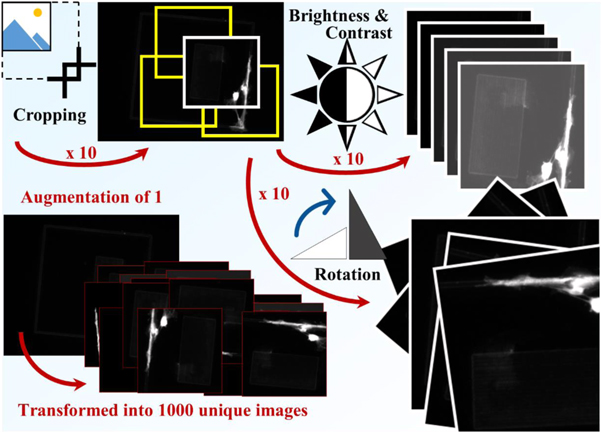
Importantly, these statistically significant predictions were achieved using a training set of roughly 200 images, much fewer than the thousands to tens-of-thousands that deep learning is frequently claimed to require [78, 88]. Through clever uses of data-augmentation, the equivalent of trace DNA amplification for small datasets, such as randomised cropping, rotation, and flipping of images, a few hundred images can be converted into a more appropriate dataset of several thousands of unique images (figure 11). However, just as amplification success of trace DNA can decrease with too many cycles, too much augmentation can decrease the success of a DNN. The effect of a single problematic image will be amplified, and, after training on limited data, the network will be unable to generalise to the level required for testing. Rather than discover features required for solving the desired problem in a real-world application, the network will only discover features relevant to the undiversified training data. Overfitting to the training dataset can be eliminated through conservative use of data augmentation, training and testing on varied data (easily achievable with a large dataset), and stopping the network before over-training, and over-fitting, seemingly improves accuracy while sacrificing generalisation and real-work applicability. In the case of deep learning, where networks can frequently take weeks to train, sometimes less is more. The lack of overfitting in a model capable of predicting stem cell behaviour, which was trained on relatively few images, shows that deep learning can be applied in fields that are not as data-heavy as i.e. genomics without impacting success. Another way to increase quantity of training data is to use augmentation, which does not destabilise training and can improve result quality in smaller dataset driven tasks [105].
Figure 11. Data augmentation is the process of increasing the training data through techniques such as randomised cropping, rotation and alterations to brightness and contrast. Each technique can increase the size of the dataset by orders of magnitude, so a single raw image can be augmented to produce thousands of unique images. Variation is important for training DNNs, so augmenting too heavily can lead to reduced performance. However, light augmentation can increase variety and therefore final performance.
Download figure:
Standard image High-resolution imageIntegration of machine intelligence with stem cell research is not limited to topographical response, as it has also been used to help understand differentiation potential [106, 107], determine reaction to micro-environmental cues [104], predict and detect pluripotent colony formation [93, 108], investigate immunosuppressive outcomes [109], and provide general quality control automation [110, 111]. In bioreactor-based experiments, machine learning models were capable of predicting whether final levels of cardiomyoctes, derived from human pluripotent stem cells and with therapeutic potential, would be classified as 'insufficient' with 84% accuracy after five days of feature analysis, and reaching 90% accuracy after seven days [107]. This saves both resources and time, and gave insight into which features are most relevant to predict cardiomyocyte content, as failing experiments can be interrupted and possibly corrected. However, the quality and resulting functionality and maturity were not predicted with this limited approach. Another study used machine intelligence for quality control identification, with images of good, semi-good and bad quality colonies, but accuracy was lower, with the best overall accuracy at only 62.4% (figure 12) [111]. Lower accuracy was in part due to the classification of semi-good and bad colonies, which were too subjective for the models to identify with high accuracy. Either the images did not provide enough information, there weren't enough images, or the model was not complex enough. The 83.8% accuracy for identifying the good cultures shows that there is a place for machine learning in quality control, but with a larger dataset, a more quantified approach, and a more sophisticated network model required.
