Abstract
Scaffold-based tissue engineering approaches have been commonly used for skin regeneration or wound healings caused by diseases or trauma. For an ideal complete healing process, scaffold structures need to meet the criteria of biocompatibility, biodegradability, and antimicrobial properties, as well as to provide geometrical necessities for the regeneration of damaged tissue. In this study, design, synthesis and characterization of a three dimensional (3D) printable copolymer based on polycaprolactone-block-poly(1,3-propylene succinate) (PCL-PPSu) including anti-microbial silver particles is presented. 3D printing of PCL-PPSu copolymers provided a lower processing temperature compared to neat PCL, hence, inclusion of temperature-sensitive bioactive reagents into the developed copolymer could be realized. In addition, 3D printed block copolymer showed an enhanced hydrolytic and enzymatic degradation behavior. Cell viability and cytotoxicity of the developed copolymer were evaluated by using human dermal fibroblast (HDF) cells. The addition of silver nitrate within the polymer matrix resulted in a significant decrease in the adhesion of different types of microorganisms on the scaffold without inducing any cytotoxicity on HDF cells in vitro. The results suggested that 3D printed PCL-PPSu scaffolds containing anti-microbial silver particles could be considered as a promising biomaterial for emerging skin regenerative therapies, in the light of its adaptability to 3D printing technology, low-processing temperature, enhanced degradation behavior and antimicrobial properties.
Export citation and abstract BibTeX RIS
1. Introduction
The skin plays a crucial role as the largest tissue in the body, which acts as a thermal, mechanical, and bacterial barrier. Severe skin injuries caused by burns, traumas or wounds may not heal by themselves and require skin tissue substitutes [1–4]. To mimic the network topology of the extracellular matrix (ECM) of the skin, a highly porous 3D structure with interconnected pores is essential to support cell migration and proliferation, nutrient and waste transport, and vascularization [5–8]. Control over the structural and geometrical features of 3D scaffolds, as well as the identification of candidate materials with acceptable biocompatibility and similarity of physical and mechanical properties with host tissue, have yet to be addressed [6, 7, 9]. Moreover, the degradation behavior of scaffolds can be tuned with respect to the requirements of tissue engineering applications. In this respect, novel fabrication strategies can play a significant role in producing desired scaffold architectures with tunable material properties [10, 11].
Additive manufacturing (AM), also known as 3D printing, is one of the most promising scaffold fabrication strategies due to its ability to produce complex structures with customized geometries by utilizing computer-aided design (CAD) models [12–14]. Among the AM processes, extrusion-based 3D printing is a popular and widely used technique since it can produce polymeric scaffolds with customized geometries with ease and low-cost [15–17].
Polycaprolactone (PCL) is a semicrystalline polymer widely used for tissue engineering applications due to its biodegradability, non-toxicity, ease of thermal processing, high decomposition temperature, excellent mechanical properties, and the possibility of its incorporation as a composite, physical blend, or copolymer with other biomaterials [18, 19]. However, some challenges such as low degradation rate due to a high degree of crystallinity, hydrophobicity, and relatively high melting temperature, might limit the application of PCL [20]. A common way of addressing these limitations is to blend or copolymerize PCL with other appropriate polymers to tune the rate of degradation and its hydrophilicity, as well as to lower the melting point [20]. Bio-based polymers with monomers from renewable sources have attracted a great deal of attention because of their advantages such as conservation of limited resources, low toxicity, biodegradability, availability and environmental friendliness [21, 22]. Among them, poly(propylene succinate) (PPSu), a biodegradable polyester, is a promising candidate to be copolymerized with PCL since it has a low melting point of around 44 ˚C and a glass transition temperature of around −36 ˚C with a fast degradation rate. Previous studies showed that the addition of PPSu to PCL increased the enzymatic degradation rate and its hydrophilicity [22–25].
Infection is one of the significant healing hindrances for the skin repair procedure. The functionalities of utilized scaffold such as antimicrobial activity, could significantly affect the healing process during post-implantation. Hence, the scaffolds which possess antimicrobial characteristics in addition to the desired geometrical and physicochemical properties would be advantageous [26–28]. To date, different strategies in skin tissue regeneration and wound healing applications have been implemented, including the incorporation of various types of antibiotics and silver compounds [28–32]. Due to the development of the strains of antibiotic-resistant bacteria and the environmental concerns about using antibiotics, alternatives such as incorporating silver into the biopolymers are used in skin regeneration therapies [33, 34].
Furthermore, the effectiveness of antibiotics is usually restricted to certain bacteria, while silver-containing components have shown promising results against a broad spectrum of microorganisms including fungi, viruses, and some Gram-negative and Gram-positive bacteria. Silver, in both metallic and ionic forms, possesses strong antibacterial and anti-inflammatory properties and has been used in many applications such as cosmetics, wound dressing, medical products, and wound healing scaffolds. In addition, it shows low systemic toxicity and can enhance the epithelialization of the wounds and reduce surface inflammation [35–40].
To date, there are numerous attempts at the utilization of PCL and its blends and copolymers with biodegradable polyesters for 3D printing applications [41–43]. To the best of our knowledge, there is only one study about the preparation of poly(butylene succinate)/polylactide blends and further processing by fused deposition modeling (FDM) printing [44]. In this context, this study is the first attempt at the development of 3D printable PCL-PPSu block copolymer with silver-induced biocidal properties, enhanced degradation and a relatively low-processing temperature compared to neat PCL. In this paper, a block copolymer of PCL-PPSu with different molar fractions and composites containing silver nitrate was synthesized and characterized to be used for skin tissue engineering and wound healing applications. The printability of the resultant composites was further investigated and an ideal composition was used for 3D printing of well-defined porous scaffolds. Degradation behavior and hydrophilicity of the selected copolymer were investigated by enzymatic/hydrolytic degradation studies and contact angle measurements, respectively. Biocompatibility of the composition was evaluated by viability analysis of human dermal fibroblast (HDF) cells in vitro. Furthermore, the antimicrobial activities of the scaffolds were analyzed with microbial cell attachment and zone of inhibition test for C. albicans, P. aeruginosa, E. coli, and S. aureus microorganisms.
