Abstract
A complete fingerprint of a tissue sample requires a detailed description of its cellular and extracellular components while minimizing artifacts. We introduce the application of a novel scanning electron microscope (airSEMTM) in conjunction with light microscopy for functional analysis of tissue preparations at nanometric resolution (<10 nm) and under ambient conditions. Our metal-staining protocols enable easy and detailed visualization of tissues and their extracellular scaffolds. A multimodality imaging setup, featuring airSEMTM and a light microscope on the same platform, provides a convenient and easy-to-use system for obtaining structural and functional correlative data. The airSEMTM imaging station complements other existing imaging solutions and shows great potential for studies of complex biological systems.
Similar content being viewed by others
Introduction
Imaging methods have become increasingly important for studies of biological tissue specimens, as they provide information about sample morphology, topography and content from the level of individual molecules to the whole organ in health and various pathological stages. The scanning electron microscope (SEM) is a tool commonly used for obtaining nanometer spatial resolution images of the topography of a wide range of biological specimens, including microorganisms, cells and tissues. Conventional SEM settings, however, occur at high vacuum conditions, using fixed, completely dry, conductive and electrically grounded samples1,2. Higher spatial resolution may be achieved by using a transmission electron microscope (TEM) using samples embedded in resin and cut into ~0.06–1 μm sections.
Another option is cryo-electron microscopy (cryoEM) of both SEM and TEM, in which appropriate cryo-preparation3,4,5 provides a more faithful representation of biological samples at high resolution and avoids many of the otherwise induced artifacts. This way sample shrinkage and alterations of topography and ultrastructure are prevented. However, cryoEM is not always advisable on its own, since the image contrast is largely reduced due to the low inherent contrast of cryo-preserved biological specimens. In addition, sublimation of the samples using a high-vacuum cryo-SEM may induce charging, thus decreasing the resolution. Therefore, additional consideration regarding sample preparation is required6. Charging problems of insulating biological samples can be overcome by advanced SEM techniques, such as environmental SEM (ESEM)7, variable pressure SEM8 or helium ion microscopy (HIM)9,10.
There is a growing appreciation for the fact that the combination of data derived from an identical area of a particular sample using different visualizing modalities creates a multilayered stack of information that enables close inspection of structure- composition- function relationships. To this end, correlative light and electron microscopy (CLEM) has become a widely used informative tool11,12,13,14,15. Yet, the current available techniques are time consuming, involve complex sample manipulation, unidirectional (data has to be collected in a certain order, once a sample goes to the next modality it can't go back) and has precision limitations in matching the same area of a sample under different microscopes. In addition, sample preparation for conventional SEM imaging renders the tissue non-suitable for further downstream analyses such as proteomics or genomics, which are highly relevant in characterizing tissues undergoing pathological events. In order to provide a global approach, it is important not only to keep the original tissue morphology, but also its composition, thus the sample has to be appropriately prepared for both high resolution imaging followed by further downstream analyses.
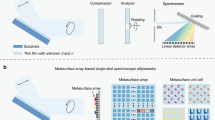
Here we introduce the use of the airSEMTM, http://www.b-nano.com/, a novel scanning electron microscope operating at ambient conditions and capable of measuring insulated, hydrated, non-coated biological samples (Figure 1a, Methods). Notably, the airSEMTM microscope may be easily integrated with various visualization modalities as part of the multimodality station (Figure 1b, Methods). This station enables one to image a specific, registered, region of interest (ROI) using correlative light and electron microscopies e.g. two photon and confocal reflection microscopes16,17. This makes the airSEMTM imaging station a powerful tool for studying biological tissues using different visualizing modalities. The use of the airSEMTM imaging station with sample preparation procedures based on our specially designed staining protocols, allows easy and fast data collection of high-quality imaging data by switching between microscopes on the same platform.
Schematic presentation of the airSEMTM and airSEMTM imaging station.
(a) A simplified description of the airSEMTM microscope. (b) AirSEMTM imaging station, describing the combination of the two visualizing modalities onto one platform. The sample is shuttled between LM and airSEMTM thus providing the continuation of magnification of the ROI.
In airSEMTM, (Figure 1a) a field-emitter column is utilized to generate, accelerate, shape and manipulate an electron beam in vacuum. This conditioned beam is consequently driven outside of the vacuum environment through a thin membrane and is allowed to interact with a sample located directly below the beam's exit point. As in conventional SEMs the beam is raster scanned across the ROI while the products of the beam-specimen interaction at each point are recorded to generate an image. Some of the detectors used to record these beam-sample interaction products are located in an evacuated environment – the same vacuum seen by the lower part of a SEM column while other detectors are located in an ambient environment (Figure 1a). The data presented in this work were collected with a solid state backscattered electron detector which resides in the evacuated environment. In contrast to other commercially existing solutions that allow EM based imaging of non-vacuum compatible or nonconductive specimens such as environmental SEM (ESEM)7, Quantomixs WetSEM® capsules18,19, JEOL's ClairScope® (Atmospheric SEM)20, or state-of-the-art imaging modality (D-EXA)21airSEMTM have a finite gap (50–200 μm in size) separating the specimen from the membrane.