Figure 12. Machine intelligence can identify cell colony quality with up to 83.8% accuracy for 'good' quality colonies. While accuracy for 'semi-good' and 'bad' requires further improvement before the system is capable of real-world application, there is possibility for real-time non-invasive stem cell quality control through the use of AI.
Download figure:
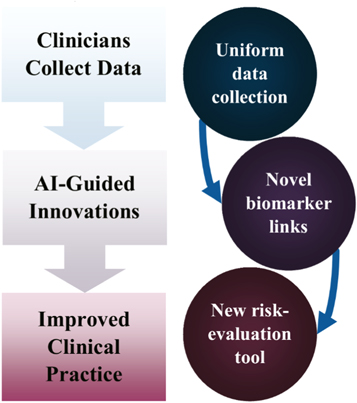
Standard image High-resolution imageOnce a high quality stem cell colony has been cultured, there is still a need to prevent chronic graft-versus-host disease (GvHD), a major cause of morbidity and mortality after stem cell transplantation [112]. The relationship between the severity of chronic GvHD and National Institutes of Health biomarkers has not been easily interpreted, and clinical outcomes are still unpredictable to clinicians, but not to AI [113]. Successfully highlighting unique chronic GvHD phenotypes, the machine learning workflow was then reversed to generate a clinically applicable decision-tree tool. Similar work has also been applied to stem cell mortality 100 days after transplantation, in the field of acute leukaemia, with a decision-tree model developed for risk evaluation of patients as a result of machined learning predictions [114]. Clinical understanding and application, in both cases, was enhanced through use of machine learning (figure 13).
Figure 13. A flow chart illustrating how collecting data for training AI can lead to improved clinical practice. It was improved through the use of new risk-evaluation decision tree models, which were designed from AI-highlighted biomarkers—the biomarkers that influenced AI predicted patient outcome. This approach can be applied to multiple medical conditions.
Download figure:
Standard image High-resolution imageBetween cell culture-based testing and clinical application, there is often necessary animal experimentation. Animal experimentation is an essential component in the progression advancing promising, successful, in-vitro experiments to clinical translation and application of biomaterials, drugs and techniques in the human medical field. It has been found that it is difficult to predict the outcome of in vivo experiments based on in-vitro data, therefore AI is a novel tool for predicting the outcome without the need for extensive animal experimentation [115]. The use of animals in research is tightly regulated and controlled by the Home Office in the UK, requiring in-depth planning, training and competency assessments with accreditation and licensing of facilities and staff. The use of the PREPARE guidelines by researchers to swiftly revise the factors, which should be considered prior to starting an animal based experiment, is highly recommended to avoid any late realisation that an experiment should have been performed differently, which would then save the frustration, time and expense of repeating experiments and, most importantly, using greater animal numbers unnecessarily [116]. The Arrive Guidelines give guidance on all the necessary information which should be published to make clear to the audience the methods and statistics employed, and of all the factors considered prior to, and during, an in vivo study, to allow studies to be repeatable, built upon and transparent to other researchers [117]. The NC3Rs (National Centre for the Replacement, Refinement and Reduction of Animals in Research) are an organisation dedicated to the '3Rs'; replacement (of animal use or animal type), refinement (of methods applied to aid welfare) and reduction (of animal number) used in research.
The use of AI has been applied to different areas of animal-based research which will aid the 3Rs. The use of Multiple Particle Path Dosimetry (MPPD) models to determine micro- and nano-particulate distribution in animal models is an important aspect of toxicity modelling when using nanomaterials [118]. While toxicity studies using machine learning are not yet able to recapitulate the in vivo scenario, they can be used as an adjunct [119]. The use of deep learning has been applied to assess posture in response to stimuli in neurology research, and the use of computational neuroethology as a tool to assess behaviour in many species [120, 121]. The mapping of behaviour is very important to allow a non-invasive observation using programs such as DeepLabCut and LEAP [122, 123]—a field recently reviewed by Pereira et al [124]. This current work on decreasing reliance on purely animal-based experimentation is a promising step in increasing application of the 3Rs without being detrimental to experimental advances in lab settings progressing to clinical application in humans.