2. Materials and methods
2.1. Materials
Succinic acid (98%) and sodium nitrate (99.5%) were purchased from Fluka. Stannous octoate, ε-caprolactone (97%), 1, 3-propanediol (99%), tetrabutyl titanate (TBT, 97%), methanol (99%), chloroform (97%), sodium chloride (NaCl), silver nitrate (AgNO3), yeast extract, peptone from casein, pancreatic digest, ethanol (≥99.8%), Lipase Pseudomonas cepacia, and dimethyl sulfoxide (DMSO) were obtained from Sigma-Aldrich. Chloroform, nitric acid (HNO3), and sodium borohydride were purchased from Merck. HDF cells and all the microorganisms used in the antibacterial studies were purchased from ATCC (UK). Dulbecco's modified Eagle medium (1X DMEM, 4.5 g l−1 D-Glucose, L-glutamine, sodium pyruvate), fetal bovine serum (FBS), HyClone phosphate-buffered saline (10 X PBS, w/o calcium, magnesium), and Pen-Strep (10 000 Units ml−1 penicillin, 10,000 μg ml−1 streptomycin) were obtained from Gibco (UK). Cell proliferation reagent WST-1 was purchased from Roche. PBS tablets were obtained from MP Biomedicals, LLC (France).
2.2. Synthesis of polycaprolactone
PCL was synthesized via ring-opening polymerization of ε-caprolactone. Briefly, ε-caprolactone (50 g) and stannous octoate (20 μl) monomers were placed in a round bottom flask and heated to 180 °C and maintained for 24 h. After completion of the reaction, the product was dissolved in chloroform (50 ml) and precipitated in cold methanol. Then, the solution was kept at −20 °C overnight and followed by a filtration process. The product was dried in a vacuum oven for two days for solvent removal.
2.3. Synthesis of poly(1, 3 propylene succinate)
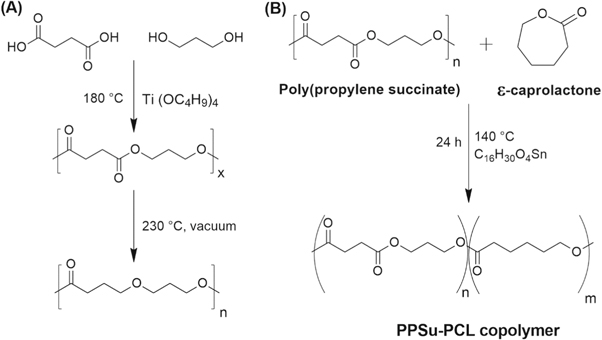
PPSu was synthesized based on a two-stage melt polycondensation polymerization method. In the first step, succinic acid (30 g, 0.254 mol) and 1, 3-propanediol (20.24 g, 0.2794 mol) were charged into a reactor in the presence of TBT (26 μl, 76.2 μmol) as the catalyst. The reaction mixture was heated up to 180 °C under nitrogen atmosphere through refluxing for 90 min. During this step, succinic acid monomers were esterified to form oligomers. In the second stage, the reaction temperature was increased up to 200 °C while a low vacuum (0.8 bar) was applied for 1 h and then a high vacuum (1.5 mbar) was used for 2 h at 220 °C. The reaction was kept overnight at 230 °C under a low vacuum (0.8 bar) to complete the second stage of the reaction and obtain PPSu polymer. The reaction product was dissolved into 200 ml methanol and placed at 4 °C for 24 h. The white solid precipitates were separated from the solvent by filtration and dried for 2 d under vacuum. Figure 1(A) depicts the reaction route of PPSu polymerization, and table 1 summarizes the reaction conditions and molecular weight of PPSu.
Figure 1. Reaction routes of (A) PPSu, and (B) PCL-PPSu block copolymer.
Download figure:
Standard image High-resolution imageTable 1. The number of reactants in the polymer synthesis and molecular weight of PPSu.
| Polymer type | Succinic acid (g) | 1, 3-propanediol (g) | Ti(OBu)4 (μl) | Yield (%) | Mw (g mol−1) | Mn (g mol−1) |
|---|---|---|---|---|---|---|
| PPSu | 30 | 20.24 | 26 | 54 | 4201 | 3388 |
2.4. Synthesis of PCL-PPSu block copolymers
PCL-PPSu block copolymer was synthesized through a bulk polymerization technique. PPSu to ε-caprolactone monomer was in the weight ratio of 1:10 by using 2 g of PPSu and 20 g of ε-caprolactone monomers in the presence of stannous octoate (0.05 wt% of ε-caprolactone) at 140 °C for 24 h. Afterward, the resultant product was dissolved into chloroform (50 ml) and was precipitated by adding cold methanol (300 ml). The resultant polymer was separated from the solvent and unreacted monomers were removed by filtration and then the material was kept drying in vacuum for three days at room temperature. In addition to 1:10 weight ratio, PPSu was undergone polymerization with ε-caprolactone monomer in the ratio of 1:5. PPSu-PCL having 1:10 weight ratio showed better printing behavior due to the differences in number average molecular weight and polydispersity index (PDI) of these two synthesized polymers. The details about experimental conditions and characterization results are given in table S1 and S2 is (available online at stacks.iop.org/BMM/15/035015/mmedia) in the supplementary document.
Functional groups of the polymers were investigated by Nicolet iS 10 Fourier transform infrared spectroscopy (FTIR). FTIR spectra were obtained in %Transmittance mode in the spectral region of 400 to 4000 cm−1 using a resolution of 2 cm−1 and 32 co-added scans. 1H- and 13C- nuclear magnetic resonance spectroscopy (1H-NMR and 13C-NMR) were used to evaluate the composition and structure of the synthesized polymers (Varian Unity Inova, 500 MHz spectrometer). The thermal behavior of the polymers was analyzed by differential scanning calorimetry (DSC) using a TA-Q2000 instrument within the temperature range of −50 to 100 °C at heating and cooling ramp of 10 °C min−1 under nitrogen atmosphere. The molecular weight and PDI of polymers were determined by Viscotek-VE2001 gel permeation chromatography (GPC) in DMF. Among all the combinations of the synthesized copolymers, the initial printability tests were performed to screen the suitable candidate material for 3D printing (data not shown). The addition of PPSu at higher fractions resulted in increased elasticity of copolymer, which resulted in poor printability. For 3D printing, antibacterial and in vitro experiments, the copolymer with PPSu:PCL ratio of 1:10 was selected.