Thus, in airSEMTM the sample is not encapsulated and does not touch the microscope. Instead it is free to be scanned under the beam at ambient conditions (Figure 1b). The airSEMTM is, therefore, similar in that sense to an optical microscope: the imaged object resides in an ambient environment and is free to be brought for imaging to a focal plane below a “lens” (Figure 1b), the role of which is played by the airSEMTM membrane.
Notably, in the airSEMTM imaging station the sample is mounted on a single motion control system (Figure 1b). The position of the optical axis and the airSEMTM imaging axis are both reachable by this single motion control system and their location is fixed and pre-calibrated. Therefore, correlation between light microscope (LM) images and airSEMTM images is achieved naturally without the need for any markers to be present on the sample. Correlative microscopy is therefore simple and easy in the airSEMTM imaging station as the registration of images is intrinsic to the system's architecture.
Here we demonstrate the advantages of this innovative technique for biological applications. These advantages help to extract multifaceted information from the same biological specimen.
Results
Integrative visualization of collagen type 1 fibrils
Fibrillar collagen type 1 is the major structural protein in the extracellular matrix (ECM) of many tissues such as tendons, cornea, bones and skin. The organization of collagen type 1 is hierarchical and its detailed structure at the level of microfibrils, fibrils and fibers in different tissues is of interest for scientists of various fields such as biophysics, structural biology and system biology. Collagen fibrils are long quasi-crystalline aggregates, which may be visualized by SEM, but specific structural characteristics are usually studied by TEM22.
Figures 2a,b demonstrate integrative LM and airSEMTM imaging of native collagen fibrils. Combining LM with airSEMTM allowed the selection of a ROI to be followed by high magnification imaging of collagen type 1. Remarkably, the image of collagen type 1 fibrils by airSEMTM (Figure 2b) shows classical 67 nm D-periodicity in a similar manner to that of TEM (Figure 2c). They both exhibit a characteristic bright and dark periodicity, indicating the regions of high and low electron densities of overlap and gap structural regions. This is a result of collagen molecule arrangement in a staggered parallel alignment along the fibril axis23. Moreover, microfibrillar structures of 5 triple-helical collagen molecules24 within the collagen fibril are visible in images acquired by both airSEMTM and TEM. Thus, airSEMTM imaging depicts structural features of individual collagen fibrils. Obviously, the visualization of collagen fibrils by conventional SEM in the secondary electron mode (Figure 2d, Supplementary Fig. S1 online) provides complementary information about the rough topographical appearance of the sample.
Integrative visualization of collagen type 1 fibrils by LM and airSEMTM complemented by TEM and conventional SEM images.
(a) The collagen fibrils were visualized by LM and a ROI was chosen for further inspection by airSEMTM. (b) High resolution airSEMTM image of the ROI. The beam energy is 30 keV. (c) TEM image of collagen fibril. Accelerating voltage is 120 keV. (d) Image of collagen fibril obtained by conventional SEM. The beam energy is 1.5 keV. Scale bar: (a) 50 μm. (b,c,d) 1 μm.
Application of dual metal staining for detailed imaging of a normal lung tissue and ECM scaffolds using airSEMTM
AirSEMTM may be successfully applied for studies of biological samples, especially for visualization of tissues and their ECM scaffolds. Since the main channel used in airSEMTM is backscattered electron imaging (BSE), the scattered electrons provide an image contrast as a function of elemental composition. Because the higher-atomic-mass elements backscatter the incident beam of electrons more strongly compared to the lower-atomic-mass ones, the sample regions rich in heavier elements appear brighter than regions poorer in such elements or richer in lighter elements. Based on this principle, we developed sample preparation protocols that enhance sample contrast by incorporating pairs of heavy metal stains with affinity to various tissue components. This concept allows the imaging of multiple anatomical features of an intact hydrated and non- coated tissue section, as well as its native ECM scaffolds.
We applied ruthenium red in combination with uranyl acetate stains for the study of anatomical structures and ECM architecture of mouse lungs. Ruthenium red is a polycationic dye that is routinely used for tissue histological stains. This metal-based dye stains acidic polysaccharides25,26 of the ECM proteins and cell surface components1.
Importantly, previous reports prove that following ordinary fixation, 20–70% of the proteoglycans in the tissue may be lost27,28 and this loss may be prevented by the use of cationic compounds29 such as ruthenium red. To protect the natural content of mucopolysaccharide or glycosaminoglycan (GAGs) binding proteins in different tissue samples we incorporated ruthenium red in the fixation process as suggested in studies of articular cartilage proteoglycan localization30. The contrast was further improved by application of uranyl acetate, a widely used stain in EM which binds to the free carboxyl and amino groups of proteins, while maintaining their antigenicity31. This multi-tone staining combination increased the contrast of the resulting images, compared to non- stained tissues (see Supplementary Fig. S2 online).