7. Applying deep learning
7.1. Where things Go wrong
Alongside overfitting, there are several areas that can lead to unsuccessful integration of deep learning. It is important to understand exactly what a network is designed to do, and how data could be biased before and during training. Both are major considerations when applying deep learning, as the ramifications of giving a network an incorrect task, or utilising a badly chosen loss function, will probably not be uncovered until lengthy data capture, processing and network training has been completed. And, if the data is biased, even the correct task can lead to an incorrect result and an unusable network. A good example of giving a network an incorrect task is when AI was used to automate the play of several Nintendo Entertainment System (NES) games [125]. The task given across multiple games was to win. This can be interpreted in multiple ways—should the network be coded to achieve a high score, to play for as long as possible, to reach a higher level, or combine multiple parameters? While being tasked to win resulted in successful, if inhumanly bug exploiting, play for NES games such as Super Mario Bros., the same algorithm was unable to play Tetris. Instead of learning how and where to successfully place blocks, it instead learned to pause the game the moment before game-over (figure 14). It would then leave the game paused. The algorithm had implemented a win applied by angry younger siblings everywhere, where 'the only winning move is not to play'.
Figure 14. When a simple AI system was given the task of not reaching 'game over', or losing, in the game Tetris, it learned the most efficient way of accomplishing this—it paused the game. While unexpected solutions can be a benefit of applying AI, it also shows the importance of instructing detailed and relevant task functions.
Download figure:
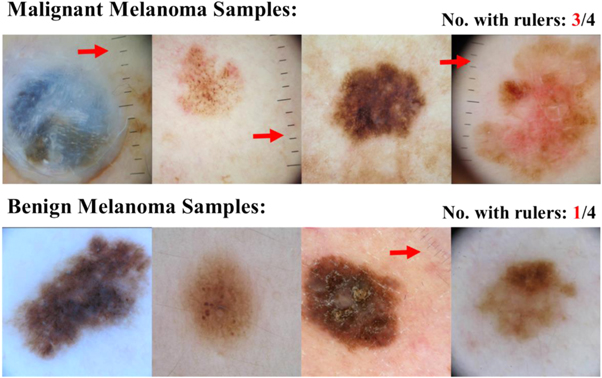
Standard image High-resolution imageData bias is an equally important consideration [126]. A 2017 Nature article with several thousand citations claimed to have developed a network capable of 'augmenting clinical decision-making for dermatology specialists' [127]. In the paper, sample images of malignant cancers used for training and testing the network were collected from a wide source of patients. However, it is worth noting that some images contain a measuring ruler, while others do not. The network certainly noted this variance as, after publication, the authors discovered that simply placing a ruler in the image increased the likelihood of the network predicting malignant cancer. This is not an example of a network that has trained incorrectly, but an example of a network that has trained correctly on biased data. Dermatologists are more likely to record the size of a skin lesion with a ruler if they suspect there is reason for concern, which leads to a higher likelihood of rulers in malignant melanoma images, seen when sampling the International Skin Imaging Collaboration (ISIC): Melanoma Project archive (figure 15) [128]. With a more balanced dataset, where rulers were proportionately represented in both malignant and benign lesion images, the feature would not have been extracted by the network as an important marker, and applications in dermatology and oncology would not have been stalled. When applying AI, researchers must be diligent—a relatively minor feature can bias a dataset and corrupt a network, turning a cancer detecting algorithm (cited thousands of times) into a simple ruler detector.
Figure 15. Images used to train a network subsequently published in Nature, which was discovered to be unusable in clinical application due to training bias. Randomly sampling 160 malignant and 160 benign melanoma images for 4 of each from the ISIC Archive, 3 of the 4 malignant images had rulers while only 1 of the benign images had a ruler visible. While not statistically valid for determining %-bias across the dataset used for the network, it shows the bias towards visible rulers within malignant melanoma images within the ISIC database [128].