2.5. Contact angle measurement
Apparent change in the wettability of the copolymers compared with PCL was assessed using water contact angle measurements (Terra Lab, Turkey). Samples of PCL and PCL-PPSu were prepared by solution casting, and a deionized water droplet was placed onto the surface of polymer films (4 samples per each polymer), and the calculated contact angle values were presented as mean ±SD.
2.6. Enzymatic degradation
PCL and PCL-PPSu based polymeric films with the same size and mass were prepared by casting. PBS (0.01 M, 1×) with a pH of 7.4 and 1X concentration was prepared by dissolving tablets in Milli Q water. Lipase Pseudomonas cepacia enzyme was dissolved in PBS at the concentration of 0.15 mg ml−1 of PBS, and the enzyme concentration for the polymer was 0.4 unit mg−1.
Degradation tests were conducted in two different groups of PCL and PCL-PPSu copolymer films. Samples were kept in vials containing medium and were incubated at 37 °C at 100 rpm for up to 10 d. The medium was changed every 3 d. At specific time points, the polymer films were removed from the media and washed three times with deionized water followed by drying in a vacuum oven at room temperature until reaching a constant weight. Each sample was weighed before and after soaking in media. The percentage mass loss of the samples was calculated using equation (1) as an indicator of the degree of biodegradability, where Mi and Mf are the initial and the final mass of the samples, respectively.
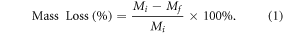
The surface morphology of the polymer films at different time points was observed using Zeiss Leo Supra VP 35 field emission scanning electron microscope (FE-SEM) at 4 kV acceleration voltage. Samples were sputter-coated with a thin layer of gold-palladium by a Denton Vacuum Desk V sputter before SEM imaging.
2.7. Hydrolytic degradation
Films of PCL and PCL-PPSu block copolymer were prepared by solution casting with prolonged drying time to ensure complete solvent removal. Hydrolytic degradation tests were conducted in two different mediums, including PBS, with a pH of 7.4 and DMEM cell culture media. Polymer films with the same dimensions were kept at an equal amount of media and maintained at 37 °C with constant shaking of 180 rpm in an incubator shaker for up to 21 d. On specific days, the polymer films were removed from the media and washed with deionized water and dried in a vacuum oven at 30 °C. The same protocol as mentioned in section 2.6 was followed to measure the degree of degradability.
2.8. Analysis of silver ion release and silver distribution
Silver release behavior of PCL-PPSu/AgNO3 composites was studied by soaking them in PBS at 37 °C and shaking a rate of 50 rpm over 21 d. The samples were selected at the same size and shape to eliminate the effect of their size and shape for ionic release. PBS solution was not changed during the analysis to measure the cumulative release amount. At specific time points, silver ion concentration was measured by inductively coupled plasma optical emission spectrometry (ICP-OES) (Agilent Technologies, model 5110). Polymer samples used for these measurements were impregnated with 1% (wt/wt) silver nitrate.
Both copolymer films and solutions were characterized to measure the amount of silver released in PBS solution and total silver within the polymer. Acid digestion was performed for sample preparation. Briefly, copolymer films were dissolved in 5 ml of HNO3 and kept on a hot plate for 10 min for complete dissolution. Afterward, the solution was diluted with a 20-fold dilution factor and filtered for further analysis. The same protocol was followed for PBS solutions with a dilution factor of 20. To evaluate the homogeneity of silver distribution in the polymer matrix, samples were analyzed with FE-SEM equipped with energy dispersive x-ray spectroscopy (EDS). Polymer films were sputter-coated with gold-palladium before imaging.
2.9. 3D printing process
A custom-built computer numerical control (CNC) 3D printer setup was utilized in this work explained elsewhere [45]. Briefly, the motorized printer head was connected to a computer and controlled by MACH3 CNC software. Rhinoceros 5 (Robert McNeel & Associates) was used to generate G-codes for desired printing patterns. The printer head mounted on the z-axis consisted of a 10-ml metal syringe and a nozzle with an inner diameter of 150 μm (Musashi Engineering Inc., Japan) heated by a heating jacket (New Era Pump Systems, Inc. NY, USA). A pneumatic dispensing system (Nordson EFD Performus V) was connected to the syringe. Figure 2 represents a schematic view of the 3D printer machine and the printing setup during printing. The printing temperature was selected based on thermal properties obtained by DSC measurements. The synthesized copolymer impregnated with silver nitrate was printed at 55 °C (the selection of temperature was based on the offset temperature of melting point in DSC data and the corresponding viscosity) while the samples were thermally equilibrated at 55 °C for 2 h prior printing. The applied pressure and print speed were set as 2 bars and 210 mm min−1, respectively. The extruded molten polymer was solidified on a collector at room temperature by the aid of a small cooling fan with the current of 80 mA placed 2.5 cm far from the nozzle. Cuboid structures with dimensions of 10 × 10 × 2 mm (15 consecutive layers) and gap size of 350 μm were 3D printed. Increments along the Z-axis of each layer was selected the same as the nozzle size (150 μm). Morphology of the printed scaffolds was investigated by FE-SEM. Pore size, filament diameter, and the variation of deposition profiles and shape fidelity of the printed samples were analyzed by using ImageJ software on the obtained SEM images. Measurements were done from 20 random regions of the scaffolds at different layers of each SEM picture. The data were reported as mean ±standard deviation.
Figure 2. (A) Schematic view of the custom-made 3D printer, (B) real image of the printing setup during the scaffold printing.
Download figure:
Standard image High-resolution image2.10. Biocompatibility evaluation
Cell viability and biocompatibility of the 3D printed constructs was evaluated by cell viability and morphology analysis after treatment of HDF cells with the extracts of the scaffolds. The extract samples were prepared based on the instructions at ISO 10993–12 with small modifications. Briefly, a 20 mg ml−1 of the 3D printed PCL, PCL/AgNO3, PCL-PPSu, and PCL-PPSu/AgNO3 scaffolds were incubated in fibroblast basal media for 72 h at 37 °C at 150 rpm. After the incubation period, the liquid portion was taken and filtered through a 0.22 μm filter. Previous studies showed that embedding 5% of AgNO3 into polymer scaffolds improved the antibacterial properties of the scaffolds with no significant cytotoxic effect on the cells [46, 47]. The optimum concentration was assessed by incorporation of 5% (wt/wt) AgNO3 in the polymer matrix and further examination of cell viability.