Lung anatomy has been extensively studied before, using conventional SEM32. Notably, applying airSEMTM imaging using our protocols depicted detailed lung structures, such as bronchi, alveolar ducts (AD), alveolar sacs (AS), large blood vessels (VS) and fine capillaries (C) (Figure 3). Higher magnification images of a bronchus (Figure 3b), alveolar ducts (Figure 3d) and alveolar sacs, distinctly compartmentalized by septa (S), as well as capillaries accompanying the alveolar walls are presented in Figure 3a,c,e. Importantly, in addition to 30 keV operating mode, 10 keV were applied to underline topographical features (see Supplementary Fig. S3 online). This mode allowed the observation of the mucus layer excreted by the secretory cells that line the bronchial lumen (see Supplementary Fig. S3c,d online).
(a-e) A detailed imaging of normal lung tissue and ECM scaffold using airSEMTM. (a–e) Normal lung tissue. (a) Multiple lung alveolar ducts (AD) and alveolar sacs (AS) are observed. Capillaries (C) shown as small openings following the alveolar cavities. Red blood cells are visible inside a small blood vessel (VS). (b) Bronchus (c) alveolar sacs compartmentalized by septa (S). (d) Alveolar duct (e) A large blood vessel containing numerous biconcave red blood cells. (f–j) Cell-free ECM scaffold of mouse lungs. (f) Bright fibrillar structures emphasized by uranyl acetate embedded in darker proteoglycan-rich environment stained by ruthenium red. Dense fibrillar boundaries (arrows) are separated from thin fibrils (asterisks). Interwoven mesh of delicate fibers is traversing interstitial spaces. (g) High-density outermost wall of a bronchus and the thinner fibers lining the openings into secondary smaller bronchi. (h, i) Alveolar ducts and sacs. (j) Thick bundle of fibers marks the opening of a big blood vessel. Operation voltage is 30 keV. Scale bar: (a) 100 μm, (b–e) 20 μm.
The physiological function of the lungs is highly dependent on the ECM architecture and composition. By imaging decellularized lung sections33 stained with a pair of metals, we obtained detailed information about the lung scaffold architecture, consisting mainly of fibrillar proteins and GAGs containing macromolecules (Figure 3f–j). This image collection shows a spatial and compositional mapping of ECM by highlighting the brighter fibrillar structures stained mostly by uranyl acetate immersed in a darker, GAGs enriched, environment that is stained by ruthenium red. The contrast in EM images in general and in airSEMTM BSE is affected by both material contrast and topography. Our images show that heavy metal staining in airSEMTM BSE imaging greatly improves the image contrast by enhancing the material contrast, which is otherwise very poor in ECM samples (see Supplementary Fig. S4 online).
Visualization using airSEMTM allowed demarcating the dense fibrillar boundaries of several lung components (Figure 3f–j arrows) and separating them from thin fibrils (Figure 3f–j asterisks). The outermost walls of a big bronchus (Figure 3g) were identified due to the densely packed fibers lining it (Figure 3g arrow) while distinguishing it from the intraluminal branches diverging into secondary smaller bronchi, which are lined by thinner fibers (Figure 3g asterisks). The fibrillar pattern was also detected in the alveolar ducts and sacs that are discriminated from each other on the bases of fiber density and thickness (Figure 3h, i). Finally, a thick bright looking bundle of fibers defined the opening of a large blood vessel in Figure 3j and bright looking delicate fibers (Figure 3f–j asterisks) were clearly seen as an interwoven mesh traversing interstitial spaces in all images. Thus, detailed inspection of the lung by airSEMTM using BSE signal can be a substantial tool for deriving detailed information from both intact tissue and decellularized sections. Importantly, sample preparation for airSEMTM imaging does not interfere with other downstream analyses, such as conventional SEM (see Supplementary Fig. S5 online), allowing for detailed inspection of the topography of the same specimen. To prove this concept, immediately after visualization by airSEMTM, the specimen underwent the standard procedure for SEM sample preparation, initiated by alcohol dehydration (Methods). Indeed, the topographical features of lung ECM obtained by SEM, complemented those observed by airSEMTM (Supplementary Figure 5a and Figure 3f) as well as the morphology of densely packed collagen fibrils around the alveolar wall (Supplementary Figure 5b and Figure 3i).
Application of airSEMTM imaging station for correlative LM-SEM microscopy
The main fibril-producing proteins in the ECM are collagens, elastins, fibronectins and laminins34. Since collagen type 1 is the most dominant component responsible for both the mechanical and tensile strength of the lung ECM35, it is important to be able to distinguish it from the fibrils of other proteins and evaluate its distribution. Furthermore, collagen degradation and deposition are processes that accompany multiple pathologic conditions and therefore are important hallmarks of diagnosis and characterization. Mapping of collagen distribution was achieved by using the multimodal station combining fluorescent (FM) and airSEMTM microscopes (Figure 4)5.
Correlative LM-airSEM microscopy.
(a) Fresh frozen cryo-sectioned lung slice was labeled using anti collagen type I Cy3 staining showing localization to collagen fibrils around alveolar cavities. The collagen lining pattern was discontinuous (asterisks) in places where the thin basement membrane of capillaries comes into close contact with the alveolar basement membrane. In addition, a continuous collagen pattern demarcated a large blood vessel (arrowheads). The dispersal of fine collagen fibers in the interstitial space is depicted. (b) AirSEMTM imaging of the same ROI, taken at the same field of view as the fluorescent images. (d) DAPI staining of cell nuclei. (c,e) Superimposition of fluorescently labeled collagen and cell distribution, respectively, with airSEMTM image. (f) Correlative microscopy image combining three stains: Cy3, DAPI and uranyl acetate. Operation voltage for airSEMTM imaging is 30 keV. Scale bar: 50 μm.
