Abstract
Human enterovirus B106 (EV-B106) is a new member of the enterovirus B species. To date, only three nucleotide sequences of EV-B106 have been published, and only one full-length genome sequence (the Yunnan strain 148/YN/CHN/12) is available in the GenBank database. In this study, we conducted phylogenetic characterisation of four EV-B106 strains isolated in Xinjiang, China. Pairwise comparisons of the nucleotide sequences and the deduced amino acid sequences revealed that the four Xinjiang EV-B106 strains had only 80.5–80.8% nucleotide identity and 95.4–97.3% amino acid identity with the Yunnan EV-B106 strain, indicating high mutagenicity. Similarity plots and bootscanning analyses revealed that frequent intertypic recombination occurred in all four Xinjiang EV-B106 strains in the non-structural region. These four strains may share a donor sequence with the EV-B85 strain, which circulated in Xinjiang in 2011, indicating extensive genetic exchanges between these strains. All Xinjiang EV-B106 strains were temperature-sensitive. An antibody seroprevalence study against EV-B106 in two Xinjiang prefectures also showed low titres of neutralizing antibodies, suggesting limited exposure and transmission in the population. This study contributes the whole genome sequences of EV-B106 to the GenBank database and provides valuable information regarding the molecular epidemiology of EV-B106 in China.
Similar content being viewed by others
Introduction
The genus Enterovirus, within family Picornaviridae, order Picornavirales, consists of 12 species: enterovirus A–H, J, and rhinovirus A–C (www.picornaviridae.com). Based on genomic characteristics, human enteroviruses (EV) can be divided into four species: EV-A, EV-B, EV-C, and EV-D, including more than 100 serotypes of polioviruses (PVs), Coxsackie A viruses (CVAs), Coxsackie B viruses (CVBs), echoviruses, and enteroviruses named with digital serial numbers since EV-D68 was designated1,2,3,4. Human enteroviruses are small, non-enveloped, single-stranded, and positive-sense RNA viruses. The genome, composed of approximately 7500 nucleotides (nt), contains a long open reading frame (ORF) flanked by a 5′-untranslated region (UTR) and a 3′-UTR. The ORF can be translated into a 2200-amino acid-long polyprotein, and then cleaved into the 3 polyprotein precursors P1, P2, and P3, which encode the structural proteins VP4, VP2, VP3, and VP1, non-structural proteins 2A, 2B, and 2C, and non-structural proteins 3A, 3B, 3C, and 3D. The mature proteins and precursors, such as 2BC, 3AB, 3CD, participate in replication. The 5′-UTR is roughly 740-nucleotides in length and includes a structure known as the internal ribosome entry site, which is vital for the initiation of translation. The 3′-UTR, consisting of approximately 100 nucleotides containing a long poly (A) stretch, is important in RNA replication5,6.
EVs can cause a wide range of diseases such as acute flaccid paralysis (AFP), acute aseptic meningitis, encephalitis, myocarditis, hand-foot-and-mouth disease, and others. However, most EV infections are asymptomatic or lead to only mild symptoms resembling the common cold or slight febrile diseases. Classification of EVs is now based on variations in the VP1 coding region containing a serotype-specific sequence and is known as the “molecular serotyping” method. In this method, strains possessing more than 75% VP1 nucleotide similarity (85% amino acids similarity) are considered to belong to the same serotype. In contrast, strains with less than 70% VP1 nucleotide similarity are considered to belong to different serotypes. VP1 nucleotide similarity between 70–75% is considered the “grey zone”, for which additional information such as neutralization testing with specific antiserum or sequences of other regions to assist with the identification is required2,7,8,9,10. Because the “molecular serotyping” method has gradually replaced the traditional labour-intensive and time-consuming neutralization test method, it is now the major EV serotyping criterion used in the laboratory.
Enterovirus B106 (EV-B106) is a newly identified serotype within EV-B. The prototype strain (BAN2001-10634) was isolated in Bangladesh in 2001; however, sequences of the prototype strain were not released in the GenBank database. To date, there is only one EV-B106 full-length genome sequence in the GenBank database (strain 12148/YN/CHN/12, hereafter referred to as strain 12148/YN), isolated from an AFP patient in Yunnan Province, China in 201211. Apart from that, only one complete P1 sequence of EV-B106 (strain PAK_NIH_SP1202B, hereafter referred to as strain SP1202B) from Pakistan in 201012 and a partial VP1 sequence of EV-B106 (strain BOL/03-10665A, hereafter referred to as strain 10665A) from Bolivia in 200313 were deposited in the GenBank database. In this study, we determined the full-length genome sequences of four EV-B106 strains (strain HTPS-QDH11F/XJ/CHN/2011, strain KS-KSH28F/XJ/CHN/2011, strain KS-MGTH90F/XJ/CHN/2011, and strain AKS-AWT-AFP2F/XJ/CHN/2011, hereafter referred to as HTPS-QDH11F, KS-KSH28F/XJ, KS-MGTH90F, and AKS-AWT-AFP2F, respectively) isolated in Xinjiang Uygur Autonomous Region of China in 2011. These strains were characterised by phylogenetic analysis, antigenic analysis, and seroepidemiology survey.
