Abstract
Malaria remains a major challenge to global health causing extensive morbidity and mortality. Yet, there is no efficient vaccine and the immune response remains incompletely understood. Apical Membrane Antigen 1 (AMA1), a leading vaccine candidate, plays a key role during merozoite invasion into erythrocytes by interacting with Rhoptry Neck Protein 2 (RON2). We generated a human anti-AMA1-antibody (humAbAMA1) by EBV-transformation of sorted B-lymphocytes from a Ghanaian donor and subsequent rescue of antibody variable regions. The antibody was expressed in Nicotiana benthamiana and in HEK239-6E, characterized for binding specificity and epitope, and analyzed for its inhibitory effect on Plasmodium falciparum. The generated humAbAMA1 shows an affinity of 106–135 pM. It inhibits the parasite strain 3D7A growth in vitro with an expression system-independent IC50-value of 35 μg/ml (95% confidence interval: 33 μg/ml–37 μg/ml), which is three to eight times lower than the IC50-values of inhibitory antibodies 4G2 and 1F9. The epitope was mapped to the close proximity of the RON2-peptide binding groove. Competition for binding between the RON2-peptide and humAbAMA1 was confirmed by surface plasmon resonance spectroscopy measurements. The particularly advantageous inhibitory activity of this fully human antibody might provide a basis for future therapeutic applications.
Similar content being viewed by others
Introduction
Malaria remains a major challenge to global healthcare and is one of the major causes of morbidity and mortality in childhood, especially in Sub-Saharan Africa. Of the six plasmodium species which are pathogenic to humans, Plasmodium falciparum is the most frequent. It brings about the severest form of malaria, malaria tropica, affecting mostly children by cerebral malaria and severe malarial anaemia1,2. Natural occurring premunition to malaria develops slowly and wanes without frequent exposure2,3,4,5. Several lines of defense of the human immune system contribute to the successful control of plasmodial infections. Besides cellular mechanisms, e.g. by T cell-mediated killing of blood and liver stages6,7,8, the defense mediated by antibodies plays a critical role. The spectrum of anti-plasmodial antibodies usually increases with age, i.e. with cumulative exposure9,10,11 and the majority of the antibodies is directed against merozoites11, the invasive forms of the erythrocytic replication cycle. Antibodies, representing the adaptive arm of humoral immune defense, are all-round talents which can exert their functions by mere binding (primary function) and by recruiting effector cells and/ or complement factors (secondary functions). In case of plasmodial infections the modes of action during the blood stage thus include (1) the blocking of erythrocyte invasion by merozoites, (2) the neutralization of merozoites by agglutination, (3) the initiation of the complement cascade resulting in further opsonization and lysis, and (4) the recruitment of neutrophilic granulocytes and monocytes/macrophages12,13,14,15,16,17,18,19,20.
The invasion of merozoites into erythrocytes is a complex process, which can be subdivided into a pre-invasion phase, the ‘classical’ invasion phase, and an echinocytosis phase21,22. One of the key proteins during the ‘classical’ invasion is Apical Membrane Antigen 1 (AMA1). In the human host this protein is mainly expressed in the late plasmodial stages, i.e. in late trophozoites and schizonts23,24,25. Initially, the 83-kDa precursor of AMA1 (AMA183) is localized in the micronemes26,27. By the time of schizont rupture and release of young merozoites AMA183 is processed to give the mature 66-kDa form (AMA166) which remains membrane-bound24,28,29. AMA166 then translocates to the merozoite’s apical end to fulfill its function in the invasion process by interacting with Rhoptry Neck Protein 2 (RON2)24,27. RON2 is secreted from the rhoptries just prior to invasion and inserts into the erythrocyte membrane30,31,32. The interaction of AMA1 and RON2 takes place between the hydrophobic trough of AMA1 and a small extracellular hydrophobic domain of RON231,33,34. This interaction is critical since the AMA1:RON2 complex is part of the sealing moving junction and constitutes the anchor for the actin-myosin motor which pulls the merozoite into the red blood cell to be invaded32,35,36,37. Several studies showed that AMA1-specific antibodies can inhibit invasion38,39.
Many anti-plasmodial monoclonal antibodies (mAbs) have been generated in mice or other rodents which helped to gain valuable insights into the functions of a plethora of plasmodial proteins40,41,42,43,44. However, such mAbs do not necessarily reflect the naturally acquired anti-plasmodial immunoglobulin repertoire in humans which takes years – or even decades – to develop. So far, only few human anti-plasmodial monoclonal antibodies (humAbs) have been generated. Among these are humAbs directed at Merozoite Surface Protein 1 (MSP1), MSP2, MSP3, MSP10, NPNA1, Pfs48/45, and VAR2CSA45,46,47,48,49,50,51,52. However, to the best of our knowledge, no humAb specific to AMA1 has been described yet. Here, we report the isolation, expression and characterization of the first human monoclonal antibody recognizing P. falciparum AMA1, called humAbAMA1.
Results
Selection of PBMC donor and EBV-transformation and screening
In order to choose a promising candidate for the generation of an AMA1-specific human monoclonal antibody, plasma of 31 adult Ghanaian blood donors were screened by indirect ELISA for IgG reactivity against different recombinant variants of AMA1, including the allelic variant of Plasmodium falciparum strain 3D7, as well as a mixture of three artificial diversity covering variants of AMA1 (described by Remarque et al.53). Against the single allele recombinant antigen AMA1 (3D7), 23 out of 31 (74%) of the Ghanaian plasma samples showed a positive immune response (Fig. 1A). The reactivity of the plasma against AMA1 (DiCo1-3) has been shown before54. Here, 30 out of 31 samples showed a positive reaction (reactivity superior of the negative control plus 2 standard deviations), corresponding to 97%.
Plasma IgG reactivities against AMA1.
Using samples from 31 Ghanaian blood donors (G01–31) as well as a European control pool of two donors which had never been exposed to Malaria (NIP), ELISA was performed to estimate the reactivity against 3D7 AMA1. As positive control, a pool of four highly reactive Ghanaian donors was used. Values reflect relative reactivity in comparison to this positive control’s reactivity, which was set to 1 (A). The reactivity of the samples to AMA1 (3D7) and AMA1 (DiCo 1-3) (previously published in Feller et al.)54 were compared and the degree of correlation was estimated using the Spearman rank sum test (B). The reactivity of the plasma samples against both the native antigen and the reduced/alkylated antigen was estimated by ELISA, and shown as a box plot (C). The p-values were estimated using the Wilcoxon matched pairs test.
The reactivities of the Ghanaian plasma samples against the variants AMA1 (3D7) and AMA1 (DiCo1-3) showed a strong positive correlation (p < 0.0001, Fig. 1B). Next, these plasma IgG were tested for recognition of conformational epitopes of AMA1. Therefore, ELISA using the same plasma samples were carried out similarly, but additionally using reduced/alkylated version of AMA1 (3D7). The results indicate that the majority of the antibodies are directed against conformational epitopes. The median reactivity of the plasma samples to reduced vs. the reactivity to the native antigen decreased by a factor of 17 (p < 0.0001, Wilcoxon matched pairs test, Fig. 1C). Only 2 of the 31 samples showed a reactivity against the reduced antigen AMA1 (3D7) and reactivities of these two were only slightly above the threshold of two standard variations above the negative control reactivity.
The sample of donor G10 had the highest reactivity against AMA1 (DiCo1-3) and is also among the samples with highest reactivity against AMA1 (3D7). This is why PBMCs of donor G10 were selected for the following steps of antibody generation. The PBMCs were thawed, antigen-specific B-cells were stained (IgG+/CD22+/anti-AMA1 (DiCo1-3)+) and sorted by flow cytometry. The supernatants of the cultured lymphoblastoids were tested for specific antibody production after two weeks of culture, the supernatant of well 1E4 showed high reactivity against AMA1 (DiCo1-3). Subsequently, the sequences coding for variable domains of the antibody light and heavy chain were amplified from cDNA from these cells, sequenced and cloned into expression vectors for recombinant production.