Correlative FM-airSEMTM visualization was performed on lung tissue sections using fluorescent Cyanine 3 (Cy3) conjugate for collagen type 1 labeling as well as nuclear staining using DAPI followed by uranyl acetate (Methods). It is noteworthy to mention that examining other metal stains such as osmium tetroxide, sodium silicotungstate, potassium permanganate and tannic acid, caused significant fluorescence quenching and only uranyl acetate maintained 88% and 75% of the initial fluorescent signal intensity for collagen and DAPI, respectively (see Supplementary Fig. S6 online). Figure 4a demonstrates the distribution of collagen type 1 in 10 μm thick sections of normal lung. The high intensity of the fluorescent signal shows the localization of collagen fibrils around alveolar cavities. The pattern of lining is typically discontinuous (Figure 4a asterisks) in places where the thin basement membrane of capillaries comes into close contact with the alveolar basement membrane.
In addition, a large blood vessel is shown (Figure 4a arrow head) in which the collagen is continuous. The dispersal of fine collagen fibers in the interstitial space is also shown. Figure 4b shows registration of the same ROIs by airSEMTM. It is taken at the same field of view as Figure 4a and therefore allows overlaying of the information from the two imaging techniques (Figure 4c). This superimposition marks the localization of distinct collagen type 1 fibrils among other fibrillar structures (Figure 4c cross) in the lung as well as the arrangement of cells within the ECM environment (Figure 4d, e). The superimposition of LM and airSEMTM images (Figure 4f) represents the complexity and integrity of the lung tissue.
Importantly, although airSEMTM scanning resulted in variable quenching in the ROI, areas that were not scanned maintained their fluorescence and were suitable for further imaging by correlative microscopy (see Supplementary Fig. S7 online).
Discussion
In this work we showed that the airSEMTM imaging station (Figure 1) together with developed and adopted sample preparation protocols is a convenient and user-friendly device for the study of tissues and ECMs. In contrast to commercially existing solutions, the specimen is free to be imaged under the LM with following translation under the beam at ambient conditions. Importantly, the images from different modalities are accurately registered and automatically matched. Also noteworthy is that high-resolution airSEMTM can be easily integrated with any visualizing modality such as confocal, two- photon excitation, infrared or atomic force microscopes.
Since the airSEMTM microscope may visualize samples at ambient conditions, one can significantly simplify the sample preparation protocols. Thus, the time consuming procedures of dehydration or coating necessary for conventional SEM, are not needed for visualization by airSEMTM. Moreover, we showed that airSEMTM provides complementary information for other imaging modalities (Figure 2, Supplementary Fig. S1 online).
Importantly, we developed dual metal staining protocols, based on the combination of metal-containing histological and EM stains for tissue imaging by airSEMTM. Our images prove that the application of different metal-composing protocols significantly enhances the image contrast of tissue samples (Figure 2,3), where the topographical and shadowing effects had significantly less contribution (see Supplementary Fig. S2,S4 online). Moreover, we showed that the combined use of ruthenium red and uranyl acetate provides spatial and compositional mapping of ECM. This mapping could not have been achieved by the inherently poor ECM scaffold contrast (Figure SI4) or by using each metal stain separately. Importantly, the use of other metal-containing histological or EM stains may underline other features of biological samples using airSEMTM.
Samples prepared for airSEMTM imaging are further appropriate for downstream analysis by SEM. In the case of ECM scaffold imaging, the detailed information about ECM architecture obtained by airSEMTM (Figure 3) was complemented by the topographical information obtained by SEM (see Supplementary Fig. S5 online). Importantly, different information could be obtained by using airSEMTM at variable beam energies. Reducing the beam energy from 30 to 10 keV was used to visualize the mucus layer on the bronchial epithelial surface (see Supplementary Fig. S3c, d online). Notably, this mucus layer was not observable when the samples were prepared by standard SEM procedure in a previous study32.
As our images show, unstained samples can be imaged by airSEMTM (see Supplementary Fig. S2, S4 online). Thus, these samples are applicable for additional analyses, such as proteomics and genomics in a manner as recently described for dried, fixed or histologically stained samples36,37,38.
Notably, specimens of thicknesses ranging from hundreds of nanometers (Figure 2b) to millimeter (see Supplementary Fig. S1a online) can be placed on any support and the preparation procedure can be completed within 1 hour. Importantly, specimens with a varying extent of hydration are applicable for airSEMTM imaging.