Download figure:
Standard image High-resolution imageThe most reliable way to overcome incorrect task functions and biased datasets is to minimise a difference in conditions between training and target application. This mismatch can be characterised into several subsections [86]:
- I.Target mismatch (the user wants to know what biomaterial promotes differentiation but the network is trained on predicting proliferation);
- II.Loss function mismatch (the training loss function is squared error in predicting total cell numbers, but the bioengineer only needs the colony to be above a specific density);
- III.Data mismatch and selection bias (only donor cells from a single patient group were used);
- IV.Nonstationary environments (the model does not adjust for the change in cell behaviour as the colony size increases);
- V.Reactive and adversarial environments (culture of the colony is changed as a result of using the model and therefore a second proliferation prediction would not be valid);
- VI.Confounding variables and causality (proximity of a ruler does not cause a lesion to be malignant).
However, these issues can be overcome by designing the network with, and using data collected by, the end-user(s) in a full collaboration. Or, for simpler tasks that do not require novel data combinations or computer hardware beyond a standard gaming/image processing PC, the end-user applying AI to their research and training the network themselves, using publicly available deep learning architectures, such as pix2pix (https://github.com/phillipi/pix2pix) [129]. This has the added benefit of establishing trust between the results of the network and the end-user, where the user has a detailed knowledge of expectations and limitations to the network, easing integration.
7.2. How to integrate AI
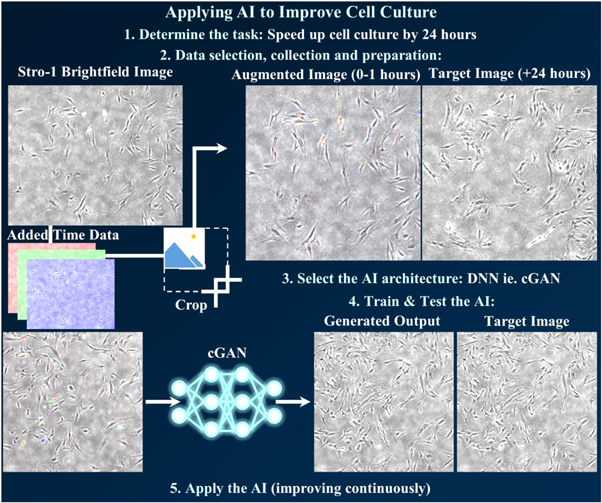
There are several important and interwoven steps in integrating deep learning, where the first is to determine the exact task that the network is going to undertake (figure 16). This needs to be a task that can be accomplished using the data, while also providing a benefit to the field, such as through reducing time, resources, or increasing scientific understanding. Data selection is important. While it is easy to obtain many selfies, a network is not going to be able to accurately determine whether a patient has COVID-19 from a smartphone selfie image, but it is likely possible to determine cases from CT images of lungs, which are less readily available [130]. However, chest x-rays are easier in some hospital settings to collect, so that could become a compromise between containing relevant features for task completion with accessibility of data. Not only is accessibility important for developing a large, well varied training dataset, it is also important for eventual ease of application in a real-world setting.
Figure 16. Step-by-step example of applying AI to improve a time-consuming and resource-expensive task. Step 1 is determining a task which balances improvement with ease of data-acquisition, such as reducing time needed for cell culture through AI-prediction of Stro-1 selected HBMSCs after an additional 24 h. Step 2 is selecting appropriate data (a brightfield image at time-points before and after 24 h), collecting the data (a continual time-lapse experiment at multiple positions with multiple cell cultures), and preparation (augmentation to provide maximum relevant data per input image and maximum images without overfitting). Images were processed here by combining 3 time points and then performing randomised cropping. Step 3 is to decide on AI architecture(s), depending on both the task and the data acquired. For image data, a cGAN is the starting-off architecture used here, as it has proven successful for similar tasks and code is readily available and open source. Step 4 is to train and then test the architecture, making multiple changes to both hyperparameters and the architecture until results can be trusted. The final step is to apply the AI, as the inclusion of new input and output data will reveal current limitations and provide improvement to the AI, while simultaneously saving 24 h of cell culture time.