Recombinant expression of humAbAMA1
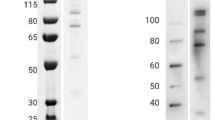
The expression cassettes for the antibody production in plants and mammalian cell culture are shown in Fig. 2A. HumAbAMA1, the recombinant antibody of EBV-clone 1E4, was produced as a full-size human IgG1:κ by Agrobacterium-mediated transient protein expression in N. benthamiana plants. After harvest and homogenization of the plant material, the antibodies were purified by Protein A chromatography. The final yield of the pure and intact plant produced antibody was 70 mg/kg fresh leaf material. Similarly, secretory production of the antibody was performed by transient transfection of HEK293-6E cells. The cell culture supernatants were purified by Protein A chromatography. Here, the productivity of the antibody after 6 days of antibody production was approx. 10 mg/l cell culture supernatant. Integrity and purity of both antibodies was assessed by SDS-PAGE (Fig. 2B).
Cloning, expression and binding specificity of humAbAMA1.
HumAbAMA1 was cloned as full length expression construct in both vectors pTRAkt and pTT5 for transient expression in plants and in HEK293-6E cells, respectively (A). The expression and purity of the full length antibodies expressed both in plants (N. benthamiana) and in mammalian cells (HEK293) was controlled by SDS-PAGE. Both non-reduced and reduced samples of the antibodies were loaded on the gel (B). The binding of humAbAMA1to AMA1 (3D7) was tested by indirect ELISA. Dilutions of the antigen (AMA1 (3D7)) were coated onto ELISA plates, then human antibodies were applied followed by goat anti-human antibody coupled to alkaline phosphatase (Dianova, Hamburg, Germany). Black diamonds: Semi-immune (premune) plasma pool; green circles: humAbAMA1 (N. benthamiana); Red open squares: humAbAMA1 (HEK293); orange inverted triangles: mock control antibody (2G12).
HumAbAMA1 recognizes a conformational epitope and binds to it with high affinity
The binding specificity of the antibody humAbAMA1 was first tested in indirect ELISA. The reactivity of the antibody was concentration dependent, with a detection limit of approx. 6 ng/well (Fig. 2C). AMA1 possesses eight disulfide bridges, which strongly stabilize its tertiary structure55. Rodent-derived anti-AMA1 mAbs, such as the murine mAb 1F9 and the rat mAb 4G2 recognize conformational, reduction-sensitive epitopes43,56. This is also the case for humAbAMA1, which does not react with reduced/alkylated AMA1 in indirect ELISA (data not shown). Surface plasmon resonance (SPR) spectroscopy measurements were used to compare the affinity of humAbAMA1 produced in plants and in HEK293-6E cells towards the recombinant antigen AMA1 (3D7), revealing very high affinities (KD) of 135 ± 8 pM and 106 ± 5 pM for the plant and HEK-derived antibodies, respectively (Fig. 3). Additionally, the affinity of the antibody humAbAMA1 produced in N. benthamiana against two further AMA1 antigen variants representing the strains HB3 and FCR3 have been measured. The binding towards AMA1 (HB3) is in the similar range (667 ± 7 pM), whereas the binding to AMA1 (FCR3) is two orders of magnitude lower (73 ± 0.2 μM). The kinetics of the antibodies are more precisely summarized in Table 1.
Sensorgrams of surface plasmon resonance (SPR) measurement for the determination of humAbAMA1 affinity to AMA1 (3D7).
The affinity of the antibody humAbAMA1 was estimated by SPR spectroscopy. For each cycle recombinant and purified humAbAMA1 either produced in N. benthamiana (A, green sensogramms) or produced in HEK293-6E cells (B, red sensograms) was captured on a Protein A surface (65 RU for N. benthamiana produced or 32 RU for HEK produced humAbAMA1). Subsequently, the antigen AMA1 (3D7) was injected at concentrations of 500 nM, 250 nM, 125 nM, 62, 5 nM, 31.25 nM, 15.6 nM, 7.6 nM, 3.9 nM, 1.95 nM and 0.975 nM for 180 s, followed by a buffer injection step for 420 sec. To determine kinetic constants, the data was fitted using a 1:1 Langmuir interaction model using the software BIAEval 4.0. Fit curves are shown in light grey.
HumAbAMA1 recognizes AMA1 in its native context on the surface of merozoites and schizonts
Next, humAbAMA1 was tested for binding native AMA1 located on merozoites and schizonts. To this end, immunofluorescence assays (IFAs) were carried out using double staining with the humAbAMA1, as well as the MSP4-specific antibody 2.44 (Fig. 4, upper panel). We observed a characteristic punctiform staining at one end of ripening and free merozoites, which is in line with the apical localization of AMA1. In some cases we also saw a staining of the outer membrane of free merozoites, indicating colocalization with antibody 2.44 (Fig. 4, lower panel). This is consistent with the partial redistribution of AMA1 once the merozoites have been released upon schizont rupture57. The data obtained strongly resemble immunofluorescence images using AMA1-specific antibodies published earlier38,57.
Immunofluorescence imaging of humAbAMA1.
The specific binding of the recombinant antibody humAbAMA1 to both schizonts (upper panel) and free merozoites (lower panel) was analyzed by immunofluorescence assays. Parasites were co-stained with humAbAMA1 (red, first column), murine anti-MSP4 antibody (green, second column) and the nuclear stain Hoechst 33342 (blue, third column). The last column shows the overlay image. Human and murine antibodies were detected by fluorescently labelled antibodies goat-anti-human IgG (H + L)-Cy3 (Dianova) and goat-anti-mouse IgG (H + L)-Alexa Fluor® 488 (Life Technologies), respectively. Images were taken at a 630-fold magnification with a Leica TCS SP8 microscope. The right panel shows the overlay of the three fluorescence images. Scale bar 7.5 μm.
Plasmodium falciparum strains 3D7A, HB3 and FCR3 are strongly inhibited by humAbAMA1
The inhibitory potential of humAbAMA1 was tested in in vitro growth inhibition assay (GIA) with P. falciparum 3D7A, HB3 and FCR3. We found that P. falciparum strain 3D7A was reproducibly inhibited by plant-produced humAbAMA1 with an IC50 value of 35 μg/ml (95% confidence interval (CI): 33–37 μg/ml). Inhibition assays using the antibodies produced in mammalian cell culture showed the same IC50 value of 35 μg/ml (95% CI: 34–37 μg/ml) (Fig. 5A). As controls, we expressed the rodent-derived antibodies described in the literature (4G2 and 1F9) as chimeric antibodies. The IC50 values of these antibodies were 105 μg/ml (95% CI: 99–111 μg/ml) and 292 μg/ml (95% CI: 250–342 μg/ml) for the chimeric antibodies 4G2 and 1F9, respectively, thus being three to eight times higher than the IC50 value of humAbAMA1. The IC50-value of the humAbAMA1 against the parasite strains HB3 and FCR3 was also 3-fold and 4-fold lower than the chimeric antibody 4G2, respectively. The second chimeric control antibody inhibited the parasite strains HB3 and FCR3 at values above 1000 μg/ml, thus the estimated values were 7 to >13-fold higher than the antibody humAbAMA1. The assays are represented in Fig. 5 and the estimated IC50-values summarized in Table 2.
In vitro growth inhibition activity of humAbAMA1.
The anti-parasitic effect was estimated using a standardized Growth Inhibition Assay (GIA). Purified antibodies were added to the parasite strain Plasmodium falciparum 3D7A (A), HB3 (B) and FCR3 (C) at schizont stage and co-incubated for 40–46 h. Growth of the parasites was estimated by a pLDH assay. All tests were carried out in technical triplicates; error bars correspond to the standard error of the mean. Antibodies tested were the humAbAMA1 produced in plants (green circles), humAbAMA1 produced in HEK293 (red open squares, only tested in strains 3D7A), chimeric antibody 4G2 (blue closed squares) and chimeric 1F9 (purple triangles).