As previously mentioned, the combination of different visualization modalities using automatic registration of ROIs greatly simplifies correlation microscopy by providing structural-compositional insights from complex tissues (Figure 4). We showed that the specially adopted protocols do not cause significant quenching of the fluorescent signal arising from Cy3 and DAPI incorporated into the tissue, when overlaid with uranyl acetate (Figure 4, Supplementary Fig. S6 online). This immuno-EM staining demarcated the distribution of collagen type 1 and cells within the tissue. Moreover, we were able to recognize collagen type 1 among other fibrillar molecules of the ECM. Importantly, easy switching between modalities allowed quick inspection of the sample, choosing of ROIs and scanning by airSEMTM. Thus, airSEMTM imaging station provided correlative microscopy that is based on a continuous preparation step and imaging procedure. Typically, in CLEM, samples have to be transferred between apparatuses and set ups, making them susceptible to contamination and damage and prolonging the time required for imaging11,12,13,14,15. In addition, AirSEMTM imaging station settings do not require sample placement or cell growth on a specialized membrane19,20,21. No specific sample holders, navigation markers or ROI retrieval are needed, thus significantly decreasing the time required for correlative imaging. In addition, airSEMTM imaging is not restricted to a confined sample region of hundreds of microns, but rather offers imaging of different regions across the entire sample19,20,21,39.
Thus, the main advantages of applying an airSEMTM imaging station to biological studies include: (1) Quick sample sliding between modalities due to the sample being maintained at ambient conditions. (2) Easy choosing of ROIs as well as correlative microscopy due to integration of several imaging modalities on the same platform using automatic registration. (3) Fast and easy-to-use sample preparation protocols adopted specifically for airSEMTM imaging station. The sample may be imaged in variable hydration states, without critical point drying and coating. (4) Possibility to facilitate integrative studies using consecutive downstream analyses on the same sample.
The results presented here show the high potential and broad scope of applications of the airSEMTM imaging station. It is particularly relevant to advanced multidisciplinary studies that require the combination of different levels of information in a robust and easily reproducible manner. We believe that the airSEMTM imaging station is an effective and easy-to-use tool, which complements other existing imaging solutions.
Methods
Operation principles of airSEMTM microscope and airSEMTM imaging station
In airSEMTM (Figure 1a), as in any other electron microscope, electrons are emitted from an electron source in vacuum (a field emitter, which requires ultra-high vacuum, was used in the work described here) and are then collimated and focused using a standard electron optics apparatus to form an ultra-sharp beam. In contrast to standard EM techniques, where the lower part of the column (typically consisting of a final lens and its pole-piece) is hermetically connected to a vacuum chamber residing in the same, typically, high vacuum conditions, in airSEMTM the lower part of the column is sealed off yet leaving a small area transparent to the beam electrons in the form of a thin membrane. This membrane is typically tens of nanometers thick and hundreds of micrometers wide. The sample, in turn is brought to a plane close to, but lower than, the membrane plane. The electron beam thus travels through the membrane and through the small gap between the membrane and the sample. This gap is typically 50–200 micrometers thick. Importantly, this gap allows free translation of the sample top surface relative to the imaging axis. The beam-sample interactions generate a multitude of products that can be detected using a variety of detectors. In airSEMTM, some of these detectors are located in the same vacuum environment as the lower parts of the column, while others reside outside the vacuum, immersed in the same ambient as the sample. The key technological challenge of avoiding primary beam electron scattering is met by a multitude of strategies. These include using a thin membrane, allowing only a small membrane-sample gap and using fairly high beam energies (5–30 keV). Backscattered electrons, carrying both material contrast and topography contrast also have high energies, rendering them natural candidates for the main imaging channel in airSEMTM. The BSE detector used in this work resides on the vacuum side of the membrane. The primary differentiator of airSEMTM – the ability to image a free sample in air, further facilitates easy integration of an airSEMTM scope with other modalities (e.g. confocal, micro Raman, AFM to name a few) (Figure 1b). This seemingly simple statement in fact has major consequences; the most prominent of which is the opportunity to construct an architecture whereby the coordinates of different modalities are interlinked by the use of a single motion control system. Thus, in an airSEMTM imaging station, the relative (fixed) position of multiple imaging axes is calibrated into the controlling software facilitating both quick and easy repetitive intra-modality imaging and fast and accurate correlative microscopy without artificial alignment marks. In addition, standard image processing techniques may be used to improve the overlay of multiple images down to single pixel accuracy.
The airSEM resolution for this work is defined to be better than 10 nm at 10–30 keV as determined by standard gold on carbon surface reference images.
No background subtraction, contrast stretching or gamma correction was applied to the SEM images.
Collagen preparation and staining
Rat-tails from adult (3–6 months) Norwegian rats were obtained from other experiments. The protocol for preparation of collagen fibers have been previously described40.
For visualization of individual collagen fibrils single collagen fibers from tendon (approximately 0.2 to 0.6 mm in diameter) were dissected and extensively washed in TNC buffer (50 mM TRIS, pH 7.4, 150 mM NaCl, 10 mM CaCl2, 0.02% NaN3) to remove the macroscopic pieces of tissue. The collagen fibers were then disrupted mechanically, mounted on SuperFrost Plus (Thermo- Scientific) glass slides and stained by 4% sodium silicotangstate (EM grade Agar Scientific), pH 7 for 1 min followed by DDW washing.