Download figure:
Standard image High-resolution imageOnce the data type has been obtained, and the task ascertained, the next step is to acquire an appropriate network architecture. The methodology for using deep learning to predict adult skeletal stem cell response to micro-patterned topographies [35] is the exact same method as used in the 3D labelling of cells in placenta with nanoscale resolution [92], and in the super-resolution transformation of 20× optical microscope images into 1500× SEM images [131]. In total, the architecture used (the previously mentioned open-source pix2pix [129]) has been cited almost seven thousand times, implying it has been used for hundreds, if not thousands, of different purposes in dozens of varied fields. However, it is worth noting that, as the field of biological applications of deep learning grows, there are many competing architectures providing slightly different precision and accuracy results under the same conditions [75]. Application of several different architectures, which are increasingly available with Open Access publishing and desire for real-world usage and application from the machine learning researchers, is recommended for maximum optimisation [132]. Using multiple different architectures and multiple different data types is a good way of confirming that the desired task can be accomplished, if the application is novel.
Training the chosen network architecture on obtained data is time consuming but not challenging. There are many hyperparameters that need to be adjusted to achieve optimal network performance, which all intertwine due to how each effects the network. Mathematical understanding will help with choosing optimiser and activation functions, affecting how data is transformed through the network, but sticking with those suggested by the network architecture creator will still produce good results. However, the number of layers, the number of filters per layer, the learning rate, and some other hyperparameters will likely need several adjustments before the network is producing optimal results. An increase in the number of layers and filters will increase the complexity of the network, but also the time taken for the network to train and the size of GPU and computing power required to run the network. The learning rate, unlike previous hyperparameters, can be changed throughout training. This is the size of each error-correcting step that the network takes in development of accurate data-transformation. Too large a step size and the network will continually step over the optimal solution, and either fluctuate around it (sometimes detectable and therefore fixable) or miss it entirely (the network simply will not train successfully). Too small also produces problems, as the network could become stuck on a sub-optimal solution without being able to step through to a better solution, or simply waste time (imagine an adult trying to cover a large distance while using the stride length of a small child).
Testing is the most important step for both validation and accountability, as promises of success determined by high training accuracy do not automatically transfer to high accuracy in real world usage. The more similar testing data is to training data, the better the testing results will be, but it will not give any indication for applicability (figure 17). Testing must therefore be vigorous, with data not used in training the network (to detect possible overfitting), and preferably with substantial features not accounted for in the training data (to determine applicability). Random extraction of data from the training dataset for testing is commonplace but not always the best practice. For example, if a dataset contains x-ray images of lungs from multiple patients and collected by multiple clinicians across three hospitals, two hospitals should be used for training, which preferably contain the least biased data (gender, age, race, etc., in appropriate levels), and one hospital should be used for testing. This would contain differences in equipment, clinicians and patients from the training data, allowing for vigorous testing in how well the AI could be applied to different settings, while still leaving enough variety in the training dataset to successfully train the network. If a dataset is less varied, such as using a single time-lapse experiment to train a network into predicting cell behaviour over time, extracting frames randomly throughout or from the centre of the data would pose no challenge to the network, which has already seen similar data both before and after the prediction point in question. A better challenge would be testing on data extracted from the end of the time-lapse experiment, where conditions such as cell density, colony health, and lighting are subtly different, and a prediction would be without any training data at future points to guide the network.