Next, combinations of the human monoclonal antibodies humAbAMA1 and humAb10.1 (anti-MSP10, previously isolated in our laboratory)52 were tested for synergistic effects. The antibodies were mixed in five different ratios, added in serial dilutions to the parasite culture (P. falciparum strain 3D7A) and the IC50 of each combination was determined. Generally, two calculations are performed to evaluate the degree of synergy, the sum of FIC and HEWLETT’s S. Values corresponding to sum of FIC below 1 and HEWLETT’s S above 1 are regarded as synergistic, FIC below 0.5 and HEWLETT’s S above 2 reflect strongly synergistic effects. As shown in the isobologram, the combination of the AMA1-specific antibody and the MSP10 antibody humAb10.1 shows synergistic inhibition of the parasite strain 3D7A (Fig. 6). The calculated FIC and HEWLETT’s S are 0.68 and 1.64, respectively. In comparison, two AMA1-specific antibodies (humAbAMA1 and 4G2) rather act in an additive way, as shown in Fig. 6 (sum of FIC = 0.92 and HEWLETT’s S = 1.11).
Synergistic effect of monoclonal antibodies on parasitic growth inhibition.
The combinations of the humAbAMA1 with either the MSP10-specific humAb10.1 (pink diamonds) or the chimeric antibody 4G2 (blue circles), respectively, were tested for synergistic growth inhibition. Antibodies were applied singly as well as in combinations. Antibodies were mixed in different ratios (1:4, 1:2, 1:1, 2:1, 4:1) proportional to their respective estimated IC50 values. Titrations of each of the mixtures were added to the parasites. The IC50 value of each of the antibody mixes was estimated and is depicted as relative inhibition. Curves were fitted using the One phase Decay function of GraphPad Prism 5.0.
HumAbAMA1 partially shares an epitope with mAb 1F9
After the confirmation of the strong inhibitory response in GIAs, humAbAMA1 was tested for potential epitope overlapping with the well-characterized inhibitory antibodies 1F9 or 4G2. To this end, a “pairwise epitope mapping” was carried out. In a multiple capture setup AMA1 (3D7) was first caught on a mAb1D7 (AMA1 domain III-specific antibody) surface. Then, the antibodies to be tested were injected singly or consecutively. It was found that humAbAMA1 and 1F9 partially share an epitope, i.e. 1F9 could not bind efficiently anymore if humAbAMA1 had been injected first. However, their epitopes appeared not to be completely identical since a small proportion of 1F9 could still recognize AMA1 (3D7) (Fig. 7, cycle 6). In cycle 3 (Fig. 7), it became clear that humAbAMA1 and 4G2 could bind simultaneously. Thus, their corresponding epitopes neither overlap nor show signs of allosteric competition. Subsequently, the epitope was more specifically determined using the CLIPS (Chemical Linkage of Peptides onto Scaffolds) technology (Pepscan Presto BV, Lelystad, The Netherlands), in which peptides covering the complete sequence of AMA1 (3D7) were screened. The advantage of using this method in comparison to standard peptide-screening methodology is that the three-dimensional structures of the original protein are simulated by introducing various peptide loop patterns58. The results suggest that the antibody specifically binds to the loop of the peptide 194-TLDEMRHFYKDNK-206 within the domain I of AMA1, which is in close proximity to the hydrophobic RON2 binding groove (Fig. 8). The binding site of humAbAMA1 overlaps largely with the epitope of 1F956.
Competition measurements of humAbAMA1 with the 4G2 and 1F9.
Using SPR spectroscopy, pairwise epitope mapping was performed. The chip was functionalized with antibody mAb1D7, which binds to domain III of AMA1. This chip was regenerable. Subsequently, in each cycle AMA1 (3D7) (1750 RU) was caught. Then, humAbAMA1, 1F9, or 4G2 were injected singly (cycles 1, 2, 4 and 5) or consecutively (cycles 3 and 6). In cycles 1–3, 1 μg/ml of each antibody was applied, whereas in cycles 4–6, the concentration of each antibody was 10 μg/ml. In case spectral resonance is increasing with the binding of the secondary antibody, no competition is detectable, in case the signal is not increased after binding of the secondary antibody, the epitopes either overlap or binding is compromised by allosteric competition.
Epitope Mapping.
The epitope of humAbAMA1 was mapped using the CLIPS-constrained peptides. Its localization pertains to domain I of AMA1 (yellow) and is highlighted in red. The published epitope of 1F9 within domain I is highlighted in cyan, the region in which the epitopes of humAbAMA1 and 1F9 overlap is highlighted in magenta56. The domain II of AMAI is shown in blue. The RON2 peptide (green) is bound to its specific hydrophobic binding groove in domain I. The structural data was obtained from the protein data base PDB, accession number 3ZWZ102. The structure in the second image is a 90° rotation of the first image.
RON2sp1 and humAbAMA1 compete for binding to AMA1 (3D7)
HumAbAMA1 was capable of inhibiting P. falciparum 3D7A with an IC50 value of 35 μg/mL (Fig. 5) and competed with 1F9 for binding to AMA1 on the same epitope. We were thus interested if humAbAMA1 might as well interfere with the binding of RON2, the natural ligand of AMA1, which had been previously described for both 1F9 and 4G242,43,56,59. AMA1 (3D7) and RON2sp1 peptide, the portion of RON2, which binds to the hydrophobic trough of AMA160, were pre-incubated in different molar ratios. In SPR measurements it was then analyzed to which extent humAbAMA1 could still bind AMA1. As can be seen in Fig. 9 the more RON2sp1 had been binding to AMA1 (3D7), the lower was the proportion of humAbAMA1 recognizing AMA1 (3D7). This indicates that humAbAMA1 binds to an epitope on AMA1 which is at least partially obscured once RON2sp1 has bound to AMA1.
Competition of humAbAMA1 and RON2sp1 peptide.
The determination of the epitope region recognized by humAbAMA1 using the peptide array, as well as the binning experiments suggests a possible steric interference between humAbAMA1 and RON2sp1. To investigate the potential competition between RON2sp1 and humAbAMA1 for AMA1-binding AMA1 (3D7) was preincubated with various concentrations of the peptide RON2sp1 for 1 hour. Subsequently, the AMA1 (3D7):RON2sp1 solutions were injected over a surface featuring a defined amount (400 RU) of humAbAMA1 caught on a Protein A-functionalized chip using a BIAcore T200 SPR instrument. For visualization of the results the measured response units were plotted against the concentration of RON2sp1 (μM) used as competitor.
Discussion
The presented work describes the first human antibody recognizing the malaria vaccine candidate AMA1. HumAbAMA1 reflects a significant degree of affinity maturation in vivo (88–96% germ line homology), recognizes a conformational variable epitope lying in very close proximity to the RON2 binding site, and inhibits various P. falciparum strains with IC50 values ranging from of 35 μg/ml for the strain 3D7A to 195 μg/ml for the strain FCR3.
Most individuals who are premune to malaria have developed antibodies directed at AMA110,54,61,62 and the recognition of conformational determinants of AMA1 is believed to be crucial for a protective AMA1-specific response63,64,65. Rodent antibodies such as 1F9 and 4G2 with inhibitory potential had been reported about fifteen to twenty years ago66,67. Here, for the first time, a similar human monoclonal antibody is described. The natural affinity maturation of humAbAMA1 resulted in a binding strength of <150 pM, which is close to the range of 10−11 M that is considered the lower limit to be achievable in vivo68,69,70. Only few human antibodies have been disclosed which possess KD values of as low as ≤10−10 M71,72. Moreover, as compared to 1F9 and 4G2 exhibiting KD values of 1.3 × 10−9 M and 3.3 × 10−9 M, respectively73, the affinity of humAbAMA1 to AMA1 (3D7) is 7- to 18-times stronger.
In analogy to the higher affinity, the inhibitory potential of humAbAMA1 described by its IC50 value of 35 μg/ml is well below the IC50 values reported for 1F9 (ca. 850 μg/ml)42 and AMA1-specific polyclonal human IgG (70–240 μg/ml)38,74 and to our knowledge, it is the lowest IC50-value for a blood-stage antibody described so far. Differences of the values we obtained here and the inhibitory concentrations described in the literature might be based on differences in the assays (parasitemia, hematocrit, readout) as well as the fact that we used recombinant chimeric antibodies, whereas in the publications stated above, the original rodent antibodies have been used.