Extracted collagen fibers were stained with 4% sodium silicotangstate (pH = 7) for 45 min and dehydrated through an ethanol series increasing in concentration to 100% ethanol. After that the samples were dried in a critical point dryer and mounted onto SEM stubs with double-sided adhesive tape. The samples were then kept in a desiccator. Prior to observation by conventional SEM, the samples were coated with gold/palladium alloy. Examination was carried out using an Ultra 55 Feg Zeiss SEM operating at 3 keV using a 10 μm aperture.
Dual metal staining of intact tissues
Lungs were inflated with 300 μL of 4% PFA prior to removal from the body and placed in OCT for 10 min. Lungs were then placed on dried ice and finally frozen at −80°C until analyzed. Tissues were cryo-sectioned at a thickness of 300 μm. Sections were washed three times in a large volume of PBS to remove OCT remnants, followed by three DDW washes. For tissue imaging, the slices were gently placed on SuperFrost Plus glass slides, washed by DDW and stained with 0.1% ruthenium red (EM grade, Sigma-Aldrich) in a 0.1 M sodium cacodylate buffer pH 7.4 (analytical standard, Sigma-Aldrich) for 15 min. The sections were then thoroughly washed with DDW and stained with a 2% uranyl acetate solution for 10 min. The samples then were washed with DDW and allowed to dry in the air at room temperature for 5–7 min before airSEMTM imaging.
For the observation of unstained tissues the lung sections were washed from OCT by PBS and DDW as described above and mounted on SuperFrost Plus glass slides.
Dual metal staining of ECM scaffolds
Lungs were inflated with air prior to removal from the body and placed in OCT for 10 min. Lungs were then frozen at −80°C until analyzed. Tissues were cryo-sectioned at a thickness of 300 μm, washed three times in a large volume of PBS, followed by rinsing with DDW. For ECM imaging, the sections were first de-cellularized using a previously described procedure33. Sections were gently placed on SuperFrost Plus glass slides and further stained with 0.1% ruthenium red in a 0.1 M sodium cacodylate buffer (pH 7.4) (analytical standard, Sigma-Aldrich) containing 2% Paraformaldehyde (EM grade, Bar-Naor)/2.5% Glutaraldehyde (EM grade I, Sigma-Aldrich) for 15 min. The sections were then thoroughly washed with DDW and stained with a 2% uranyl acetate solution for 10 min.
Remarkably, we have observed (data not shown) that the fixation of ECM scaffolds may be excluded from the protocol of sample preparation.
To provide airSEMTM imaging with further analysis by conventional SEM the lung ECM scaffolds were placed on mica, modified according to the improved procedure of Vandenberg et al.41. Shortly thereafter, fresh cleaved mica was incubated with 0.5% APTES (99% purity, Sigma -Aldrich) in toluene (analytical grade, Frutarom, Israel) for 10 min and then rinsed with absolute ethanol (Sigma-Aldrich) 4×, 200 μL, acetone (Sigma -Aldrich) 4×, 200 μL and absolute ethanol 4×, 200 μL. The modified mica surface was gently dried in the open air for 5 min. The amine surface was activated by incubation with 6% gluteraldehyde (EM grade I, Sigma-Aldrich) in DDW water (pH 9.0) for 30 min. The mica was then washed by DDW (12×, 200 μL) and gently dried in the air for 3 min. The slices of decellarized lungs were then placed on the modified mica. Immediately after the observation by airSEMTM the sample was further dehydrated by ethanol substitution, critical point dehydration method and coated by Au/Palladium according to standard sample preparation procedure for SEM (see above).
Staining procedure for correlative microscopy
For collagen type 1 staining, 10 μm fresh frozen cryo-sectioned lungs were fixed for 10 min using 4% FPA, followed by blocking with 3% BSA (min.98%, Sigma-Aldrich) in PBS for 45 min. The slices were sequentially incubated with rabbit anti-collagen type I primary monoclonal antibody (Abcam), diluted 1:200, for 1 h at room temperature, followed by incubation with a goat anti-Rabbit-HRP conjugated secondary antibody (Abcam), diluted 1:300 in blocking solution, for 45 min. HRP expansion using an anti-HRP antibody was followed by TSA PLUS Cyanine 3/Fluorescein System HRP amplification kit (Perkin Elmer) conjugate incubation for 1 h at room temperature, followed by incubation with DAPI (BioLegend) (diluted 1:200 in PBS) for 10 minutes. After thoroughly washing with DDW, samples were stained with 2% uranyl acetate for 5 min, washed by DDW and allowed to dry for 5–7 min at room temperature in the dark without a cover slip before airSEMTM imaging.
Optical images
Optical images in this study were obtained using the upright airSEMTM imaging station microscope, using a 5× (MPLNX/0.1) and 40× (MPLNX/0.75) air objective. Epifluorescent images were acquired using standard filter cubes, DAPI [Ex': BP330–385; Em ':BA420] and Cy3 [Ex' BP530–550; Em' 575–625].
Fluorescence quantification
Loss of fluorescence intensity was monitored using ImageJ. Using a point-selection tool, the intensity of 3–4 fluorescent spots from 2–3 sections in each condition was measured. The obtained values were averaged and compared (see Supplementary Fig. S6 online).