Results
Isolation and molecular typing of the four Xinjiang strains
All four viral strains (HTPS-QDH11F, KS-KSH28F/XJ, KS-MGTH90F, and AKS-AWT-AFP2F) were isolated from human rhabdomyosarcoma (RD) cells. After a complete EV-like cytopathic effect (CPE) was observed, the cell cultures were harvested. The four strains were confirmed to belong to EV-B when the VP4 sequences were amplified using the species-specific primer pairs EVP4 and OL68-1 and analysed using an online enterovirus genotyping tool14,15. The entire VP1 sequences of these four EV strains were analysed using the Basic Local Alignment Search Tool (BLAST) server by comparison with sequences available in the GenBank database; EV serotypes were determined using the online enterovirus genotyping tool15. The results indicated that the VP1 sequences of the four Xinjiang strains showed the highest similarity with those of the EV-B106 strain in the GenBank database, which reached a percentage of as high as 92.8%; therefore, based on the EV molecular typing criteria, the four strains were identified as EV-B106.
Full-length genomic analysis of the four Xinjiang EV-B106 strains
The full-length genome sequences of the four Xinjiang EV-B106 strains were determined. The results showed that they were all 7421–7422 nucleotides in length with a single ORF of 6579 nucleotides encoding a single polypeptide of 2192 amino acids. The sequences were flanked by a non-coding 5′-UTR of 741–742 nucleotides and non-coding 3′-UTR of 101 nucleotides, with the latter preceding a long poly(A) tail. The overall compositions of the four stains (HTPS-QDH11F, KS-KSH28F/XJ, KS-MGTH90F, and AKS-AWT-AFP2F) were 28.39–28.68% A, 23.73–24.01% C, 24.43–24.70% G, and 22.93–23.16% T, respectively. Because the full-length sequence of the prototype strain of EV-B106 was unavailable, we aligned the full-length genomes of the four Xinjiang strains with the Yunnan strain (12148/YN). The results showed that all four Xinjiang EV-B106 strains contained three nucleotide insertions at positions 7318, 7332, and 7425 and had one or two deletions compared to the Yunnan strain (12148/YN). Strain HTPS-QDH11F had two deletions at positions 95 and 7317; strain KS-KSH28F/XJ had one deletion at position 7317; strain KS-MGTH90F had two deletions at positions 95 and 7347; and strain AKS-AWT-AFP2F had one deletion at position 7347.
Pairwise comparisons of the nucleotide sequences and the deduced amino acid sequences were conducted among the four Xinjiang EV-B106 strains with the Yunnan EV-B106 strain and Pakistan EV-B106 strain, as well as of other prototype strains with the EV-B species (Table 1). The complete genome nucleotide sequence and amino acids similarities among the four Xinjiang strains were 93.3–98.1% and 98.7–99.2%, respectively. Furthermore, the four Xinjiang strains showed 80.5–80.8% nucleotide identity and 95.4–97.3% amino acid identity with the Yunnan EV-B106 strain. For the P1 coding sequences of the four Xinjiang strains, 91.6–92.4% nucleotide identity and 96.9–97.5% amino acid identity were observed when compared with the Pakistan strain.
Phylogenetic analysis of Xinjiang EV-B106 strains with other EV-B genomes
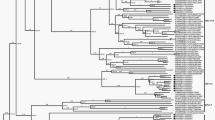
A phylogenetic tree was constructed based on the partial VP1, P1, P2, and P3 coding region nucleotide sequences of the four Xinjiang EV-B106 strains, Yunnan EV-B106 strain, Pakistan EV-B106 strain (VP1 and P1 region sequences only), Bolivia EV-B106 strain (partial VP1 region sequence only), and other prototype strains of EV-B in the GenBank database (Figs 1 and 2). Based on the phylogenetic trees constructed from the 303-nucleotide (nt 2585–2887) partial VP1 sequences of the four Xinjiang strains, all Xinjiang strains clustered with the Pakistan strain and showed significant genetic similarity to the Bolivia strain, but did not cluster with the Yunnan strain (Fig. 1).
Four Xinjiang EV-B106 strains (indicated by circles) and 56 other EV-B prototype strains were analysed by nucleotide sequence alignment using the neighbour-joining algorithms implemented in the MEGA 5.0 program. Numbers at the nodes indicate bootstrap support for that node (percentage of 1000 bootstrap replicates). The triangle and square indicates the Pakistan EV-B106 strain and Yunnan EV-B106, respectively. The scale bars represent the genetic distance. All panels have the same scale. (a) VP1 coding sequences; (b) P1 coding sequences; (c) P2 coding sequences; and (d) P3 coding sequences.