Very efficient murine mAbs directed at other plasmodial targets such as reticulocyte-binding protein homologue 5 (Rh5) show IC50 values of 50–100 μg/ml44, and AMA1-specific polyclonal rabbit IgG generated by hyperimmunization protocols could achieve IC50 values in a similar range of 34–180 μg/ml73,75. A direct comparison between polyclonal and monoclonal antibodies is difficult as in vivo, an entire spectrum of antibodies contributes to the defense against plasmodial infections and a successful control is likely due to synergistic effects of these antibodies. Therefore, we used humAbAMA1 in combination with humAb10.1, which showed the best IC50 value of all MSP10-specific antibodies we described52. We found a weak synergistic effect, characterized by a HEWLETT’s S of 1.6. Nonetheless, the effect is inferior to the synergistic effect that has been described for other polyclonal antibody combinations, e.g. murine Rh4- and Rh5-specific mAbs76, or combinations of anti-plasmodial chemotherapeutics77,78.
Although humAbAMA1 inhibits the parasite strains HB3 and FCR3 with IC50- values of 126 and 195 μg/ml, which is 3.6 to 5.6-fold higher than the respective IC50-value measured in the strain 3D7A, it is still superior to the inhibitory capacity of the well-described antibodies 4G2 and 1F9. It is a very interesting observation that the antibody showed a direct relation between the affinity to the antigen variant and the inhibitory capacity to the respective parasite strain. This needs to be reconfirmed with an increased number of parasite strains and antigen variants.
AMA1-specific antibodies may inhibit plasmodia by different means, i.e. (1) the blocking of epitopes that are important for the interaction with other proteins such as RON2, (2) the prevention of structural rearrangements within the AMA1 molecule required for efficient RON2-binding, (3) the impairment of the proteolytic processing of AMA1, and (4) the interference of the redistribution of the cleavage products79. Moreover, a fourth mode of action could be the premature induction of the phosphorylation of serine 610 in the cytoplasmic tail of AMA1 and the untimely initiation of downstream signalling events, e.g. by imitating the binding of its natural ligand. The phosphorylation of S610 was shown to be critical for invasion37,80,81. Based on the current data at hand we cannot conclusively say how humAbAMA1 exerts its inhibition of P. falciparum 3D7A. However, there are several clues. The first evidence is that 1F9 and humAbAMA1 competed for binding to AMA1 (3D7), whereas 4G2 and humAbAMA1 does not (Fig. 7). 4G2 binds to a region of domain II which is distant from the hydrophobic trough of AMA134,43, but until now, the crystal structure of 4G2 binding to AMA1 could not be resolved. 1F9 on the other hand binds close to the hydrophobic cleft of AMA1 and to adjacent regions in domain I, including residues P188, M190, P192, E197, H200, F201, Y202, D204, M224, I225, N228 in AMA1 (3D7)56. This region shares a significant overlap with the epitope region of humAbAMA1 identified by peptide array analysis (AA 194–206). Additionally, we showed either a clear intereference with the binding site of RON2, since humAbAMA1 could not bind efficiently to AMA1:RON2sp1 complexes in an SPR setup (Fig. 9). This implies that by the binding of RON2sp1 the epitope of humAbAMA1 was either masked or structurally rearranged by RON2sp1. The evidence given strongly argues for an interference of humAbAMA1 with the binding of RON2 to AMA1 on the parasite, thus preventing the formation of the AMA1:RON2 complex, leading to the inhibition we observed in GIAs. However, this mode of inhibition remains to be proven.
In this study we took a closer look at P. falciparum strains 3D7A, HB3 and FCR3 as well as their respective AMA1 alleles. It is well known that AMA1 is subject to diversifying and balancing selection in order to evade the host’s immune system as well as to maintain its structure and function82,83,84. Therefore, future experiments will have to address if humAbAMA1 is capable of inhibiting even more P. falciparum strains than the presented ones. Nevertheless, the data presented here already shows that the antibody has some cross-strain inhibitory activity, even though it binds to a variable region of the parasite. The determination of the structures of a Fab fragment of humAbAMA1 and of the Fab:AMA1 complex will likely help to shed more light on the nature of the epitope and the mode of action of the new antibody we presented.
Since transient expression in N. benthamiana is a well-established antibody52 and vaccine candidate85 expression platform in our lab, we used it for the production of humAbAMA1 and compared expression levels and antigen-binding properties with humAbAMA1 produced by transient transfection of HEK293-6E cells. As shown by SPR-analysis, the plant and HEK produced molecules show no significant differences in AMA1 binding.
Besides elucidating the epitope targets and parasite growth inhibitory mechanisms of naturally occurring human Pf-specific mAbs, such molecules are also of interest for therapeutic use. Of crucial importance for a therapeutic antibody is its ability to inhibit a brought set of parasites occurring naturally. The set of parasites chosen here represent a number of variants of the estimated epitope. All strains could be inhibited by humAbAMA1, albeit at varying concentrations. Nevertheless, further investigations are necessary in order to estimate the full therapeutic potential of humAbAMA1. Even though the majority of monoclonal antibodies used in therapy is targeting tumors or chronic inflammatory diseases, there are certain applications for antibodies and antibody cocktails also in other fields like the treatment of toxin mediated86 and infectious diseases. Post-exposure treatment of rabies infections includes the injection of rabies virus specific antibodies isolated from horse serum87, but various groups are working on the development of recombinantly produced humanized or human monoclonal antibodies to replace such sera. Humanized monoclonal antibodies have been proposed as therapeutics for West Nile Virus infections88 and a cocktail of three humanized antibodies89 targeting the Ebola virus EBOV glycoprotein has received some attention in the context of the latest outbreak of EBOV in several West African countries. The antibody-based Ebola-emergency treatment ZMapp used in a small number of human patients has been produced in N. benthamiana plants by transient expression90. In the case of malaria antibodies could be used for short-term prevention and/or therapy in clinical cases with drug resistant strains91. Based on antibodies such as the one presented here in combination with affordable expression systems, the cure of malaria infections or the generation of passive vaccination strategies becomes feasible.
Materials and Methods
Ethics statement and blood donors
This study was conducted according to the principles expressed in the Declaration of Helsinki. Ethical approval was given by the Committee on Human Research Publication and Ethics (CHRPE) of the Kwame Nkrumah University of Science and Technology in Ghana. After the goals of the study had been carefully explained, written informed consent was obtained from all participants. By the time of blood donation, the 31 volunteers were between 20 and 45 years old and had not undergone any clinical phase of malaria for at least two years. The Ghanaian study, as well as the inclusion and exclusion criteria had been described previously92. Each volunteer donated 50 ml of heparinized blood which was then used for the preparation of plasma and PBMCs as described below.
Preparation of peripheral blood mononuclear cells (PBMCs) from Ghanaian donors
PBMCs were prepared from 50 ml of heparinized peripheral blood from each donor. The blood was diluted in one volume of PBS and was then layered onto Ficoll (GE Healthcare, Uppsala, Sweden). After centrifugation (800× g, 20 min, 25 °C, without break) the PBMCs in the interphase were drawn off, washed once in PBS containing 1% (v/v) FCS (400× g, 10 min, 25 °C), and once in PBS containing 10% (v/v) FCS (300× g, 10 min, 25 °C). They were then taken up in freezing medium (10% (v/v) DMSO in FCS) at cellular concentrations ranging from 3–6 × 106 cells/ml and were cryopreserved. The temperature was gradually decreased to −80 °C (1 K/min) using isopropyl-containing freezing containers.
Fluorescent labeling of recombinant AMA1 (DiCo 1-3)
AMA1 (DiCo 1-3) was kindly provided by Jürgen Drossard (Fraunhofer IME, Aachen, Germany) with the kind permission of Ed Remarque (BPRC Rijswijk, The Netherlands). The proteins were chemically labeled with “AlexaFluor488-TFP” (Life Technologies, Darmstadt, Germany) as recommended by the manual’s instructions. On average, the proteins were found to be labeled with 5.9 AlexaFluor 488 moieties (mol/mol) as determined by a spectrophotometric measurement.
Reduction and alkylation of AMA1 (3D7)
For testing of conformational dependency of epitope recognition by polyclonal AMA1-specific antibodies and by humAbAMA1, AMA1 (3D7) was irreversibly reduced. To this end, it was treated with 5 mM DTT at 56 °C for 45 min while shaking. The reaction was stopped by adding 15 mM iodoacetamide. After incubation for 30 min in the dark 5 mM DTT were added to quench excessive iodoacetamide.