LM and airSEMTM image overlay
Since the registration of both LM and airSEMTM data channels is a result of the inherent system design (Figure 1b), the acquired images in both optical and SEM were of the same field of view (FOV), the same number of pixels and same pixel size. In the data presented in this article no manual adjustment was needed, as stated in the text, since the design of the system enables to reach registration accuracy down to the nm range with simple hardware modifications. The images were then overlaid using ImageJ.
References
Hayat, M. A. Principles and Techniques of Electron Microscopy: Biological Applications. 4th edn, (Cambridge University Press 1 2000).
Fischer, E. R., Hansen, B. T., Nair, V., Hoyt, F. H. & Dorward, D. W. Scanning Electron Microscopy. Curr. Protoc. Microbiol. 25:22.22.21–22B.22.47. (2012).
Adrian, M., Dubochet, J., Lepault, J. & McDowall, A. W. Cryo-electron microscopy of viruses. Nature 308, 32–36 (1984).
Leforestier, A., Lemercier, N. & Livolant, F. Contribution of cryoelectron microscopy of vitreous sections to the understanding of biological membrane structure. Proc. Natl. Acad. Sci. U. S. A. 109, 8959–8964 (2012).
Vidavsky, N. et al. Initial stages of calcium uptake and mineral deposition in sea urchin embryos. Proc. Natl. Acad. Sci. U. S. A. 111, 39–44 (2013).
Stokes, D. J., Mugnier, J. Y. & Clarke, C. J. Static and dynamic experiments in cryo-electron microscopy: comparative observations using high-vacuum, low-voltage and low-vacuum SEM. J. Microsc. 213, 198–204 (2004).
Muscariello, L. et al. A critical overview of ESEM applications in the biological field. J. Cell Physiol. 205, 328–334 (2005).
Weber, K., Delben, J., Bromage, T. G. & Duarte, S. Comparison of SEM and VPSEM imaging techniques with respect to Streptococcus mutans biofilm topography. FEMS Microbiol. Lett. 350, 175–179 (2014).
Vanden Berg-Foels, W. S., Scipioni, L., Huynh, C. & Wen, X. Helium ion microscopy for high-resolution visualization of the articular cartilage collagen network. J. Microsc. 246, 166–176 (2012).
Joens, M. S. et al. Helium Ion Microscopy (HIM) for the imaging of biological samples at sub-nanometer resolution. Sci. Rep. 3, 3514 (2013).
Giepmans, B. N., Deerinck, T. J., Smarr, B. L., Jones, Y. Z. & Ellisman, M. H. Correlated light and electron microscopic imaging of multiple endogenous proteins using Quantum dots. Nat. Methods. 10, 743–749 (2005).
Lucas, M. S., Günthert, M., Gasser, P., Lucas, F. & Wepf, R. Bridging microscopes: 3D correlative light and scanning electron microscopy of complex biological structures. Methods Cell Biol. 111, 325–356 (2012).
Gaietta, G. M. et al. Golgi twins in late mitosis revealed by genetically encoded tags for live cell imaging and correlated electron microscopy. Proc. Natl. Acad. Sci. U. S. A. 103, 17777–17782 (2006).
Biel, S. S., Kawaschinski, K., Wittern, K. P., Hintze, U. & Wepf, R. From tissue to cellular ultrastructure: closing the gap between micro- and nanostructural imaging. J. Microsc. 212, 91–99 (2003).
Watanabe, S. et al. Protein localization in electron micrographs using fluorescence nanoscopy. Nat. Methods. 8, 80–84 (2011).
Friedl, P. et al. Migration of highly aggressive MV3 melanoma cells in 3-dimensional collagen lattices results in local matrix reorganization and shedding of alpha2 and beta1 integrins and CD44. Cancer Res. 57, 2061–2070 (1997).
Gunzer, M., Kampgen, E., Brocker, E. B., Zanker, K. S. & Friedl, P. Migration of dendritic cells in 3D-collagen lattices. Visualisation of dynamic interactions with the substratum and the distribution of surface structures via a novel confocal reflection imaging technique. Adv. Exp. Med. Biol. 417, 7–103 (1997).
Thiberge, S. et al. Scanning electron microscopy of cells and tissues under fully hydrated conditions. Proc. Natl. Acad. Sci. U. S. A. 101, 3346–3351 (2004).
Thiberge, S., Zik, O. & Moses, E. An apparatus for imaging liquids, cells and other wet samples in the scanning electron microscopy. Rev. Sci. Instrum. 75, 2280–2289 (2004).
Nishiyama, H. et al. Atmospheric scanning electron microscope observes cells and tissues in open medium through silicon nitride film. J. Struct. Biol. 169, 438–449 (2010).
Nawa, Y. et al. Multi-color imaging of fluorescent nanodiamonds in living HeLa cells using direct electron-beam excitation. Chemphyschem. 15, 721–726 (2014).
Wess, T. J. Collagen fibril form and function. Adv Protein Chem. 70, 341–374 (2005).
Hodge, A. J. & Petruska, J. A. Recent studies with the electron microscope on ordered aggregates of the tropocollagen molecule. In Aspects of Protein Structure. Ramachandran G. N. editor. Academic Press, New York. 289–300 (1963).
Orgel, J. P., Irving, T. C., Miller, A. & Wess, T. J. Microfibrillar structure of type I collagen in situ. Proc. Natl. Acad. Sci. U. S. A. 103, 9001–9005 (2006).