HDF cells were cultured in fibroblast basal media (ATCC, PCS-201-030) containing fibroblast growth kit-serum-free (ATCC, PCS-201-040) and 1% Penicillin-Streptomycin Ampicillin (PSA). The cells were maintained at 37 °C under an atmosphere composed of 95% air and 5% CO2. Cells were seeded in 96-well plate at a density of 5 × 103 cells/well and incubated for 24 h. After the incubation period, several dilutions of the extracts (100%, 50%, 25%, 12.5%, 6.2%, 3.1%, 1.6%, 0.8% and 0.4%) were added into the cell culture followed by incubation for 24 h. Fibroblast basal media including 5% PBS and 5% DMSO were used as negative and positive controls, respectively. At the end of the incubation, the media was removed, and cells were rinsed with PBS. Cell viability was determined using the WST-1 colorimetric assay.
For morphological characterization, cells were seeded in 24-well plate (25 × 103 cells/well) and incubated for 24 h. Dilution of the extracts of the 3D printed samples with concentrations of 100%, 50%, and 25% were added into the wells and were incubated for 24 h. Morphological changes were monitored using a Zeiss PrimoVert light microscope.
2.11. Antibacterial activity of block copolymer impregnated with silver nitrate
The synthesized PCL-PPSu copolymer and PCL were dissolved in chloroform, and the solution of AgNO3 in ethanol was added with final concentrations of 1% and 2.5% (wt/wt). Concentrations of AgNO3 were selected based on in vitro cytotoxicity assessments. Antimicrobial activities of rectangular 5 × 5 mm polymer films were determined by examining the zone forming potential through diffusion using model microorganisms including Gram-negative P. aeruginosa and E. coli, Gram-positive bacteria S. aureus, and yeast C. albicans. A 200 μl of microorganism cultures grown overnight were added by spreading on lysogeny broth (LB) agar Petri dishes and then, rectangular polymer materials with/without AgNO3 were placed on LB agar Petri dishes and incubated at 37 °C overnight. Antimicrobial activity was determined by observing the presence of the zone of inhibition around the copolymer films after the incubation period and the diameter of each inhabitation zone was measured and reported in mm.
Adhesion of microorganisms on the copolymer surfaces was investigated. Briefly, the overnight microorganism cultures were seeded onto the PCL-PPSu and PCL-PPSu/AgNO3 copolymer films and incubated at 37 °C for 3 h. Incubation was continued overnight at 80 rpm with the addition of fresh LB medium. At the end of the incubation time, copolymer films were removed from the medium and subjected to washing with 0.9% NaCl (three times). The copolymer films taken from the saline solution (0.9% NaCl) were sonicated for 3 min to release the bacteria into the saline solution. 100 μl of saline solutions were spread on LB agar Petri dishes and incubated at 37 °C overnight. After incubation, the growth densities on LB agar Petri dishes were monitored.
2.12. Statistical analysis
The statistical evaluations for cell viability were performed using t-test, and cell viability data, hydrolytic and enzymatic degradation values and silver ion concentrations were expressed as the means ±SD with at least three experiments. Those with p values <0.01 were considered as significant differences.
3. Results and discussion
3.1. Structural analysis of PCL-PPSu block copolymers
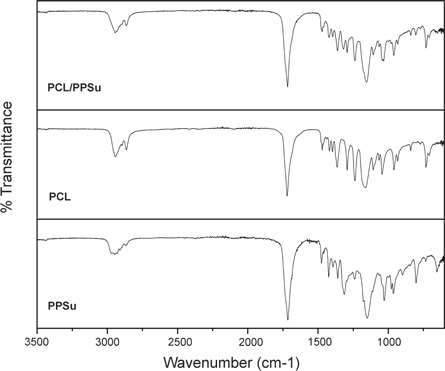
FTIR spectra of PCL, PPSu, and PCL-PPSu block copolymer are illustrated in figure 3. PCL exhibited a sharp peak at 1720 cm−1, which is due to carbonyl stretching, while the two weak peaks at 2944 cm−1 and 2863 cm−1 correspond to the asymmetric and symmetric –C–H stretching. Additionally, the peak at 1176 cm−1 is assigned to the –C–O stretching [48]. In the PPSu spectra, the peak at 1715 cm−1 indicates the presence of the carbonyl group in the polymer structure [49]. The weak peak at 2971 cm−1 corresponds to the stretching of –C–H bond related to the ester group. Furthermore, 1150 cm−1 is attributed to the bending of –C–O– ester bond [50, 51]. The presence of characteristic peaks of both PPSu and PCL in the spectrum of PCL-PPSu block copolymer confirms the formation of the copolymer structure with both PPSu and PCL segments.
Figure 3. FTIR spectra of PCL-PPSu block copolymer, PCL and PPSu polymers.
Download figure:
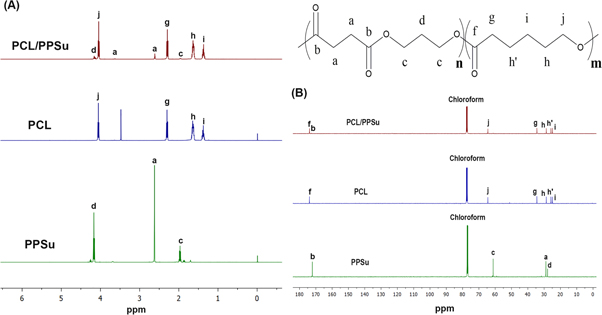
Standard image High-resolution imageNMR analysis was used to identify the molecular structure of the synthesized polymers. Figure 4(A) represents the 1H-NMR spectra of PCL, PPSu, and PCL-PPSu polymers. In the 1H-NMR spectrum of PPSu polymer, the quintuple peak at 1.94–2.01 ppm corresponds to a methylene group (c) of the 1, 3-propanediol fragment. The characteristic peak at 4.1–4.2 ppm is attributed to the protons of two similar methylene groups (a) while the protons of two methylene groups on succinic acid monomer (a) appear as a single peak at 2.6 ppm. On the other hand, PCL gives a triple peak because of protons on the methylene group in the neighbor of oxygen (j) at 4.05–4.1 ppm. The protons of methylene group (h, hʹ) appear as multiple peaks at 1.6–1.7 ppm. The (i) group protons is identified by a quintuple peak at 1.4 ppm, while (g) protons appear as a triple peak at 2.25 ppm [23]. 1H-NMR spectrum of PCL-PPSu showed peaks in the same regions without any significant shift confirming the presence of both segments in the structure of the copolymer. At 3.63 ppm, a small triple peak corresponds to methylene protons of hydroxyl terminated ends (–CH2–OH) of polyesters which also exists in the copolymer [51]. The changes are remarkable in the peak area which corresponds to the molar fraction of the two polymers of PCL and PPSu with the same ratio of 1:10 for PPSu:PCL [23]. The peak integral values and normalized values to (c) peak are summarized in table S3 in the supplementary document.