As shown in Fig. 2, the phylogenetic trees based on the P1 coding region sequences revealed that the four Xinjiang EV-B106 strains clustered together with the Pakistan strain and the Yunnan strain, demonstrating that these strains belong to the same serotype EV-B106 and confirming the preliminary molecular typing results. The nucleotide identities of the VP1 region between the Xinjiang strains (HTPS-QDH11F, KS-KSH28F/XJ, KS-MGTH90F, and AKS-AWT-AFP2F) and Pakistan strain were 92.8%, 92.1%, 91.4%, and 92.2%, respectively. Based on the phylogenetic tree, the EV-B77 prototype strain (strain USA-TX97-10394) showed the highest homology with all EV-B106 strains with a bootstrap support value of 100%. The nucleotide identities of the VP1 region between the four Xinjiang strains and EV-B77 prototype strain reached percentages as high as 69.5%, 70.2%, 69.1%, and 69.9%, respectively.
Unexpectedly, the four Xinjiang EV-B106 strains did not form a lineage with the only Yunnan EV-B106 strain in the trees based on the P2 and P3 coding regions, which suggesting that recombination between EV-B106 and other EV-B serotypes occurred. In the P2 coding region, the four Xinjiang EV-B106 strains clustered together and shared the highest similarity with the prototype strain EV-B87. In the P3 coding region, however, even the four Xinjiang strains did not cluster together, and the sequence similarities among their P3 regions varied widely from 82.9% (strain KS-KSH28F/XJ and KS-MGTH90F) to 97.8% (strain KS-KSH28F/XJ and HTPS-QDH11F). This unexpected finding suggested that the occurrence of recombination events among the four strains were different.
EV-B106 strains underwent extensive genetic exchanges with other EV-B strains in Xinjiang
To confirm the recombination events between the Xinjiang EV-B106 strains and other EV-B strains, similarity plots and bootscanning analyses were performed (Fig. 3). The four strains (HTPS-QDH11F, KS-KSH28F/XJ, KS-MGTH90F, and AKS-AWT-AFP2F) were picked as query sequences. The Yunnan EV-B106 strain was included in these analyses because it was the only available full-length sequence of EV-B106 in the GenBank database.
In the P1 coding region, the four Xinjiang strains showed the highest similarity with the Yunnan EV-B106 strain (12148/YN), as expected. However, in the other regions, the results revealed that the four Xinjiang strains clearly differed from the Yunnan EV-B106 strain. These results suggest that recombination events occurred in all four Xinjiang EV-B106 strains in the non-capsid regions. In the P2 region, no obvious donor sequence could be identified. In the P3 region, however, the four Xinjiang strains all recombined with other EV-B strains, but the donor sequences presented dissimilarities. One recombination event between three Xinjiang strains (excluding strain KS-MGTH90F) and EV-B100 prototype strain was identified from the 3′ end of the 3A region to the 5′ end of the 3C region. Bootscanning analysis also supported that strain KS-MGTH90F shared the highest similarity with EV-B86 followed by EV-B107 prototype strains in the 3D region, confirming the occurrence of recombination. Furthermore, recombination of a small fragment in the 5′-UTR region between strain AKS-AWT-AFP2F and Echovirus 9 prototype strain was also detected.
We further evaluated the recombinant structure of the four Xinjiang EV-B106 strains because of their distinct P3 region sequences. We screened the P3 region sequences of the four Xinjiang strains by BLAST, and found that two strains, HTPS-QDH11F and KS-KSH28F, shared the highest nucleotides similarity in the P3 region with the EV-B85 strain, which was also isolated from Xinjiang, China16. This outcome was unexpected because a previous study demonstrated that this Xinjiang EV-B85 strain contained an unknown serotype EV-B donor sequence. Because the serotype EV-B106 had not been identified at that time, we assumed that these four Xinjiang EV-B106 strains contained the donor sequence shared with the EV-B85 strain HYTY-ARL-AFP02F. We then conducted detailed analysis of the four Xinjiang EV-B106 strains with the Xinjiang EV-B85 strain with the P3 region sequences and observed that strains HTPS-QDH11F and KS-KSH28F/XJ showed 89.9% similarity with the EV-B85 strain HYTY-ARL-AFP02F in the 3D region.
To confirm that recombination events between the Xinjiang EV-B106 strains and Xinjiang EV-B85 strain occurred, we conducted both similarity plots and bootscanning analyses on the four Xinjiang EV-B106 strains and Xinjiang EV-B85 strain HYTY-ARL-AFP02F along with other EV-B prototype strains (Fig. 4) using the Xinjiang EV-B85 strain as a query sequence. The results revealed that recombination between the Xinjiang EV-B85 strain HYTY-ARL-AFP02F and the Xinjiang EV-B106 strains (strain HTPS-QDH11F and strain KS-KSH28F/XJ) in the 3D region were highly possible. The results also confirmed that the EV-B106 strains underwent extensive genetic exchanges with the other EV-B strains, including EV-B85 strains in Xinjiang, and that the unknown serotype EV-B donor sequence for the Xinjiang EV-B85 strain was also present in these Xinjiang EV-B106 strains.