Flow cytometric cell sorting
PBMCs were thawed gently (37 °C) and were washed once with RPMI 1640 medium (10% (v/v) FCS). The staining with human IgG- and human CD22-specific antibodies (anti-IgG-PE and anti-CD22-APC, BD Biosciences, Heidelberg, Germany) and fluorescent AMA1 (DiCo 1-3) in labeling buffer (2% (v/v) FCS, 1 mM EDTA in PBS (137 mM NaCl, 2.7 mM KCl, 8.1 mM Na2HPO4, 1.5 mM KH2PO4)) was carried out on ice for 30 min. Cells were then washed three times with cold labeling buffer and were individualized with a 30 μm-mesh before resuspension in particle-free RPMI 1640 (20% (v/v) FCS). Antigen-specific IgG+/CD22+ B cells were sorted with an FACS Influx cytometer (BD Biosciences, Heidelberg, Germany) into RPMI 1640 (20% (v/v) FCS).
Epstein-Barr virus transformation
After a viability staining with Trypan blue specific IgG+/CD22+ B lymphocytes were infected with Epstein-Barr virus (B95-8) and subjected to virocrine transformation as described52,93.
Enzyme-linked immunosorbent assays
ELISA procedures for the various parts of the study were carried out as described before to: (A) identify promising B cell cultures52, (B) assess the specificity of recombinant human antibodies52, and (C) screen the Ghanaian plasma samples for reactivity against AMA1 (3D7)54.
Rescue and cloning of immunoglobulin variable regions
cDNA of promising cultures was generated with the “SuperScript III CellsDirect” kit (Life Technologies, Darmstadt, Germany) according to the manufacturer’s instructions using oligonucleotides for the rescue and cloning of human immunoglobulin variable regions (Vh and Vl) as previously described94. Primers for the cloning into pTRAkt vectors were designed to correct any mutations introduced by the degenerate primer set used for nested PCRs – thereby reconstituting the authentic sequences of 1E4 Vh and 1E4 Vl. Vh and Vl were cloned individually into the respective pTRAkt vectors for apoplastic expression of human full-size IgG1:κ antibodies in plants95.
Expression of full-size humAbAMA1 IgG in Nicotiana benthamiana
Expression and purification of full-size IgG by Agrobacterium-mediated transient transfection of N. benthamiana plants was performed using agrobacteria transformed with the vectors pTRAkt_IgG1_H_1E4 (humAbAMA1 IgG heavy chain) and pTRAkt_Igκ_1E4 (humAbAMA1 Igκ light chain), respectively, as described elsewhere95.
Expression of full-size humAbAMA1 IgG in HEK293-6E
Expression of humAbAMA1 in HEK293-6E cells was performed with the help of vector pTT5, into which the expression cassette used in pTRAkt vectors described above was inserted. Transfection, cultivation of the cells as well as ProteinA-based purification was performed as described before96.
Plasmodium falciparum culture and growth inhibition assays
P. falciparum strains 3D7A, HB3 and FCR3 were cultured as described97. Parasites were maintained at a hematocrit of 5% in RPMI 1640, 25 mM HEPES, 2 mM L-glutamine, 50 μg/ml gentamycin, 10% Albumax II.
In vitro growth inhibition of the various antibodies was assessed with P. falciparum 3D7A, HB3 and FCR3 as described before53. HumAbAMA1 was applied in concentrations ranging from 4.35 mg/ml to 4.25 μg/ml. Polyclonal rabbit-anti-AMA1 IgG (BG98) in a concentration of 6 mg/ml was taken as a positive control98. Serum IgG of a naïve rabbit was taken as a corresponding negative control (6 mg/ml). HumAb 2G12, an HIV-1-specific IgG, was taken as a plant-expressed non-specific control. 2G12 was kindly provided by Jürgen Drossard. Prior to the setup of the assay P. falciparum strains had been synchronized three times by sorbitol treatments99. Parasites were seeded at schizont stage at a parasitemia of 0.4% in 2% hematocrit. After an incubation period of 40–46 h, the GIA was harvested and frozen at −80 °C for at least 16 h. Parasitaemia was read out by assessing the pLDH activity100. Relative growth inhibition was calculated as follows: % inhibition = 100 − 100 × [A655nm (sample) − A655nm (erythrocyte control)]/[A655nm (schizont control) − A655nm (erythrocyte control)]. In order to test for a synergy of invasion inhibition both humAbAMA1 and a MSP10-specific antibody, humAb10.1 (described in ref. 52), were employed. To this end, the antibodies were used singly and in ratios proportional to their respective IC50 values. The ratios were 1:1, 1.5:1, 4:1, 1:2, and 1:4. Data were analyzed by calculating HEWLETT’s S and the sum of FIC, a quantitative index of the extent of synergy76.
Immunofluorescence assay
For immunofluorescence assays P. falciparum 3D7A was synchronized by sorbitol treatments. Parasitized erythrocytes were washed once with RPMI 1640 before being taken up in FCS (low bovine IgG) and smeared onto object slides. Parasites were air-dried and then fixed in methanol at −20 °C for 10 min. Slides were incubated with 5 μg/ml humAbAMA1 and 20 μg/ml mouse-anti-MSP4(EGF) (mAb 2.44, kindly provided by Alexander Boes, Fraunhofer IME, Germany)95 in 1% FCS (low bovine IgG) in PBS in a moist box at 25 °C for 1 h. After five washing steps in PBS, the slides were treated with 1:100 goat-anti-mouse IgG(H + L)-AlexaFluor488 (Life Technologies, Darmstadt, Germany), 1:1000 goat-anti-human IgG(H + L)-Cy3 (Jackson ImmunoResearch/Dianova, Hamburg, Germany), and 10 μg/ml Hoechst 33342 in a moist box at 25 °C for 1 h. Five washing steps were carried out. Mounting medium (“ProLong Gold Antifade Reagent”, Life Technologies, Darmstadt, Germany) was used to conserve the staining. Images were taken with a Leica TCS SP8 microscope at a 630-fold magnification.
Surface plasmon resonance spectroscopy
In our study SPR measurements fulfilled three purposes. (A) Determination of the affinity of humAbAMA1 to AMA1 (3D7), (B) pairwise epitope mappings with monoclonal rodent antibodies 1F9 and 4G2, and (C) RON2-competition measurements. All measurements were performed by using a Biacore T200 instrument (Biacore, GE Healthcare) and CM5-S-Series sensor chips at 25 °C with HBS-EP (10 mM 4-(2-hydroxyethyl)-1piperazineethanesulfonic acid, 150 mM NaCl, 3 mM EDTA, 0.005% (w/v) polysorbat-20) as running buffer. (A) The kinetic analysis of the humAbAMA1 interaction was performed as previously described73. (B) To roughly map the epitope of humAbAMA1 in relation to the binding sites of previously described AMA1 specific monoclonal antibodies 1D7, 1F9 and 4G2, AMA1 (3D7) was captured (1600 RU) using a CM5 surface functionalized by EDC-NHS chemistry with 7000 RU of the rat monoclonal 1D7. Mab1D7 is specific for the ectodomain III of AMA1, recognizes an epitope, which is distant from the hydrophobic cleft, and thus does not interfere with 1F9 or 4G2 binding. The antibodies 1F9, 4G2 and humAbAMA1 (at concentrations of 1 μg/ml and/or 10 μg/ml) were separately and consecutively injected for 180 s and binding levels were compared. (C) In order to rule out if humAbAMA1 and RON2 compete for binding to AMA1, AMA1 (3D7) and RON2sp1 were mixed in different molar ratios and incubated for 1 h at 25 °C. (RON2sp1 corresponds to the portion of RON2 which binds to the hydrophobic through of AMA1102). The extent to which humAbAMA1 could bind to AMA1:RON2sp1 complexes was then determined. To this end, first, humAbAMA1 was caught on a Protein A-functionalized chip and then the different pre-incubated AMA1:RON2sp1 solutions were injected.