Figure 4. (A) 1H-NMR and (B) 13C-NMR spectra of PPSu, PCL and PCL-PPSu block copolymer.
Download figure:
Standard image High-resolution image13C-NMR spectra of the synthesized polymers are presented in figure 4(B). 13C-NMR spectrum of PPSu showed a peak at 172.3 ppm which attributes to the carbonyl carbon in the polymer structure (b), the peak at 61.4 ppm is assigned to the (c) carbon in PPSu molecular structure. In addition, the peaks at 29.1 and 28.9 ppm correspond to methylene carbons of succinic acid fragment (a), and the methylene group of 1,3-propanediol (d) fragment, respectively[52]. In the 13C-NMR spectrum of PCL, the peak at 173.7 ppm is attributed to carbonyl group (f), 64.2 ppm peak is assigned to (j) carbon, the peak at 34.2 ppm corresponds to methylene group adjacent to carbonyl group (g), 27.9, 25.6, and 24.6 ppm peaks correspond to methylene groups (h), (hʹ) and (i), respectively [53]. However, overlapping of PPSu and PCL peaks and the dominance of PCL peaks in 13C-NMR spectra limits the ability of structural identification of PCL-PPSu through 13C-NMR.
3.2. Thermal behavior and 3D printing of PCL-PPSu block copolymers
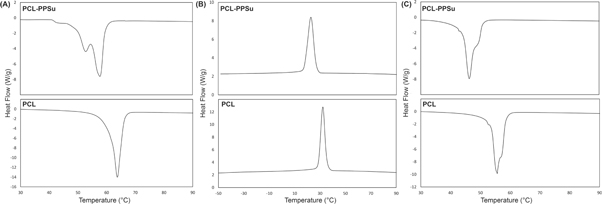
DSC curves for the heating and cooling cycles of PCL and PCL-PPSu copolymer are presented in figure 5. Both polymers underwent crystallization during corresponding cooling cycles. The onset of the crystallization peak for PCL was at 38 °C (Tc), and the maximum value was at 32 °C. On the other hand, the crystallization of PCL-PPSu started at 28 °C, and the maximum value was set at 23 °C. The heat of crystallization (ΔHc) obtained from cooling cycles showed the state of the crystallization and its kinetics. The heat of crystallization (ΔHc) for PCL was 72 J g−1 while the heat released during the crystallization of the copolymer was 32 J g−1. The higher crystallization of PCL stems from its higher PDI providing higher chain mobility compared to PCL-PPSu block copolymer [54]. The copolymer's melting point in the second heating cycle occurred at a lower temperature than in the PCL sample (42 °C and 50 °C for the copolymer and PCL, respectively). The PCL-PPSu graph represents a semicrystalline behavior with a double melting point in the first heating cycle which could be attributed to the different blockchains of the copolymer [23]. In addition, the heat of melting (ΔHm) showed a similar trend as ΔHc as presented in table S2, due to the difference in kinetics of crystallization of PCL-PPSu.
Figure 5. DSC curves of the PCL-PPSu and PCL; (A) the first heating cycle, (B) the first cooling cycle, (C) the second heating cycle. The heating and cooling cycles were conducted with a rate of 10 °C min−1.
Download figure:
Standard image High-resolution imageProcessing temperature plays a critical role in the 3D printing of materials in the melt phase. The mutual dependence of melt viscosity and chemical stability of the polymers on process temperature have been already discussed in several studies [54–56]. The processing temperature of the thermoplastics would also impact the thermal degradation and accordingly, chemical stability of the polymer would be compromised upon increasing the temperature [57]. On the other hand, the feasibility of embedding drugs and nanoparticles in the polymer is strongly affected by the temperature in which the inclusions could be utilized without degradation and any further effect on the drug release [58]. Hence, lowering the temperature could broaden the application of the candidate drugs and nanoparticles. Moreover, the lower processing temperature during 3D printing would extend the lifetime of the embedded drugs. In the last few decades, many studies have been done to investigate the thermal stability of the drugs, and they are categorized as resistant to low, moderate, high temperatures, or non-heat resistant [58, 59]. However, these studies have mostly explored the effect of temperature for a short period of time [60]. In this respect, lower melting temperature observed in PCL-PPSu copolymer compared to PCL might be an advantage in extending the stability and application window of both the polymer and possibly the embedded drugs.
The heating and cooling profiles during 3D printing were designed by considering DSC results. In this way, polymer melt in the syringe reservoir was slowly cooled down below the crystallization temperature between each run to allow the complete development of a crystalline structure. Uniformity of the flow properties and repeatability of the 3D printing process significantly depend on thermal history and the processing conditions. The development of a fully crystalline structure in the syringe reservoir before each printing trial minimized the alteration of flow profile at the same processing conditions.
The structure and morphology of the 3D printed scaffolds were observed using FE-SEM (figure 6). The interconnected porous structure and structural uniformity of a 3D printed scaffold are of great importance. Hence, to evaluate the structural uniformity and the material's printability, filament diameter, and inter-layer gap distance of the printed structures were measured from SEM images using ImageJ software. The samples were cut in the z-axis and the gap distance were measured form different layers. Each measurement was reported as an average of 20 random locations in the 3D printed scaffold. The structure stability was assessed by the determination of standard deviation (SD) of the calculated values. The measured printed filament diameter was 272.40 μm ±5.89 (SD), and the inter-layer gap distance was calculated as 330.80 μm ±4.04 (SD). The low values of the standard deviations demonstrated that 3D printed scaffolds possessed structural uniformity in terms of filament diameter and pore size. This indicates that the selection of process parameters based on the physical characteristics of the synthesized copolymer resulted in good adaptation with the 3D printing process. Designed gap size was selected to be 350 μm and due to filament expansion during printing, it reduced to 330 μm. By selecting a low pressure with a high print speed, we could optimize the resulting structure in terms of filament length and gap distance. Figure 6(D) represents a cross-section image of the scaffold to evaluate the interconnectivity of the pores. SEM images and the subsequent image analysis showed that the synthesized copolymer composite could be 3D printed with high structural uniformity and controlled geometry at the examined process conditions. It has been shown that interconnected porous structure of the 3D printed scaffolds can provide proper nutrient and oxygen flow for the seeded cells and support the formation of new tissue upon implantation.