Xinjiang EV-B106 shows temperature sensitivity
The four Xinjiang EV-B106 strains were compared based on replication capacity at an elevated temperature (39.5 °C). A Xinjiang EV-B85 strain (strain HYTY-ARL-AFP02F, a non-temperature sensitive strain) was used for an experiment control. The results showed that all EV-B106 strains were temperature-sensitive with titre reductions of more than 2 logarithms at 36 °C/39.5 °C (Fig. 5).
A Xinjiang EV-B85 strain (strain HYTY-ARL-AFP02F, showing non-temperature sensitivity) was used as an experiment control. The blue and red lines represent the growth trend of the viruses on RD cells at 36 °C and 39.5 °C, respectively. (a) strain HTPS-QDH11F (EV-B106); (b) strain KS-KSH28F/XJ (EV-B106); (c) strain KS-MGTH90F (EV-B106); (d) strain AKS-AWT-AFP2F (EV-B106); (e) strain HYTY-ARL-AFP02F (EV-B85).
Seroprevalence of EV-B106 in Xinjiang
A total of 50 serum samples were collected from newborns to 4-year-old children from Xinjiang, with 25 were collected from Kashgar prefecture and 25 from Hotan prefecture. From the samples surveyed, 21 were seropositive for EV-B106 (>1:8), with a total positive rate of 42%. The geometric mean titre (GMT) among the seropositive samples was 1:21.5. The composition ratios for the EV-B106 neutralization antibody titres of <1:8, 1:8–1:64, and >1:64 were 58%, 36%, and 6%, respectively. This suggests that compared with the seroepidemiology studies of other widely endemic EVs in China, such as EV-A71 and CV-A16, the positive rate and GMT of EV-B106 was lower17; however, the prevalence of EV-B106 was greater than that of other novel EVs isolated in Xinjiang, such as EV-B81 and EV-A894,18.
There were some differences between samples collected from the Kashar and Hotan prefectures in regards to the seroprevalence rates and GMTs. In the Kashar prefecture, the positive rate of EV-B106 neutralization antibody and GMTs were 44% and 1:12.4, respectively, while in the Hotan prefecture, they were 40% and 1:39.4 respectively. Although there was no significant difference between the positive seroprevalence rates of the two regions, the GMT in the Kashar prefecture was significantly higher than that in the Hotan prefecture (seroprevalence rate: χ2 = 0.082, p > 0.05, GMTs: p < 0.01).
Discussion
The prototype strain of EV-B106 was isolated in Bangladesh in 2001, but no sequences were available in the GenBank database until recently. Approximately a decade later, three EV-B106 sequences were reported in the GenBank database from Pakistan, the Yunnan Province of China, and Bolivia in 2010, 2012, and 2013, respectively, and only one full-length genome sequence is available to date11,12,13. Interestingly, the three reported EV-B106 strains were all isolated from AFP patients, indicating possible connections between EVB-106 infections and AFP. However, few studies have evaluated EV-B106 in China or worldwide, and thus additional research is needed to elucidate the possible correlation.
In our study, four Xinjiang EV-B106 strains were described. These EV-B106 strains were isolated in the Hotan, Kashar, and Aksu prefectures, which are all located in the Southwest region of Xinjiang. Compared with the Yunnan EV-B106 strain, the first and only EV-B106 strain previously reported in China, the Xinjiang EV-B106 strains display significantly different characteristics despite belonging to the same EV serotype. Based on the phylogenetic trees, the four Xinjiang EV-B106 strains clustered with the Yunnan strain only in the VP1 and P1 coding regions, while forming different lineages in both the P2 and P3 coding regions.
Recombination is a well-known phenomenon for enteroviruses19,20,21,22,23. In our study, similarity plots and bootscanning analyses for the Xinjiang EV-B106 strains were performed to investigate intertypic recombination between EV-B86, EV-B100, and EV-B107 with the P3 non-capsid region and Echovirus 9 5′-UTR. Additionally, the P3 region sequences of the four strains were diverse, indicating that EV-B106 had been circulating in the environment for an extended time before isolation24. As EV-B86, EV-B100, and EV-B107 are all newly discovered recombinant EVs, this indicates that recombination is a driving force in the formation of novel EV serotypes.