Epitope mapping using Pepscan screening
The epitope mapping of humAbAMA1 was carried out by Pepscan Presto BV (Lelystad, The Netherlands) as described before58. For the screening, four sets of peptides were used, all based on the AMA1 sequence of the parasite strain 3D7. Set 1 consisted of linear peptides of the length of fifteen amino acids. Set 2 consisted of constrained peptides of the length of 17 amino acids, with cysteine residues inserted at positions 1 and 17, and amino acids 2–16 representing AMA1 sequences. Cysteine residues on positions 1 and 17 were linked using the CLIPS technology as described58. Set 3 was designed to mimic α-helices of AMA1. These peptides consisted of 16 amino acids with cysteine residues inserted at positions 1 and 5. Set 4 constituted peptides with lengths of 22 amino acids and with cysteine placed on position 1 and 22. In this peptide set 4, amino acids on positions 11 and 12 had been replaced by proline and glycine, respectively. In all non-linear peptide sets described, cysteine residues occurring naturally the AMA1 sequence were chemically protected by acetamidomethyl in order to prevent unintentional reaction.
Additional Information
How to cite this article: Maskus, D. J. et al. Characterization of a novel inhibitory human monoclonal antibody directed against Plasmodium falciparum Apical Membrane Antigen 1. Sci. Rep. 6, 39462; doi: 10.1038/srep39462 (2016).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Fendel, R. et al. Hemolysis is associated with low reticulocyte production index and predicts blood transfusion in severe malarial anemia. PLoS One 5, e10038 (2010).
Ramasamy, R. Molecular basis for evasion of host immunity and pathogenesis in malaria. Biochim Biophys Acta 1406, 10–27 (1998).
Day, K. P. & Marsh, K. Naturally acquired immunity to Plasmodium falciparum. Immunology today 12, A68–71 (1991).
Dzikowski, R. & Deitsch, K. W. Genetics of antigenic variation in Plasmodium falciparum. Current genetics 55, 103–110 (2009).
Langhorne, J., Ndungu, F. M., Sponaas, A.-M. & Marsh, K. Immunity to malaria: more questions than answers. Nature immunology 9, 725–732 (2008).
Elloso, M. M., van der Heyde, H. C., vande Waa, J. A., Manning, D. D. & Weidanz, W. P. Inhibition of Plasmodium falciparum in vitro by human gamma delta T cells. Journal of immunology (Baltimore, Md.: 1950) 153, 1187–1194 (1994).
Costa, G. et al. Control of Plasmodium falciparum erythrocytic cycle: γδ T cells target the red blood cell-invasive merozoites. Blood 118, 6952–6962 (2011).
Corradin, G. & Levitskaya, J. Priming of CD8(+) T Cell Responses to Liver Stage Malaria Parasite Antigens. Frontiers in immunology 5, 527 (2014).
Baird, J. K. Host age as a determinant of naturally acquired immunity to Plasmodium falciparum. Parasitol Today 11, 105–111 (1995).
Chelimo, K. et al. Antibodies to Plasmodium falciparum antigens vary by age and antigen in children in a malaria-holoendemic area of Kenya. Pediatr Infect Dis J 24, 680–684 (2005).
Osier, Faith H. A. et al. Breadth and magnitude of antibody responses to multiple Plasmodium falciparum merozoite antigens are associated with protection from clinical malaria. Infection and immunity 76, 2240–2248 (2008).
Ramasamy, R. & Rajakaruna, R. Association of malaria with inactivation of alpha1,3-galactosyl transferase in catarrhines. Biochimica et biophysica acta 1360, 241–246 (1997).
Kumaratilake, L. M., Ferrante, A., Jaeger, T. & Rzepczyk, C. GM-CSF-induced priming of human neutrophils for enhanced phagocytosis and killing of asexual blood stages of Plasmodium falciparum: synergistic effects of GM-CSF and TNF. Parasite immunology 18, 115–123 (1996).
Miller, L. H. et al. Monoclonal antibodies to a 140,000-m.w. protein on Plasmodium knowlesi merozoites inhibit their invasion of rhesus erythrocytes. Journal of immunology (Baltimore, Md.: 1950) 132, 438–442 (1984).
Ramasamy, R., Jones, G. & Lord, R. Characterisation of an inhibitory monoclonal antibody-defined epitope on a malaria vaccine candidate antigen. Immunology letters 23, 305–309 (1990).
Blackman, M. J., Heidrich, H. G., Donachie, S., McBride, J. S. & Holder, A. A. A single fragment of a malaria merozoite surface protein remains on the parasite during red cell invasion and is the target of invasion-inhibiting antibodies. The Journal of experimental medicine 172, 379–382 (1990).
Groux, H. & Gysin, J. Opsonization as an effector mechanism in human protection against asexual blood stages of Plasmodium falciparum: functional role of IgG subclasses. Research in immunology 141, 529–542 (1990).
Bouharoun-Tayoun, H., Oeuvray, C., Lunel, F. & Druilhe, P. Mechanisms underlying the monocyte-mediated antibody-dependent killing of Plasmodium falciparum asexual blood stages. J Exp Med 182, 409–418 (1995).
Jafarshad, A. et al. A novel antibody-dependent cellular cytotoxicity mechanism involved in defense against malaria requires costimulation of monocytes FcgammaRII and FcgammaRIII. Journal of immunology (Baltimore, Md.: 1950) 178, 3099–3106 (2007).
Kapelski, S., Klockenbring, T., Fischer, R., Barth, S. & Fendel, R. Assessment of the neutrophilic antibody-dependent respiratory burst (ADRB) response to Plasmodium falciparum. Journal of leukocyte biology 96, 1131–1142 (2014).
Dvorak, J. A., Miller, L. H., Whitehouse, W. C. & Shiroishi, T. Invasion of erythrocytes by malaria merozoites. Science (New York, N.Y.) 187, 748–750 (1975).
Gilson, P. R. & Crabb, B. S. Morphology and kinetics of the three distinct phases of red blood cell invasion by Plasmodium falciparum merozoites. International journal for parasitology 39, 91–96 (2009).
Agnandji, S. T. et al. Induction of Plasmodium falciparum-specific CD4+ T cells and memory B cells in Gabonese children vaccinated with RTS,S/AS01(E) and RTS,S/AS02(D). PLoS One 6, e18559 (2011).
Narum, D. L. & Thomas, A. W. Differential localization of full-length and processed forms of PF83/AMA-1 an apical membrane antigen of Plasmodium falciparum merozoites. Molecular and biochemical parasitology 67, 59–68 (1994).
Peterson, M. G. et al. Integral membrane protein located in the apical complex of Plasmodium falciparum. Molecular and cellular biology 9, 3151–3154 (1989).
Donahue, C. G., Carruthers, V. B., Gilk, S. D. & Ward, G. E. The Toxoplasma homolog of Plasmodium apical membrane antigen-1 (AMA-1) is a microneme protein secreted in response to elevated intracellular calcium levels. Molecular and biochemical parasitology 111, 15–30 (2000).
Healer, J., Crawford, S., Ralph, S., McFadden, G. & Cowman, A. F. Independent translocation of two micronemal proteins in developing Plasmodium falciparum merozoites. Infection and immunity 70, 5751–5758 (2002).
Howell, S. A., Withers-Martinez, C., Kocken, C. H., Thomas, A. W. & Blackman, M. J. Proteolytic processing and primary structure of Plasmodium falciparum apical membrane antigen-1. The Journal of biological chemistry 276, 31311–31320 (2001).
Howell, S. A. et al. A single malaria merozoite serine protease mediates shedding of multiple surface proteins by juxtamembrane cleavage. The Journal of biological chemistry 278, 23890–23898 (2003).
Besteiro, S., Michelin, A., Poncet, J., Dubremetz, J.-F. & Lebrun, M. Export of a Toxoplasma gondii rhoptry neck protein complex at the host cell membrane to form the moving junction during invasion. PLoS pathogens 5, e1000309 (2009).
Tonkin, M. L. et al. Host cell invasion by apicomplexan parasites: insights from the co-structure of AMA1 with a RON2 peptide. Science (New York, N.Y.) 333, 463–467 (2011).
Lamarque, M. et al. The RON2-AMA1 interaction is a critical step in moving junction-dependent invasion by apicomplexan parasites. PLoS pathogens 7, e1001276 (2011).
Bai, T. et al. Structure of AMA1 from Plasmodium falciparum reveals a clustering of polymorphisms that surround a conserved hydrophobic pocket. Proceedings of the National Academy of Sciences of the United States of America 102, 12736–12741 (2005).