Figure 6. SEM images of 3D printed copolymer impregnated with silver nitrate scaffolds at different magnifications; scale bars: 200 μm.
Download figure:
Standard image High-resolution image3.3. Degradation behavior
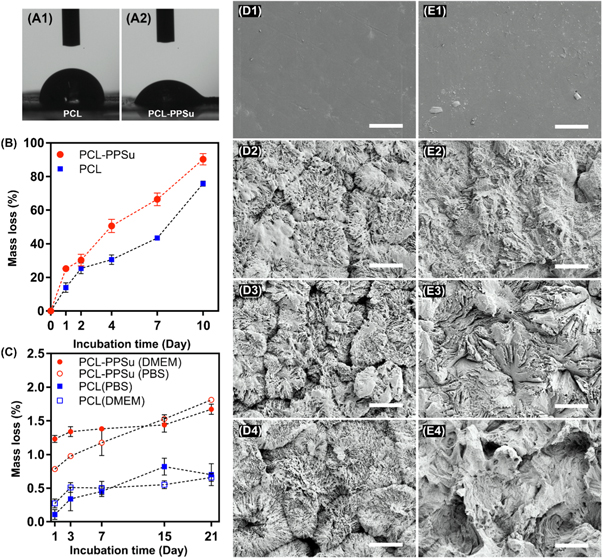
Surface wettability and the degree of hydrophilicity of a biomaterial is correlated with its cell attachment properties and possibly the hydrolytic degradation behavior [61]. Results of the water contact angle measurement for PCL and PCL-PPSu showed a significant increase in the wettability of copolymer compared with PCL. The measured contact angle values for PCL and PCL-PPSu samples were 87.43° ± 4.7 and 61.23° ± 3.5, respectively (figure 7(A)). As the figure implies, the wettability and hence the hydrophilicity of the copolymer enhanced significantly in comparison with pure PCL.
Figure 7. Wettability and degradation of PCL-PPSu and PCL in different media. Water contact angle measurements for (A1) PCL and (A2) PCL-PPSu. (B) Enzymatic degradation as a function of incubation time at 37 °C for PCL-PPSu and PCL. (C) Hydrolytic degradation as a function of incubation time at 37 °C for PCL-PPSu and PCL in PBS and DMEM. SEM micrographs of degraded PCL-PPSu films (D1–D4) and PCL films (E1–E4) at day 0, day 2, day 7, and day 10 of enzymatic degradation; scale bars: 20 μm.
Download figure:
Standard image High-resolution imageEnzymatic hydrolysis of the PCL and PCL-PPSu samples in PBS solutions containing Pseudomonas cepacia lipase, which degrades polyesters by breaking the ester bonds [21, 62], at 37 °C were investigated by monitoring mass loss and morphology of the samples during 10 d. The percentage of mass loss versus time shows much higher values of mass loss with a steeper slope for the copolymer compared with PCL (figure 7(B)). After 1 d incubation, the mass loss percentage for the copolymer was 25.19%, while this value was almost half (13.90%) for the PCL. The values of mass loss percentage for PCL-PPSu copolymer and PCL after 10 d were 90.2% and 75.8%, respectively. The same behavior were observed, as the incubation of copolymer samples more than 10 d resulted in complete physical destruction and since the accurate measurement of mass loss was not further possible, this point was considered as the 100% degradation state. The copolymer was completely degraded after 11 d while the corresponding time for PCL was at day 14 (data for 100% degradation was not shown in the graph). The extent of degradation at every time points and also the slope of the graph were both higher for the copolymer compared with PCL which is in agreement with previous studies [25, 63].
The cleavage of ester end groups of the PCL and PCL-PPSu samples during enzymatic degradation would be affected by the degree of crystallinity. Due to the greater number of ester groups in the copolymer, more available sites for bond cleavage would be expected [62, 64, 65]. Knowing the fact that the degree of crystallinity is related to the polymer's molecular weight, the lower degradation rate of PCL compared to PCL-PPSu was in conformity with the molecular weight data obtained from GPC [66]. The number of methylene groups between the ester groups is another important key factor in the hydrolysis of polyesters, and as it rises, degradability increases [67]. This was in agreement with the observed higher degradation rate of PCL-PPSu copolymer.
Figures 7(D) and (E) illustrate SEM images of surface morphology of the polymer films before and after hydrolysis at different time points. Before the experiments, the polymer films demonstrated smooth surfaces (figure 7(D1) and (E1)). Further incubation resulted in the formation of a flower-like morphology in PCL-PPSu surface (figure 7(D2)–(D4)) corresponding to spherulites formation during crystallization [68]. After 2 d of hydrolysis and attaching the lipase to polyester surfaces [21], portions of these spherulites were seemed to be degraded and dissolved in the solution which is suggested to be related to the amorphous regions, while the crystalline segments remained intact (figure 7(D2)) [65, 68]. By further increasing the hydrolysis time, increased surface roughness was observed in both PCL and copolymer samples. Enzymatic degradation is a heterogeneous process that initiates from the surface, and the crystallinity of the polymer plays a vital role in localized degradation progress, which was observed in both samples. Higher crystallinity of PCL samples resulted in slower rates of hydrolysis at the same time intervals, which was evident in morphological features of degraded samples as well as mass loss values throughout the incubation.