Furthermore, a previously reported Xinjiang EV-B85 strain (strain HYTY-ARL-AFP02F, a representative strain of Xinjiang EV-B85 isolated from an AFP patient) showing high nucleotide similarity with the Xinjiang EV-B106 strains in the P3 region was screened. A previous study of strain HYTY-ARL-AFP02F indicates that recombination occurred with an unknown EV-B donor sequence16. Interestingly, a similar donor sequence was found in two Xinjiang EV-B106 strains, HTPS-QDH11F and KS-KSH28F. Moreover, the EV-B85 strains analysed in the previous study and four EV-B106 strains analysed in this study were all isolated from AFP patients and their healthy contacts in the same region (Southern Xinjiang) in the same year (2011). These conditions are favourable for recombination because recombination between different EV serotypes typically occurs when distinct viruses infect and replicate in the same cell25,26. Although recombination between EV-B85 and EV-B106 was possible, it is difficult to determine whether the EV-B106 strain was exact the “unknown serotype” EV-B donor for the Xinjiang EV-B85 strains. However, we predict that Xinjiang EV-B106 strains and EV-B85 strains co-circulated with other EV-Bs and underwent extensive genetic exchanges with other EV-B strains; thus, it is likely that related P3 sequences of Xinjiang EV-B85 and EV-B106 originated from a common unknown EV-B ancestor through recombination.
The four Xinjiang EV-B106 strains were all temperature-sensitive, suggesting that their virulence is currently not high enough to have strong transmissibility16,27. However, strain AKS-AWT-AFP2F was isolated from an AFP patient, and thus the virulence of EV-B106 cannot be underestimated. Furthermore, because recombination occurred between the four strains and other EV-B strains, particularly with some non-temperature-insensitive strains such as Xinjiang EV-B85 strains, it is difficult to predict whether EV-B106 will evolve and heighten its virulence in the future.
A seroepidemiological analysis of EV-B106 in the Xinjiang Autonomous Region of China was conducted. The results showed that GMTs were higher in the Kashar prefecture than in the Hotan prefecture (p < 0.05), suggesting that these EV-B106 strains widely circulated in the Kashar prefecture, particularly because two of the four EV-B106 strains were isolated in the Kashar prefecture. However, compared to the other prevalent EVs in China, such as EV-A71 and CV-A1617,28, the seropositive rate and GMT of the EV-B106 neutralization antibody in the two regions of Xinjiang were relatively low, demonstrating that the extent of transmission and exposure of the viral population to the novel EV-B106 serotype were highly limited. The low rate of virus isolation also supports this.
In conclusion, we reported the full-length genome sequences of four EV-B106 strains during AFP surveillance in Xinjiang Uygur Autonomous Region, China. To date, only one EV-B106 strain (Yunnan strain) has been reported worldwide, indicating that this serotype is not widespread in China or in the world. Sequence analysis suggested that these four strains have high genetic diversity compared with the Yunnan EV-B106 strain, intertypic recombination in the non-structural protein region of all the four strains, and extensive genetic exchanges with the other EV-B circulating in Xinjiang. The high divergence among the EV-B106 strains reflects that this virus is not newly emergent, but has circulated in the environment for many years. In addition, the differences among the P3 region sequences of four Xinjiang EV-B106 strains indicate that they have been circulating separately for some time. This study expands the number of whole genome sequences of EV-B106 in the GenBank database and provides valuable information regarding the molecular epidemiology of EV-B106 in China, which can be utilized to evaluate the association between EV-B106 and EV-B106-related diseases such as AFP.
Methods
Sample collection
This study did not involve human participants or human experimentation; the only human materials used were stool samples collected from one AFP patient and three healthy children at the instigation of the Ministry of Health P. R. of China for public health purposes. Written informed consent for the use of their clinical samples was acquired from all individuals involved in this study. This study was approved by the Ethics Review Committee of the National Institute for Viral Disease Control and Prevention (NIVDC), Chinese Center for Disease Control and Prevention, all experimental protocols were approved by NIVDC, and the methods were carried out in accordance with the approved guidelines.
The four EV-B106 strains (strain HTPS-QDH11F, strain KS-KSH28F, strain KS-MGTH90F, and strain AKS-AWT-AFP2F) used in this study were isolated from stool specimens from one AFP patient and three healthy children in Hotan, Kashgar, and Aksu Prefectures of Xinjiang Autonomous Region, China. The samples were collected in 2011 during poliovirus surveillance activities.
In the seroprevalence study of EV-B106 antibodies, 50 healthy children ≤5 years of age were surveyed. Fifty serum samples were collected in 2013 for seroepidemiological analysis of enteroviruses, with informed parental consents, by the Xinjiang Center for Disease Control and Prevention: 25 samples from Hotan and 25 samples from Kashgar, the two regions where the strains were isolated. The same serum samples were used previously for a EV-A89 seroepidemiology study4. None of the children had any signs of disease at the time of sample collection.
Viral isolation
Stool samples from the AFP patient and three healthy children were collected and processed according to standard procedures29,30. The processed samples were then inoculated into two cell lines for isolation: RD cell and a mouse cell line with the human poliovirus receptor (L20B). Both cell lines were provided by the WHO Global Poliovirus Specialized Laboratory in the US and originally purchased from the American Type Culture Collection (Manassas, VA, USA). Infected cell cultures were harvested after complete CPE was observed.