Pizarro, J. C. et al. Crystal structure of the malaria vaccine candidate apical membrane antigen 1. Science 308, 408–411 (2005).
Baum, J. et al. A conserved molecular motor drives cell invasion and gliding motility across malaria life cycle stages and other apicomplexan parasites. The Journal of biological chemistry 281, 5197–5208 (2006).
Jewett, T. J. & Sibley, L. D. Aldolase forms a bridge between cell surface adhesins and the actin cytoskeleton in apicomplexan parasites. Molecular cell 11, 885–894 (2003).
Srinivasan, P. et al. Binding of Plasmodium merozoite proteins RON2 and AMA1 triggers commitment to invasion. Proceedings of the National Academy of Sciences of the United States of America 108, 13275–13280 (2011).
Hodder, A. N., Crewther, P. E. & Anders, R. F. Specificity of the protective antibody response to apical membrane antigen 1. Infection and immunity 69, 3286–3294 (2001).
Silvie, O. et al. A role for apical membrane antigen 1 during invasion of hepatocytes by Plasmodium falciparum sporozoites. The Journal of biological chemistry 279, 9490–9496 (2004).
McBride, J. S. & Heidrich, H. G. Fragments of the polymorphic Mr 185,000 glycoprotein from the surface of isolated Plasmodium falciparum merozoites form an antigenic complex. Molecular and biochemical parasitology 23, 71–84 (1987).
Uthaipibull, C. et al. Inhibitory and blocking monoclonal antibody epitopes on merozoite surface protein 1 of the malaria parasite Plasmodium falciparum. Journal of molecular biology 307, 1381–1394 (2001).
Coley, A. M. et al. The most polymorphic residue on Plasmodium falciparum apical membrane antigen 1 determines binding of an invasion-inhibitory antibody. Infection and immunity 74, 2628–2636 (2006).
Collins, C. R. et al. Fine mapping of an epitope recognized by an invasion-inhibitory monoclonal antibody on the malaria vaccine candidate apical membrane antigen 1. The Journal of biological chemistry 282, 7431–7441 (2007).
Douglas, A. D. et al. Neutralization of Plasmodium falciparum merozoites by antibodies against PfRH5. Journal of immunology (Baltimore, Md.: 1950) 192, 245–258 (2014).
Cheng, X.-J. et al. Production of high-affinity human monoclonal antibody fab fragments to the 19-kilodalton C-terminal merozoite surface protein 1 of Plasmodium falciparum. Infection and immunity 75, 3614–3620 (2007).
Sowa, K. M. et al. Isolation of a monoclonal antibody from a malaria patient-derived phage display library recognising the Block 2 region of Plasmodium falciparum merozoite surface protein-1. Mol Biochem Parasitol 112, 143–147 (2001).
Stubbs, J. et al. Strain-transcending Fc-dependent killing of Plasmodium falciparum by merozoite surface protein 2 allele-specific human antibodies. Infection and immunity 79, 1143–1152 (2011).
Lundquist, R. et al. Human recombinant antibodies against Plasmodium falciparum merozoite surface protein 3 cloned from peripheral blood leukocytes of individuals with immunity to malaria demonstrate antiparasitic properties. Infection and immunity 74, 3222–3231 (2006).
Chappel, J. A., Hollingdale, M. R. & Kang, A. S. IgG(4) Pf NPNA-1 a human anti-Plasmodium falciparum sporozoite monoclonal antibody cloned from a protected individual inhibits parasite invasion of hepatocytes. Hum Antibodies 13, 91–96 (2004).
Roeffen, W. F. et al. Recombinant human antibodies specific for the Pfs48/45 protein of the malaria parasite Plasmodium falciparum. J Biol Chem 276, 19807–19811 (2001).
Barfod, L. et al. Human pregnancy-associated malaria-specific B cells target polymorphic, conformational epitopes in VAR2CSA. Mol Microbiol 63, 335–347 (2007).
Maskus, D. J. et al. Isolation, production and characterization of fully human monoclonal antibodies directed to Plasmodium falciparum MSP10. Malaria journal 14, 276 (2015).
Remarque, E. J., Faber, B. W., Kocken, Clemens, H. M. & Thomas, A. W. A diversity-covering approach to immunization with Plasmodium falciparum apical membrane antigen 1 induces broader allelic recognition and growth inhibition responses in rabbits. Infection and immunity 76, 2660–2670 (2008).
Feller, T. et al. Plant-based production of recombinant Plasmodium surface protein pf38 and evaluation of its potential as a vaccine candidate. PLoS One 8, e79920 (2013).
Hodder, A. N. et al. The disulfide bond structure of Plasmodium apical membrane antigen-1. The Journal of biological chemistry 271, 29446–29452 (1996).
Coley, A. M. et al. Structure of the malaria antigen AMA1 in complex with a growth-inhibitory antibody. PLoS pathogens 3, 1308–1319 (2007).
Dutta, S., Haynes, J. D., Moch, J. K., Barbosa, A. & Lanar, D. E. Invasion-inhibitory antibodies inhibit proteolytic processing of apical membrane antigen 1 of Plasmodium falciparum merozoites. Proceedings of the National Academy of Sciences of the United States of America 100, 12295–12300 (2003).
Timmerman, P., Puijk, W. C. & Meloen, R. H. Functional reconstruction and synthetic mimicry of a conformational epitope using CLIPS technology. Journal of molecular recognition: JMR 20, 283–299 (2007).
Collins, C. R., Withers-Martinez, C., Hackett, F. & Blackman, M. J. An inhibitory antibody blocks interactions between components of the malarial invasion machinery. PLoS pathogens 5, e1000273 (2009).
Bonsignori, M. et al. Analysis of a clonal lineage of HIV-1 envelope V2/V3 conformational epitope-specific broadly neutralizing antibodies and their inferred unmutated common ancestors. Journal of virology 85, 9998–10009 (2011).
Thomas, A. W. et al. Aspects of immunity for the AMA-1 family of molecules in humans and non-human primates malarias. Memorias do Instituto Oswaldo Cruz 89 Suppl 2, 67–70 (1994).
Polley, S. D. et al. Human antibodies to recombinant protein constructs of Plasmodium falciparum Apical Membrane Antigen 1 (AMA1) and their associations with protection from malaria. Vaccine 23, 718–728 (2004).
Crewther, P. E., Matthew, M. L., Flegg, R. H. & Anders, R. F. Protective immune responses to apical membrane antigen 1 of Plasmodium chabaudi involve recognition of strain-specific epitopes. Infection and immunity 64, 3310–3317 (1996).
Anders, R. F. et al. Immunisation with recombinant AMA-1 protects mice against infection with Plasmodium chabaudi. Vaccine 16, 240–247 (1998).
Cortes, A. et al. Allele specificity of naturally acquired antibody responses against Plasmodium falciparum apical membrane antigen 1. Infection and immunity 73, 422–430 (2005).
Kocken, C. H. et al. Precise timing of expression of a Plasmodium falciparum-derived transgene in Plasmodium berghei is a critical determinant of subsequent subcellular localization. The Journal of biological chemistry 273, 15119–15124 (1998).
Coley, A. M. et al. Rapid and precise epitope mapping of monoclonal antibodies against Plasmodium falciparum AMA1 by combined phage display of fragments and random peptides. Protein engineering 14, 691–698 (2001).
Foote, J. & Milstein, C. Kinetic maturation of an immune response. Nature 352, 530–532 (1991).
Batista, F. D. & Neuberger, M. S. Affinity dependence of the B cell response to antigen: a threshold, a ceiling, and the importance of off-rate. Immunity 8, 751–759 (1998).
Klein, F. et al. Somatic mutations of the immunoglobulin framework are generally required for broad and potent HIV-1 neutralization. Cell 153, 126–138 (2013).
Wrammert, J. et al. Rapid cloning of high-affinity human monoclonal antibodies against influenza virus. Nature 453, 667–671 (2008).
Wu, X. et al. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science (New York, N.Y.) 329, 856–861 (2010).
Boes, A. et al. Detailed functional characterization of glycosylated and nonglycosylated variants of malaria vaccine candidate PfAMA1 produced in Nicotiana benthamiana and analysis of growth inhibitory responses in rabbits. Plant Biotechnol J 13, 222–234 (2015).