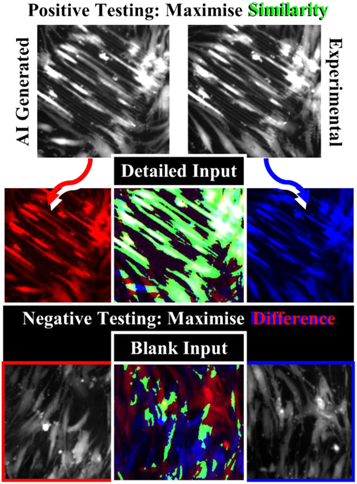
Figure 17. Comprehensive testing includes both positive and negative testing to determine both reliability and applicability. For cGANs, a successful positive test would include large areas of similarity (green) between the network-generated prediction (red) and experimentally obtained result (blue) to a detailed input. A negative test (blank input) should result in different results due to random network output, or the network has overfit to training data and is not applicable in real-world usage.
Download figure:
Standard image High-resolution image8. Future of AI integration
With the versatility of deep learning, it is hard to predict which direction will be the most influential in the future. One interesting use of deep learning progression is in the pharmaceutical field, specifically in drug development. Deep learning is expected to dramatically shorten drug product development timelines as pharmaceutical research transforms from experience-dependent and experiment-heavy studies, at great material and economic expense, to data-driven methodologies [133]. Able to identify complex correlations on small data (less than 300 formulations), and generalize without losing accuracy, deep learning has the potential to predict both in vivo and in vitro characteristics, helping control the product quality throughout development. The methodology of this study could be applied to the regenerative medicine field to discover new materials with the correct biochemical and biophysical characteristics for both in-situ and in vitro tissue engineering. It follows a similar process to the method used in stem cell topographical response prediction, where a premade machine learning framework (DeepLearning4j from https://deeplearning4j.org/) was adopted, and hyper parameters adjusted, for optimal performance.
Another interesting use of AI is a study that utilised an ensemble machine intelligence model, based on the random forest model, where hyper-parameters were semi-autonomously determined. This model predicted the dissolution behaviour (time-dependent normalised critical ion mass loss) of various glasses with the Pearson correlation coefficient as high as 0.99 (it was highly successful) [134]. The success of this model was analysed to be due to the inherently nonlinear relationship between dissolution rate of silicate glasses and the pH of the contacting solution, alongside other variables in the system. As well as having implications for the design of new glasses for healthcare, this work could also have implications for biochemical design of implants with controlled degradation rates. Alongside work on the biodegradation of collagen scaffolds using statistical modelling approaches [135], a future parallel study should include modern deep learning architectures, as they are more successful at complex non-linear tasks such as biochemical discovery than older machine learning approaches [136], and a focus on biomaterials for tissue engineering, to discover (or narrow the parameter space to aid discovery of) optimal compositions.
Once optimal compositions have been identified using AI, they must be tested thoroughly before clinical use. The Chorioallantoic Membrane (CAM) Assay, an ex vivo, preliminary model, is frequently used to determine biocompatibility, angiogenic/anti-angiogenic effects of drugs or biomaterials, which are applied to the vascular CAM. It is also used to grow tissues and tumours for applications such as oncology research. The use of the CAM has been reviewed over the years, with increasing interest in the application for bone tissue engineering to screen materials prior to in vivo studies, as reviewed by Marshall et al [137]. The angiogenic response of the CAM can be quantified using the Chalkley Scoring method with dots on a microscope eyepiece being positioned over an observed sample, and the dots counted when they overlie a blood vessel. Due to the variability of this method, calculations using computer modelling, which removes the need for a Chalkley graticule eyepiece for the microscope, have been investigated to allow calculation of a microvessel area estimate for solid tumour samples [138]. A photograph of CAM blood vessels can be taken and manipulated using computer software programs to delineate the blood vessels, allowing for easier automated counting, as shown in a recent publication by Manger et al [139]. Application of an AI to streamline this lengthy process would aid screening of options from different materials without the use of further CAM assays. Mathematical modelling of a scaffold with different cell types interacting enables extension of results for a longer experimental period than is practical, due to chicken eggs hatching at day 21 of gestation, which allows further prediction of progression in a simulated environment [140]. DNNs could perform these predictions with data collected from necessary previous and current animal studies to predict outcomes of in vivo usage of various materials, and perhaps aid the design of new materials. The materials found to be biocompatible could then be taken forward to rodent studies and the unsuccessful candidates discounted at an earlier stage of the research pathway, leading to reduction in the number of animals used, refinement of materials being investigated to be more biocompatible, and simultaneously allowing modifications to enhance angiogenesis and osteogenesis results. This would effectively replace rodents with the CAM assay and computer simulations.