Molecular typing
Viral RNA was extracted from the cell culture using a QIAamp Viral RNA Mini Kit (Qiagen, Hilden, Germany). Reverse transcription polymerase chain reaction (RT-PCR) was conducted to amplify the partial VP1 coding region using the PrimeScript One Step RT-PCR Kit Ver.2 (TaKaRa, Dalian, China) with primer pairs 490 and 4922. The PCR products were purified using the QIAquick PCR purification kit (Qiagen, Germany) and sequenced in both directions at least once from each strand using ABI 3130 Genetic Analyser (Applied Biosystems, Foster City, CA, USA). The acquired partial VP1 sequences were analysed with the BLAST server by comparing with the sequences available in the GenBank database and were determined using the EV Genotyping Tool15.
Primer designing and whole genomic sequencing
The 5′ end of the genome sequence was amplified using the 5′-Full RACE Kit (Takara, Shiga, Japan) according to the manufacturer’s instructions. The 3′ end sequence was obtained by using an oligo-dT primer (primer 7500A)31 as the downstream primer for amplification. The primers used for PCR amplification and sequencing for the rest of the genome were based on the primer walking method (Table 2).
Phylogenetic and recombination analyses
The nucleotide and deduced amino acid sequences of the four Xinjiang EV-B106 strains were compared with those of the prototype EV-B strains by pairwise alignment using the MEGA program (version 5.03)32. Phylogenetic trees were constructed by the neighbour-joining method implemented in the MEGA program and Kimura 2-parameter model. Bootstrap values of greater than 80% were considered statistically significant for grouping. The SimPlot program (version 3.5.1) was used for similarity plots and bootscanning analyses, with a 200-nucleotide window moving in 20-nucleotide steps. Bootscanning analyses were run with the neighbour-joining method33.
Assay for temperature sensitivity
Temperature sensitivities of the four Xinjiang EV-B106 strains were assayed on monolayer RD cells in 24-well plates27. A Xinjiang EV-B85 strain (strain HYTY-ARL-AFP02F, showing non-temperature sensitivity)16 was used for an experiment control in order to ensure that cells and thus overall viral replication were not affected at high temperature in experimental conditions. The 24-well plates were inoculated with 50 μL of undiluted virus stocks and placed in two different CO2 incubators: 36 °C as the optimal temperature for virus propagation and 39.5 °C as the supraoptimal temperature. After absorption at 36 °C or 39.5 °C for 1 h, the unabsorbed virus inoculum was removed and 100 μL of maintenance medium was added to each well. The plates were incubated again at 36 °C and 39.5 °C and harvested at 8 time points post-infection (4 h, 8 h, 16 h, 24 h, 48 h, 72 h, 96 h, and 120 h) in succession. The 50% cell culture infectious dose (CCID50) was calculated by the end-point dilution method on monolayer RD cells in 96-well plates at 36 °C. Virus isolates showing more than a two-logarithm reduction in titre at different temperatures were considered to be temperature-sensitive34.
Neutralizing test
Neutralizing antibodies against EV-B106 were detected in a neutralization test using the human RD cell line as previously described17 with some modifications. Strain AKS-AWT-AFP2F was selected as the attack virus strain in the neutralizing test because the four Xinjiang EV-B106 strains appeared to be antigenically equivalent (VP1 amino acid identity among the four EV-B106 strains was 98.9–99.6%) and showed the highest TCID50 value among the four Xinjiang EV-B106 strains. Serum samples were inactivated at 56 °C for 30 min, and sample dilutions from 1:4 to 1:1024 were assayed. The mixture of virus samples (50 μL) with a CCID50 of 100 and appropriate serum dilution (50 μL) was then incubated at 36 °C in a CO2 incubator. After incubation for 7 days, the highest dilution of serum that protected 50% of the cultures was recorded according to the EV-like CPE. A serum sample was considered positive if the neutralization antibody titre was observed at a dilution of 1:8, and GMT was subsequently calculated.
Statistical analysis
All statistical analyses were performed using SPSS Statistics software (version 19.0) (SPSS, Inc., Chicago, IL, USA). Chi-square test was used to compare the seroprevalence rates of the viral strains between Kashgar prefecture and Hotan prefecture. Mann-Whitney U test was used to compare the differences in GMTs. Titres below 1:8 were assumed 1:4 for calculation. Differences with an error probability of p < 0.05 were considered statistically significant.
Nucleotide sequence accession numbers
The full-length genomic sequences of the four EV-B106 strains (strain HTPS-QDH11F, strain KS-KSH28F, strain KS-MGTH90F, and strain AKS-AWT-AFP2F) described in this study were deposited in the GenBank database under the accession numbers KX171334-KX171337, respectively.