Miura, K. et al. Anti-apical-membrane-antigen-1 antibody is more effective than anti-42-kilodalton-merozoite-surface-protein-1 antibody in inhibiting plasmodium falciparum growth, as determined by the in vitro growth inhibition assay. Clin Vaccine Immunol 16, 963–968 (2009).
Boes, A. et al. Analysis of a Multi-component Multi-stage Malaria Vaccine Candidate–Tackling the Cocktail Challenge. PloS one 10, e0131456 (2015).
Williams, A. R. et al. Enhancing blockade of Plasmodium falciparum erythrocyte invasion: assessing combinations of antibodies against PfRH5 and other merozoite antigens. PLoS Pathog 8, e1002991 (2012).
Kerschbaumer, G. et al. Synergism between mefloquine and artemisinin and its enhancement by retinol in Plasmodium falciparum in vitro. Wiener klinische Wochenschrift 122 Suppl 3, 57–60 (2010).
Wurtz, N. et al. Synergy of mefloquine activity with atorvastatin, but not chloroquine and monodesethylamodiaquine, and association with the pfmdr1 gene. The Journal of antimicrobial chemotherapy 65, 1387–1394 (2010).
Dutta, S. et al. Mode of action of invasion-inhibitory antibodies directed against apical membrane antigen 1 of Plasmodium falciparum. Infection and immunity 73, 2116–2122 (2005).
Treeck, M. et al. Functional analysis of the leading malaria vaccine candidate AMA-1 reveals an essential role for the cytoplasmic domain in the invasion process. PLoS pathogens 5, e1000322 (2009).
Leykauf, K. et al. Protein kinase a dependent phosphorylation of apical membrane antigen 1 plays an important role in erythrocyte invasion by the malaria parasite. PLoS pathogens 6, e1000941 (2010).
Polley, S. D. & Conway, D. J. Strong diversifying selection on domains of the Plasmodium falciparum apical membrane antigen 1 gene. Genetics 158, 1505–1512 (2001).
Polley, S. D., Chokejindachai, W. & Conway, D. J. Allele frequency-based analyses robustly map sequence sites under balancing selection in a malaria vaccine candidate antigen. Genetics 165, 555–561 (2003).
Cortes, A. et al. Geographical structure of diversity and differences between symptomatic and asymptomatic infections for Plasmodium falciparum vaccine candidate AMA1. Infection and immunity 71, 1416–1426 (2003).
Boes, A. et al. A Plant-Based Transient Expression System for the Rapid Production of Malaria Vaccine Candidates. Methods in molecular biology (Clifton, N.J.) 1404, 597–619 (2016).
Chow, S.-K. & Casadevall, A. Monoclonal antibodies and toxins–a perspective on function and isotype. Toxins 4, 430–454 (2012).
Both, L. et al. Passive immunity in the prevention of rabies. The Lancet. Infectious diseases 12, 397–407 (2012).
Oliphant, T. et al. Development of a humanized monoclonal antibody with therapeutic potential against West Nile virus. Nature medicine 11, 522–530 (2005).
Pettitt, J. et al. Therapeutic intervention of Ebola virus infection in rhesus macaques with the MB-003 monoclonal antibody cocktail. Science translational medicine 5, 199ra113 (2013).
Davidson, E. et al. Mechanism of Binding to Ebola Virus Glycoprotein by the ZMapp, ZMAb, and MB-003 Cocktail Antibodies. Journal of virology 89, 10982–10992 (2015).
Pleass, R. J. & Holder, A. A. Opinion: antibody-based therapies for malaria. Nature reviews. Microbiology 3, 893–899 (2005).
Addai-Mensah, O. et al. Acquired immune responses to three malaria vaccine candidates and their relationship to invasion inhibition in two populations naturally exposed to malaria. Malaria journal 15, 65 (2016).
Fraussen, J. et al. A novel method for making human monoclonal antibodies. Journal of autoimmunity 35, 130–134 (2010).
Tiller, T. et al. Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J Immunol Methods 329, 112–124 (2008).
Kapelski, S. et al. Fast track antibody V-gene rescue, recombinant expression in plants and characterization of a Pf MSP4-specific antibody. Malaria journal 14, 50 (2015).
Durocher, Y., Perret, S. & Kamen, A. High-level and high-throughput recombinant protein production by transient transfection of suspension-growing human 293-EBNA1 cells. Nucleic acids research 30, E9 (2002).
Trager, W. & Jensen, J. B. Human malaria parasites in continuous culture. Science 193, 673–675 (1976).
Faber, B. W. et al. Diversity covering AMA1-MSP119 fusion proteins as malaria vaccines. Infection and immunity 81, 1479–1490 (2013).
Mordmüller, B. et al. Plasmodia express two threonine-peptidase complexes during asexual development. Mol Biochem Parasitol 148, 79–85 (2006).
Kapelski, S., Almeida, M. de, Fischer, R., Barth, S. & Fendel, R. Antimalarial activity of granzyme B and its targeted delivery by a granzyme B-single-chain fv fusion protein. Antimicrobial agents and chemotherapy 59, 669–672 (2015).
Greco, W. R., Bravo, G. & Parsons, J. C. The search for synergy: a critical review from a response surface perspective. Pharmacological reviews 47, 331–385 (1995).
Vulliez-Le Normand, B. et al. Structural and functional insights into the malaria parasite moving junction complex. PLoS pathogens 8, e1002755 (2012).
Acknowledgements
This work was supported by the “Fraunhofer Zukunftsstiftung”. Dominika Maskus was financially supported by the Jürgen Manchot Foundation. First of all, we would like to thank all blood donors from Ghana and the whole KCCR team from Kumasi/Ghana helping during the study, especially Prof. Margaret Frempong (KNUST, Kumasi, Ghana). We would also like to thank Thomas Rademacher (Fraunhofer IME, Aachen, Germany) for providing the pTRA vectors and cultivating Nicotiana benthamiana plants and Alexander Boes (Fraunhofer IME, Aachen, Germany) for providing AMA1 (3D7) and the antibody mAb 2.44. Antibody 2G12 was kindly provided by Jürgen Drossard (Fraunhofer IME, Aachen, Germany). Ed Remarque and Bart Faber (BPRC Rijswijk, The Netherlands) generously provided us with AMA1-specific polyclonal rabbit IgG (BG98) as well as AMA1 variants HB3 and FCR3. The following reagent was obtained through the MR4 as part of the BEI Resources Repository (NIAID, NIH, Bethesda, MD, USA): P. falciparum 3D7A, MRA-151, deposited by D. Walliker; P. falciparum HB3, MRA-155, deposited by T. Wellems; P. falciparum FCR3, MRA-736; hybridoma clone 1D7, deposited by CA Long (MRA 480A).
Author information
Authors and Affiliations
Contributions
D.J.Ma. created the study protocol, conceived the study, performed the experiments, analysed the data, and wrote the manuscript. H.Sp. designed and conducted all SPR measurements and contributed to writing the manuscript. M.Kr., S.Ka., M.Se., O.A.M. and S.Be. performed parts of the experiments. R.Fi., T.Kl., A.Re. and S.Ba. conceived the overall study design and contributed to writing the manuscript. R.Fe. conceived the study, created the study protocol, analyzed the data and wrote the manuscript. All authors read and approved of the final manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Maskus, D., Królik, M., Bethke, S. et al. Characterization of a novel inhibitory human monoclonal antibody directed against Plasmodium falciparum Apical Membrane Antigen 1. Sci Rep 6, 39462 (2016). https://doi.org/10.1038/srep39462
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep39462
This article is cited by
-
Structure-based design of a strain transcending AMA1-RON2L malaria vaccine
Nature Communications (2023)
-
Efficacy, T cell activation and antibody responses in accelerated Plasmodium falciparum sporozoite chemoprophylaxis vaccine regimens
npj Vaccines (2022)
-
Isolation and light chain shuffling of a Plasmodium falciparum AMA1-specific human monoclonal antibody with growth inhibitory activity
Malaria Journal (2021)
-
Heterologous protection against malaria by a simple chemoattenuated PfSPZ vaccine regimen in a randomized trial
Nature Communications (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.