Luft, J. H. Ruthenium red and violet. II. Fine structural localization in animal tissues. Anat. Rec. 171, 369–415 (1971).
Frey-Wyssling, A. The plant cell wall. Encyclopedia of Plant Anatomy, (1976).
Thyberg, J., Lohmander, S. & Friberg, U. Electron microscopic demonstration of proteoglycans in guinea pig epiphyseal cartilage. J. Ultrastruct. Res. 45, 407–427 (1973).
Engfeldt, B. & Hjertquist, S. O. Studies on the epiphysial growth zone. I. The preservation of acid glycosaminoglycans in tissues in some histotechnical procedures for electron microscopy. Virchows. Arch. B Cell Pathol. 1, 222–229 (1968).
Szirmai, J. A. Quantitative approaches in the histochemistry of mucopolysaccharides. J. Histochem. Cytochem. 11, 24–34 (1963).
Shepard, N. & Mitchell, N. The localization of articular cartilage proteoglycan by electron microscopy. Anat. Rec. 187, 463–476 (1977).
Erickson, P. A., Anderson, D. H. & Fisher, S. K. Use of uranyl acetate En Bloc to improve tissue preservation and labeling for post-Em bedding immunoelectron microscopy. Microsc. Res. Tech. 5, 303–314 (1987).
Ten Have-Opbroek, A. A. The structural composition of the pulmonary acinus in the mouse. A scanning electron microscopical and developmental-biological analysis. Anat. Embryol. (Berl). 174, 49–57 (1986).
Shamis, Y. et al. Organ-specific scaffolds for in vitro expansion, differentiation and organization of primary lung cells. Tissue Eng. Part C Methods 17, 861–870 (2011).
Alberts, B. et al. Molecular Biology of the Cell, 4th edition. (2002).
Petersen, T. H., Calle, E. A., Colehour, M. B. & Niklason, L. E. Matrix composition and mechanics of decellularized lung scaffolds. Cells Tissues Organs 195, 222–231 (2012).
Morikawa, T. et al. No evidence for interference of h&e staining in DNA testing: usefulness of DNA extraction from H&E-stained archival tissue sections. Am. J. Clin. Pathol. 138, 122–129 (2012).
Scicchitano, M. S., Dalmas, D. A., Boyce, R. W., Thomas, H. C. & Frazier, K. S. Protein extraction of formalin-fixed, paraffin-embedded tissue enables robust proteomic profiles by mass spectrometry. J. Histochem. Cytochem. 57, 849–860 (2009).
Moulédous, L. et al. Navigated laser capture microdissection as an alternative to direct histological staining for proteomic analysis of brain samples. Proteomics. 3, 610–615 (2003).
Liv, N. et al. Simultaneous correlative scanning electron and high-NA fluorescence microscopy. PLoS One 8, e55707 (2013).
Wess, T. J., Hammersley, A. P., Wess, L. & Miller, A. Molecular packing of type I collagen in tendon. J. Mol. Biol. 275 (1998).
Vandenberg, E., Elwing, H., Askendal, A. & Lundstrom, I. Protein immobilization to 3-aminopropyl triethoxy silane/glutaraldehyde surfaces: characterization by detergent washing. J. Colloid Interface Sci. 143, 327–335 (1991).
Acknowledgements
The research leading to these results has received funding from the Israeli Scientific Foundation, EU FP7 SaveMe project and Minerva. Traditional Electron microscopy studies were conducted at the Irving and Cherna Moskowitz Center for Nano and Bio-Nano Imaging at the Weizmann Institute of Science. The airSEMTM imaging was conducted at B-nano labs.
Author information
Authors and Affiliations
Contributions
I.Sa. suggested the general idea for the adaptation of airSEMTM multimodal station for tissue research, I.So. and D.T.F. designed the research, I.So., D.T.F. and A.A. prepared the samples, Y.M. and S.A. performed imaging on airSEMTM station. I.So., D.T.F. and I.Sa prepared the manuscript with contributions from A.A., Y.M. and S.A.
Ethics declarations
Competing interests
Yes, there is potential Competing Interest. The authors Yonat Milstein and Sefi Addadi work for B-nano, a privately owned company which manufactures and sells airSEMTM products.
Electronic supplementary material
Supplementary Information
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License. The images or other third party material in this article are included in the article's Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder in order to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
About this article
Cite this article
Solomonov, I., Talmi-Frank, D., Milstein, Y. et al. Introduction of correlative light and airSEMTM microscopy imaging for tissue research under ambient conditions. Sci Rep 4, 5987 (2014). https://doi.org/10.1038/srep05987
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep05987
This article is cited by
-
Calcium phosphate mineralization in bone tissues directly observed in aqueous liquid by atmospheric SEM (ASEM) without staining: microfluidics crystallization chamber and immuno-EM
Scientific Reports (2019)
-
Porosity estimation of Phyllostachys edulis (Moso bamboo) by computed tomography and backscattered electron imaging
Wood Science and Technology (2017)
-
Correlated light and electron microscopy: ultrastructure lights up!
Nature Methods (2015)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.