Additional Information
How to cite this article: Song, Y. et al. Phylogenetic Characterizations of Highly Mutated EV-B106 Recombinants Showing Extensive Genetic Exchanges with Other EV-B in Xinjiang, China. Sci. Rep. 7, 43080; doi: 10.1038/srep43080 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Picornaviridae, Virus Taxonomy: Classification and Nomenclature of Viruses: Ninth Report of the International Committee on Taxonomy of Viruses. (Elsevier, San Diego, 2011).
Oberste, M. S., Maher, K., Kilpatrick, D. R. & Pallansch, M. A. Molecular evolution of the human enteroviruses: correlation of serotype with VP1 sequence and application to picornavirus classification. J Virol. 73, 1941–1948 (1999).
Zhang, Y. et al. Molecular typing and characterization of a new serotype of human enterovirus (EV-B111) identified in China. Virus Res. 183, 75–80, doi: 10.1016/j.virusres.2014.01.002 (2014).
Fan, Q. et al. A Novel recombinant enterovirus type EV-A89 with low epidemic strength in Xinjiang, China. Sci. Rep. 5, 18558, doi: 10.1038/srep18558 (2015).
Bergamini, G., Preiss, T. & Hentze, M. W. Picornavirus IRESes and the poly(A) tail jointly promote cap-independent translation in a mammalian cell-free system. RNA 6, 1781–1790 (2000).
Zoll, J., Heus, H. A., van Kuppeveld, F. J. & Melchers, W. J. The structure-function relationship of the enterovirus 3′-UTR. Virus Res. 139, 209–216, doi: 10.1016/j.virusres.2008.07.014 (2009).
Caro, V., Guillot, S., Delpeyroux, F. & Crainic, R. Molecular strategy for ‘serotyping’ of human enteroviruses. J. Gen. Virol. 82, 79–91, doi: 10.1099/0022-1317-82-1-79 (2001).
Oberste, M. S., Nix, W. A., Maher, K. & Pallansch, M. A. Improved molecular identification of enteroviruses by RT-PCR and amplicon sequencing. J. Clin. Virol. 26, 375–377 (2003).
Oberste, M. S. et al. Comparison of classic and molecular approaches for the identification of untypeable enteroviruses. J. Clin. Microbiol. 38, 1170–1174 (2000).
Oberste, M. S. et al. Typing of human enteroviruses by partial sequencing of VP1. J. Clin. Microbiol. 37, 1288–1293 (1999).
Tang, J. et al. Complete genome characterization of a novel enterovirus type EV-B106 isolated in China, 2012. Sci. Rep. 4, 4255, doi: 10.1038/srep04255 (2014).
Shaukat, S. et al. Characterization of a novel enterovirus serotype and an enterovirus EV-B93 isolated from acute flaccid paralysis patients. PLoS One 8, e80040, doi: 10.1371/journal.pone.0080040 (2013).
Nix, W. A. et al. Diversity of picornaviruses in rural Bolivia. J. Gen. Virol. 94, 2017–2028, doi: 10.1099/vir.0.053827-0 (2013).
Ishiko, H. et al. Molecular diagnosis of human enteroviruses by phylogeny-based classification by use of the VP4 sequence. J. Infect. Dis. 185, 744–754, doi: 10.1086/339298 (2002).
Kroneman, A. et al. An automated genotyping tool for enteroviruses and noroviruses. J. Clin. Virol. 51, 121–125, doi: S1386-6532(11)00129-6 (2011).
Sun, Q. et al. Transmission of human enterovirus 85 recombinants containing new unknown serotype HEV-B donor sequences in Xinjiang Uighur autonomous region, China. PLoS One 8, e55480, doi: 10.1371/journal.pone.0055480 (2013).
Zhu et al. Retrospective seroepidemiology indicated that human enterovirus 71 and coxsackievirus A16 circulated wildly in central and southern China before large-scale outbreaks from 2008. Virol. J. 7, 300, doi: 10.1186/1743-422X-7-300 (2010).
Hu, L. et al. Phylogenetic evidence for multiple intertypic recombinations in enterovirus B81 strains isolated in Tibet, China. Sci. Rep. 4, 6035, doi: 10.1038/srep06035 (2014).
Kyriakopoulou, Z., Pliaka, V., Amoutzias, G. D. & Markoulatos, P. Recombination among human non-polio enteroviruses: implications for epidemiology and evolution. Virus Genes 50, 177–188, doi: 10.1007/s11262-014-1152-y (2015).
Lukashev, A. N. et al. Recombination strategies and evolutionary dynamics of the Human enterovirus A global gene pool. J. Gen. Virol. 95, 868–873, doi: 10.1099/vir.0.060004-0 (2014).
Domingo et al. Coxsackieviruses and quasispecies theory: evolution of enteroviruses. Curr. Top Microbiol. Immunol. 323, 3–32 (2008).
Zhang, Y. et al. Emergence and transmission pathways of rapidly evolving evolutionary branch c4a strains of human enterovirus 71 in the central plain of China. PLoS One 6, e27895, doi: 10.1371/journal.pone.0027895 (2011).