Another key area for AI integration could be in tissue-tissue interfaces, such as tendone-bone and cartilage-bone junctions, where structures and compositions are more complex than in single-tissue areas that have currently been investigated. For example, twin-screw extrusion electrospinning has been used to generate functionally graded nano-woven meshes of polycaprolactone (PCL), with incorporated beta-tricalcium phosphate (β-TCP) nanoparticles [141]. These functionally-graded scaffolds were seeded and cultured with mouse preosteoblast cells (MC3T3-E1) and, after 4 weeks, showed markers for both collagen synthesis and mineralisation. Importantly, this was similar to the bone-cartilage interface in terms of both mechanical properties and the distribution of calcium particle concentrations.
This method of scaffold generation controls the distribution of drugs/growth factors, alongside control of physical properties such as porosity, wettability and biodegradation rate. Therefore, these 3D electrospun nanofibrous and nanocomposite scaffolds could mimic the complex distributions and mechanical properties of natural tissue. Integrating AI with this method could aid tissue engineering at the bone-cartilege interface through AI-optimised distribution of TPC particles, which could lead to patient and location specific scaffolds. Additionially, different nanoparticles could be identified through AI-predictions for increased customisation, such as promoting healing-rate in one scaffold versus increased resultant bone density in another. In recent work, nano-hydroxyapatite (nHA) was added to PCL to form scaffolds with similarities to the tendon-bone interface [142]. Using AI to moderate the large parameter-space of numerous nanocomposites at varying distributions, such scaffolds could be used for other tissue-tissue interfaces throughout the body, with possible broader tissue engineering applications.
Recent work in single-cell response to nanotopographies, specifically a variety of cells from a mouse musculoskeletal system based on osteogenic-promoting patterning [33, 41, 47], has shown that a Bayesian linear regression model can predict how topographical changes will alter gene expression, quantitatively determining the effect of nanotopographies on cell function [143]. If this study was advanced to HBMSCs, with single-cell and colony behaviour investigated, it is highly likely that a DNN-based model could generate predictions for stem cell behavioural response to nanotopographies, relevant for implant and scaffold design. Combined with prior microscale studies [35], a dual-network system, a deep convolutional generative cooperative network rather than an adversarial network, where one network designs topographies and another predicts the cell response, has the potential for a completely AI-designed topographical patterning for optimal proliferation / differentiation, patient and injury specific. By splitting the workload between networks, less GPU power is required and there is a greater level of interpretability and accuracy, as the network responsible for cell response prediction, the model, can be independently tested, validated, and continuously improved, while the cooperative network, the generator, can design topographies faster than human capability. A lack of bias and the freedom of deep learning to acquire its own data-driven features would also allow for novel design solutions, similar to the new strategies and innovations developed by AlphaGo [144].
Extending this cooperative system to include the previously hypothesised network for biochemical design as another model network, an entirely autonomous system could be created where a material with the desired biochemistry, topography and cell-instructive behaviour is designed (figure 18). By linking deep learning architectures in this fashion, each network output can be scrutinised, and concerns around 'black box' processing become irrelevant, while bias in one area can be mitigated before being passed through to another. It also makes reverse-engineering of the process, and accompanying enhanced scientific understanding, possible by using the outputs of the networks at every stage as experimental data. Integration of deep learning on a step-by-step basis done by, or at least heavily involving, researchers and clinicians who collect the data used in training, and who will apply and benefit from deep learning integration, can, on its own, be time-saving and create small innovations. Through the combination of optimised experimental designs (less expensive and resource-intensive) and fewer necessary in vitro and in vivo trials and animal studies, research groups are expected to move towards a more deep learning incorporating approach in the future [145].