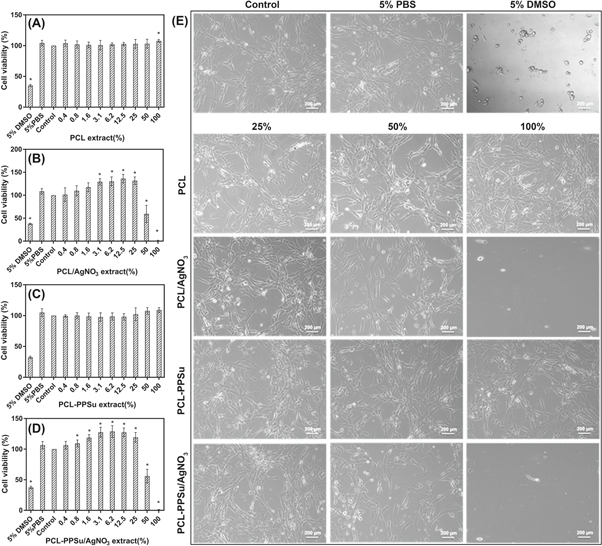
Figure 8. Cytotoxicity of the extracts of the components to HDF cells in vitro. Negative controls: PBS. Positive control: 5% DMSO in PBS. Control: Cells without scaffolds. Cytotoxicity of leachates derived from (A) PCL, (B) PCL/AgNO3, (C) PCL-PPSu and (D) PCL-PPSu/AgNO3 (*p < 0.01). Bright-field microscopy images of HDF cells after being exposed to leachates are provided in panel (E).
Download figure:
Standard image High-resolution imageBiocompatibility of PCL has been a subject of debate due to the slow rate of hydrolytic degradation which can easily span over a year [69]. As well as the higher rate of enzymatic degradation in copolymer samples, the apparent increase in wettability and the higher chance of ester cleavage in PPSu containing copolymer might have a significant impact on hydrolytic degradation. Hydrolytic degradation of the PCL and PCL-PPSu samples in DMEM and PBS media were examined by monitoring mass loss at different time points up to 21 d. The percentage of mass loss versus time shows higher values of mass loss for the copolymer compared with PCL in both media (figure 7(C)). The values of mass loss percentage for PCL-PPSu copolymer after 21 d of immersion in DMEM and PBS were 1.67% and 1.81%, respectively, while the corresponding values for PCL samples were decreased by more than half. The calculated mass loss percentage at day 21 for PCL in DMEM and PBS were 0.66% and 0.70%, respectively. A small difference in the degradation of samples in different mediums was observed. However, samples incubated in PBS showed higher mass change from the first day to the last day of the experiments. The samples incubated in DMEM medium showed a relatively high mass loss at day 1 while the rate of degradation was slower throughout the experiments. Regardless of the sample type and the medium, the mass loss percentage increased with sample incubation time.
The degree of crystallinity directly influences the hydrolytic degradation rate, which is mainly due to the cleavage of ester bonds. Since the number of ester groups in the copolymer is higher compared to PCL, the observed increase in the rate of hydrolysis was expected [64]. Considering the apparent increase in wettability of copolymers, the mutual effects of more available cleavage bonds, and the more hydrophilic nature of copolymer resulted in faster degradation rates. The reported mass loss during the three week period might seem to be negligible. However, considering the very slow rate of hydrolytic degradation of PCL which might be over several months, the observed 50% increase in the mass loss of copolymers during the short period of experiments would be promising.
3.4. Silver release and surface characterization of polymer films
The concentration of silver ion released from PCL-PPSu/AgNO3 soaked in PBS was measured by ICP-OES for 21 days of incubation. Moreover, the released ion content at each time point was compared with the total amount of silver within the bulk of the samples (table 2). The reported data belongs to the average value of 4 replicates with the corresponding standard deviations. The results demonstrate a very low released silver ions (about 4.5–7.5 mg kg−1), whereas the silver content within the the bulk of the samples were almost one thousand times higher. This confirms the hypothetis that the amount of released silver ions during the incubation period stayed below the cytotoxic level [70].
Table 2. Silver amount in PBS solutions and PCL-PPSu/AgNO3 measured with ICP-OES (The values are reported in ppm).
| Incubation time | Day 1 | Day 3 | Day 7 | Day 14 | Day 21 |
|---|---|---|---|---|---|
| Sample | Silver content (ppm) | ||||
| PBS solution | 4.35 ± 1.18 | 5.41 ± 1.92 | 6.32 ± 0.48 | 7.45 ± 1.50 | 7.56 ± 1.40 |
| Copolymer | 5581.99 ± 8.5 | 4707.92 ± 5.3 | 4396.48 ± 7.4 | 4344.17 ± 6.5 | 4210.50 ± 4.5 |
It is worth mentioning that low standard deviation of silver content in bulk samples indicates a homogeneous silver distribution through out the polymer.
Figure S1(B) shows EDS (map) of PCL-PPSu/AgNO3 EDS, which demonstrates a uniform distribution of silver on the surface of the sample.
3.5. Cell viability assessment
Cytotoxicity of PCL, PCL-PPSu, and the corresponding AgNO3 containing composites on HDF cells were investigated by using WST-1 colorimetric assay. Exposure of the extracts of PCL and PCL-PPSu samples did not affect cell viability on cultured cells compared with control groups (figures 8(A) and (C)). The results of viability assessments after exposure of several dilutions of the extracts from composite samples containing 5% (wt/wt) AgNO3 are presented in figures 8(B) and (D). The high concentration of AgNO3 caused a dramatic loss of viability, which can be due to the release of toxic Ag+ ions from the scaffold to the medium [46]. However, further dilutions of the extracts from the samples resulted in a gradual increase in the viability. Regardless of the polymer matrix, a significant increase in the viability of cells treated with 25% and less diluted media was observed.
The images in figure 8(E) show the morphology of HDF cells exposed to the polymer extracts after 24 h. Morphological characterization was performed using 100%, 50%, and 25% diluted media since no toxic effect was observed for other tested concentrations. Cells treated with PCL and PCL-PPSu extracts maintained their characteristic morphology for all tested concentrations. However, the highest concentration of PCL/AgNO3 and PCL-PPSu/AgNO3 extracts resulted in a significant loss of cell morphology and detachment of the cells from the surface due to the cytotoxic effect of Ag+ released in the medium. Although a few cells seen lost their morphology and detached from the surface, diluted extracts to 50% and 25% maintained the cell morphology. Therefore, the concentrations of AgNO3 were decreased to 2.5% and 1% in the composites for further antibacterial activity experiments.
3.6. Antibacterial activity of block copolymer impregnated with AgNO3
Antimicrobial properties of silver ions, which exhibit a high level of toxicity for a broad range of microorganisms, are promising in medical terms [36]. The polymeric structure, incorporated with AgNO3 showed an improvement in biocidal properties [40, 47]. The concentration of AgNO3 was commonly selected below 5% and both in vitro and in vivo studies possessed good antimicrobial activity with no noticeable toxicity effects [46, 47].