Muslin, C., Joffret, M. L., Pelletier, I., Blondel, B. & Delpeyroux, F. Evolution and emergence of enteroviruses through intra- and inter-species recombination: Pplasticity and phenotypic impact of modular genetic exchanges in the 5′ untranslated region. PLoS Pathog. 11, e1005266, doi: 10.1371/journal.ppat.1005266 (2015).
Zhang, Y. et al. An outbreak of hand, foot, and mouth disease associated with subgenotype C4 of human enterovirus 71 in Shandong, China. J. Clin. Virol. 44, 262–267, doi: 10.1016/j.jcv.2009.02.002 (2009).
Worobey, M. & Holmes, E. C. Evolutionary aspects of recombination in RNA viruses. J. Gen. Virol. 80 (Pt 10), 2535–2543, doi: 10.1099/0022-1317-80-10-2535 (1999).
Zhang, Y. et al. A Sabin 2-related poliovirus recombinant contains a homologous sequence of human enterovirus species C in the viral polymerase coding region. Arch. Virol. 155, 197–205 (2010).
Arita, M. et al. Temperature-sensitive mutants of enterovirus 71 show attenuation in cynomolgus monkeys. J. Gen. Virol. 86, 1391–1401, doi: 10.1099/vir.0.80784-0 (2005).
Li, W. et al. Seroepidemiology of human enterovirus71 and coxsackievirusA16 among children in Guangdong province, China. BMC Infect. Dis. 13, 322, doi: 10.1186/1471-2334-13-322 (2013).
Isolation and identification of polioviruses. WHO Polio laboratory manual, 4th edn. (World Health Organization, Geneva, 2004).
Xu, W. & Zhang, Y. Isolation and Characterization of Vaccine-Derived Polioviruses, Relevance for the Global Polio Eradication Initiative. Methods Mol. Biol. 1387, 213–226, doi: 10.1007/978-1-4939-3292-4_10 (2016).
Yang, C. F. et al. Circulation of endemic type 2 vaccine-derived poliovirus in Egypt from 1983 to 1993. J. Virol. 77, 8366–8377 (2003).
Tamura, K. et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739, doi: 10.1093/molbev/msr121 (2011).
Salminen, M. O., Carr, J. K., Burke, D. S. & McCutchan, F. E. Identification of breakpoints in intergenotypic recombinants of HIV type 1 by bootscanning. AIDS Res. Hum. Retroviruses 11, 1423–1425, doi: 10.1089/aid.1995.11.1423 (1995).
Zhang, Y. et al. Natural type 3/type 2 intertypic vaccine-related poliovirus recombinants with the first crossover sites within the VP1 capsid coding region. PLoS ONE 5, e15300, doi: 10.1371/journal.pone.0015300 (2010).
Oberste, M. S. et al. Species-specific RT-PCR amplification of human enteroviruses: a tool for rapid species identification of uncharacterized enteroviruses. J. Gen. Virol. 87, 119–128, doi: 10.1099/vir.0.81179-0 (2006).
Acknowledgements
We would like to acknowledge the staff of the national polio eradication program in the Xinjiang Uygur Autonomous Region Center for Disease Control and Prevention for stool specimens of the AFP cases and healthy children collected in this study. The study was supported by the National Natural Science Foundation of China (project nos 81672070 and 81373049), National key research and development project (project no 2016YFC1200905), and the National Key Technology R&D Program of China (project nos 2013ZX10004-101 and 2013ZX10004-202).
Author information
Authors and Affiliations
Contributions
Y.Z. and W.X. conceived and designed the experiments. Y.S., Q.F., H.C., D.Y., S.Z., H.T., Q.S. and D.W. performed the experiments. Y.S., Q.F. and Y.Z. analysed the data. Y.S. and Y.Z. wrote the main manuscript and Y.S. prepared all the figures. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Song, Y., Zhang, Y., Fan, Q. et al. Phylogenetic Characterizations of Highly Mutated EV-B106 Recombinants Showing Extensive Genetic Exchanges with Other EV-B in Xinjiang, China. Sci Rep 7, 43080 (2017). https://doi.org/10.1038/srep43080
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep43080
This article is cited by
-
Molecular characterization of echovirus 9 strains isolated from hand-foot-and-mouth disease in Kunming, Yunnan Province, China
Scientific Reports (2022)
-
Phylogenetic characteristics and molecular epidemiological analysis of novel enterovirus EV-B83 isolated from Tibet, China
Scientific Reports (2020)
-
Phylogenetic analysis and phenotypic characterisatics of two Tibet EV-C96 strains
Virology Journal (2019)
-
Identification of a new recombinant strain of echovirus 33 from children with hand, foot, and mouth disease complicated by meningitis in Yunnan, China
Virology Journal (2019)
-
Molecular characterization of echovirus 12 strains isolated from healthy children in China
Scientific Reports (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.