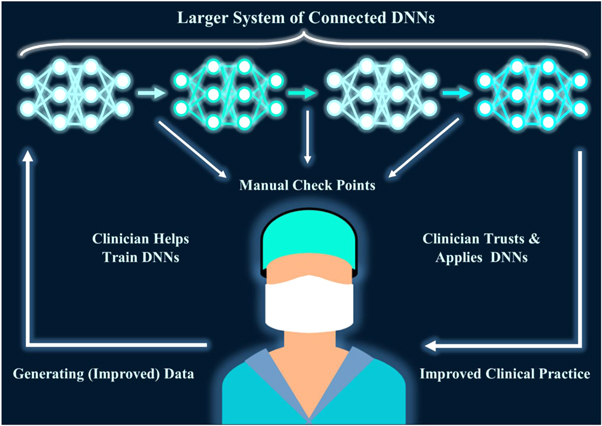
Figure 18. The Future of AI: Putting the Clinician at the Centre. In a continuous cycle, clinicians generate data used to train a system of connected DNNs. By splitting tasks into smaller sub-tasks, it creates multiple manual check points to scrutinise the DNN process, allowing for greater error correction and potential for generating further scientific understanding. As the clinician is heavily involved in training and checking the DNNs, the clinician knows the limitations and trusts the accuracy of the DNN system. Consequently, application is smoother and clinical practice is improved through DNN improvements and breakthroughs. Better data follows, leading to better DNN training, leading to even better clinical practice, in a positive feedback mechanism.
Download figure:
Standard image High-resolution imageBy joining these networks as part of a larger system, the whole tissue engineering process from laboratory bench to clinical application can be streamlined and accelerated, without losing the ability to rigorously model and validate the predictions and creations of the cooperative network. Deep learning integration with both research and clinical application will allow for an unprecedented understanding of human biology and improve tissue engineering, and regenerative medicine, to levels never seen before [79].
9. A final word
Tissue engineering is a major section of the regenerative medicine field, and becoming more important every year due to aging populations across the world. While there have been significant developments over the past decades, there are still many challenges, notably biomaterial design and understanding of stem cell behaviour. However, through use of DNNs integrated into experimental research and clinical application, many of the current challenges in medicine can be overcome, leading to a future of patient-specific and trauma-specific solutions (figure 19). The future of bone regeneration perhaps belongs to AI.
Figure 19. A collaboration of clinicians, researchers, and deep learning offers exciting possibilities for medical innovation, including in the field of tissue engineering, where there are many unknowns and parameter problems that hinder current manual experimentation.
Download figure:
Standard image High-resolution imageFunding & Acknowledgments
Ben Mills is funded by EPSRC (EP/N03368X/1) and EPSRC (EP/T026197/1). Richard O C Oreffo is funded by the BBSRC (BB/P017711/1) and the UK Regenerative Medicine Platform (MR/R015651/1). These research councils are gratefully acknowledged.
The authors would like to show appreciation to EPSRC for funding and the Faculties of Engineering & Physical Sciences and Medicine at the University of Southampton for permission to use all required resources to complete this review.
Data availability statement
All data that support the findings of this study are included within the article (and any supplementary files).
Funding and Declaration
Benita S Mackay: conceptualization, Investigation, Visualization, Data Curation, Writing - original draft. Karen Marshall: investigation, Writing - original draft. James A Grant-Jacob, Janos Kanczler, and Robert W Eason: supervision, Writing - review & editing. Richard O C Oreffo and Ben Mills: supervision, Funding acquisition, Project administration, Writing - review & editing.
Declaration of competing interest
The authors declare no conflicts of interest.