Antimicrobial properties of PCL-PPSu and PCL-PPSu/AgNO3 were determined by zone of inhibition test using E. coli, P. aeruginosa, S. aureus, and C. albicans pathogens, which are related to infections seen in implants or burn wound areas [40]. The microorganism strains were seeded on LB agar Petri dishes by spreading from overnight cultures. Then, the copolymer films with/without AgNO3 were placed carefully on the LB agar.
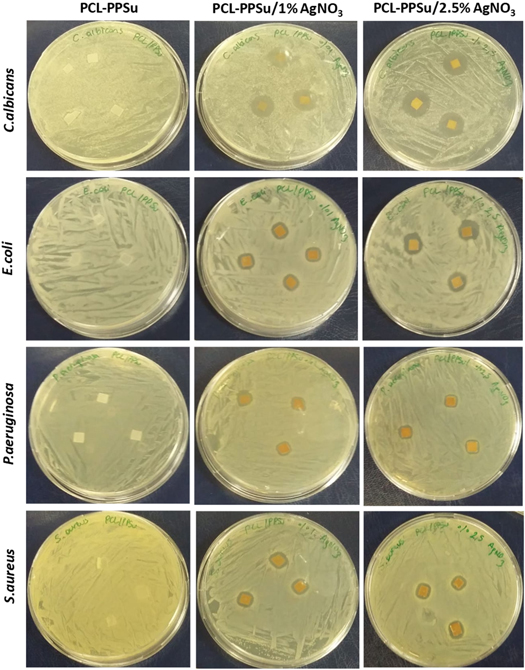
After overnight incubation, no evidence of antimicrobial effects against E. coli, P. aeruginosa, S. aureus, and C. albicans strains was observed in copolymer samples (figure 9). However, copolymers impregnated with AgNO3 exhibited antimicrobial activity against all the tested microorganisms. Evident inhibition zones were observed around the copolymer films with varying sizes corresponding to different microorganisms. As shown in figure 9, PCL-PPSu/AgNO3 films showed positive antimicrobial activity against C. albicans, a harmless member of the human microbiome which can lead to life-threatening infections under certain conditions. Similar antimicrobial activities for P. aeruginosa, E. coli and S. aureus pathogens were observed but to a lesser extent compared to C. albicans. Inhibition zones formed around the PCL-PPSu films with/without AgNO3 are presented in table 3. Data are represented as mean ± SD with 3 replicates.
Figure 9. Zone of inhibition for PCL-PPSu and PCL-PPSu/AgNO3 polymer films against different microorganisms.
Download figure:
Standard image High-resolution imageTable 3. The diameter of zone of inhibition diameter (mm) for samples with varying concentrations of AgNO3 against different microorganisms.
| Sample | Zone of inhibition diameter (mm) | |||
|---|---|---|---|---|
| C. albicans | E. coli | S. aureus | P. aeruginosa | |
| PCL | 0 | 0 | 0 | 0 |
| PCL + 1% AgNO3 | 16.3 ± 1.2 | 9.6 ± 0.6 | 9.3 ± 0.6 | 8.2 ± 0.1 |
| PCL + 2.5% AgNO3 | 17 ± 1.4 | 10.4 ± 0.6 | 10.3 ± 1.2 | 9 ± 0.1 |
| PCL-PPSu | 0 | 0 | 0 | 0 |
| PCL-PPSu + 1% AgNO3 | 14.7 ± 1.5 | 10 ± 0.1 | 9.7 ± 0.6 | 7.9 ± 0.6 |
| PCL-PPSu + 2.5% AgNO3 | 15.3 ± 1.4 | 12.6 ± 1.2 | 10.3 ± 0.6 | 8.2 ± 0.6 |
As shown in table 3, for both PCL and PCL-PPSu composite incorporated with AgNO3, the zone of inhibition diameters are as the following: C. albicans > E. coli > S. aureus > P. aeruginosa. Adhesion of microorganisms on the copolymer film surfaces was also analyzed. The results showed that the microorganism's density on PCL-PPSu copolymer films was quite high for all microorganisms (figure 10). In addition, LB medium incubated with the films became blurry due to the growth of microorganisms. However, no growth was observed in Petri dishes belongs to PCL-PPSu/AgNO3 film surfaces incubated with E. coli and P. aeruginosa, while the density of C. albicans and S. aureus decerased compared to their control groups (figure 10). Moreover, no visible turbidity in the LB media was observed.
Figure 10. Analysis of microorganism adhesion on the PCL-PPSu and PCL-PPSu/AgNO3 films.
Download figure:
Standard image High-resolution image4. Conclusions
In this study, a 3D printable block copolymer of PCL-PPSu with the adjusted composition was synthesized and characterized in detail. The effect of incorporation of PPSu within the PCL as a candidate material for skin tissue engineering applications was investigated. The assessment of structural uniformity of 3D printed structures showed that the selection of process parameters based on physical properties of synthesized copolymer resulted in 3D printing of porous structures with well-defined interconnected porosity, which could support nutrient and oxygen flow in vitro. Moreover, the synthesized PCL-PPSu copolymer showed higher enzymatic and hydrolytic degradation rates and improved hydrophilicity compared to PCL, which can be employed in designing skin regenerative scaffolds with controlled degradation behavior. To provide the antibacterial properties, silver nitrate was incorporated within the copolymers with different concentrations, and its cytotoxicity level was investigated by cell viability assay. Assured that our composite scaffolds did not show any cytoxicity for the composition of 1% (wt/wt) silver nitrate with silver releasing a very little amount of silver in the media, antimicrobial activity was utilized to investigate the effect of this antibacterial agent. It was shown that the incorporation of silver nitrate to the copolymer significantly reduced microbial cell adhesion for all microorganisms investigated. The zone of inhibition test results demonstrated that silver nitrate was more effective for E. coli and C. albicans rather than P. aeruginosa and S. aureus. We have shown that by combining the improved degradation behavior, low-processing temperature, antimicrobial properties, and adaptability with the 3D printing process, composites based on PCL-PPSu could be an attractive biomaterial for emerging skin tissue engineering and wound healing applications.
Acknowledgments
This research was supported by The Scientific and Technological Research Council of Turkey (TUBITAK) (Grant No. 213M687). We would like to thank Dr Mustafa Atilla Yazici for his help with the inductively coupled plasma optical emission spectrometry studies.