Abstract
The potential for production of chemicals from microalgal biomass has been considered as an alternative route for CO2 mitigation and establishment of biorefineries. This study presents the development of consolidated bioprocessing for succinate production from microalgal biomass using engineered Corynebacterium glutamicum. Starch-degrading and succinate-producing C. glutamicum strains produced succinate (0.16 g succinate/g total carbon source) from a mixture of starch and glucose as a model microalgal biomass. Subsequently, the engineered C. glutamicum strains were able to produce succinate (0.28 g succinate/g of total sugars including starch) from pretreated microalgal biomass of CO2-grown Chlamydomonas reinhardtii. For the first time, this work shows succinate production from CO2 via sequential fermentations of CO2-grown microalgae and engineered C. glutamicum. Therefore, consolidated bioprocessing based on microalgal biomass could be useful to promote variety of biorefineries.
Similar content being viewed by others
Introduction
Metabolic engineering, aiming for enhanced production of desired bio-products through modification of cellular metabolism, has enabled to construct microbial cell factories including engineered Escherichia coli and yeast for production of native or non-native biochemical from fermentable sugars1. However, efficient conversion of lignocellulosic biomass to fermentable sugars is critical for production of biofuels and chemicals in industrial scales2.
The recalcitrant structures of lignocellulose hamper its efficient degradation into simple sugars3. Although various methods have been developed to break the structures of lignin and crystalline cellulose prior to enzymatic hydrolysis4, the pretreatment step is a still bottleneck for fermentation of lignocellulosic biomass. On the other hand, microalgal cultivation as a potential platform for production of biofuels or chemicals has several positive aspects5, including high productivity per-acre over lignocellulosic feedstock resources6,7. In addition, microalgal lipids could be converted to biodiesel and other components of microalgal biomass including carbohydrates, polyunsaturated fatty acid and proteins can serve as CO2-derived carbon sources for biorefineries i.e. direct conversion of Spirulina to ethanol without pretreatment or enzymatic hydrolysis8. Thus, we have considered microalgal biomass as a potential feedstock to replace the lignocellulosic biomass and to produce value-added chemicals.
Corynebacterium glutamicum is a predominantly aerobic, non-pathogenic, biotin-auxotrophic Gram-positive bacterium. It is used industrially for amino acid production, in particular the flavor enhancer L-glutamate and the feed additive L-lysine9. Recently, engineering of amino acid-producing C. glutamicum have been enabled to utilize hydrolysates of rice straw, wheat bran and molasses10. Moreover, recent studies have indicated the potential of C. glutamicum as a microbial cell factory to produce other commercially relevant chemicals such as succinate, isobutanol, cadaverine and ethanol11,12,13. To broaden the substrate range, metabolic engineering of C. glutamicum has been employed to utilize non-native carbon sources such as cellobiose, N-acetylglucosamine, or starch14,15,16. Particularly, starch was used as sole carbon source for production of cadaverin17, L-glutamate18, L-lysine19 and organic acids20 in C. glutamicum via either enzyme secretion or surface-display of α-amylases. In this report, we focused on highly accumulated starch in microalgae biomass as potential carbon source for C. glutamicum. Here, we engineered a succinate-producing C. glutamicum strain to secrete starch-degrading α-amylases to produce succinate from CO2-derived microalgal biomass, as an example of consolidated bioprocessing for microalgal biomass (Fig. 1).
Scheme of CO2-derived succinate production from microalgal biomass using engineered C. glutamicum through consolidated bioprocessing.
Engineered C. glutamicum secrets α-amylase to degrade soluble starch derived from extracts of CO2-grown C. reinhardtii and produces succinate from total sugars from microalgal biomass without enzyme addition.
Results
Utilization of soluble starch by engineered C. glutamicum
Carbohydrates including starch are major constituents in the microalgal biomass of Chlamydomonas reinhardtii UTEX 90 when the algae were starved for other essential elements such as nitrogen. As C. glutamicum wild type ATCC 13032 is unable to utilize starch, we first implemented this ability by constructing strains Cg-pSbAmyA and Cg-pBlAmyS capable of secreting α-amylase into the medium (Table 1). C. glutamicum wild type harboring the empty plasmid pBbEB1c did not consume soluble starch (Sigma; no glucose detected) at all and showed no growth (Fig. 2). Strain Cg-pSbAmyA did not completely consume 0.5% (w/v) soluble starch (Sigma) within 56 hr and reached a maximal cell dry weight (cdw) of 0.5 ± 0.01 g/L. However, strain Cg-pBlAmyS completely consumed 0.5% soluble starch (Sigma) in 6 hr and reached a maximal biomass of 1.23 g ± 0.01 cdw/L, which was the same biomass of the Cg-pBbEB1c grown on 0.5% (w/v) glucose as sole carbon source (1.23 g ± 0.01 cdw/L). Thus, we measured α-amylase volume activity in the supernatants. In comparisons to Cg-pSbAmyA culture medium, 2-fold increased activities of Cg-pBlAmyS culture medium were measured at 8 hr when Cg-pBlAmyS cells reached almost the maximum cell growth (Table 2). However, low amylase activities (less than 100 U/L) of Cg-pSbAmyA were not enough to completely utilize the starch as sole carbon source. Nonetheless, we successfully constructed starch-degrading C. glutamicum strains via secreting the α-amylase and applied this system for succinate production.
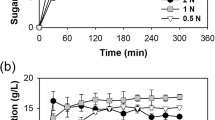
Profile of soluble starch and the growth of C. glutamicum strains.
Cg-pBbEB1c as a control strain (A) and α-amylase-secreting Cg-pSbAmyA (B) and Cg-pBlAmyS (C) strains were cultivated with 0.5% (w/v) soluble starch (Sigma) as a sole carbon source. Optical density was measured at 600 nm (closed circle; black). Soluble starch (closed square; blue) in the supernatant was quantified. Mean values and standard deviations of triplicate cultures are shown (s.d. less than 1% not shown).
Succinate production from soluble starch by engineered C. glutamicum
The succinate-producing C. glutamicum strain BL-112, which carries deletions of the genes pqo (encoding for pyruvate:menaquinone oxidoreductase), pta-ackA (encoding for phosphate acetyltransferase and aceate kinase), sdhCAB (encoding for succinate dehydrogenase complex) and cat (encoding for acetyl-CoA:CoA transferase) was transformed with the plasmids pBbEB1c-torA-SbAmyA(cg.co) and pBbEB1c-torA-BlAmyS(cg.co). As a result, we constructed BL-1-pSbAmyA and BL-1-pBlAmyS strains.
Microalgal biomass consist mainly carbohydrates (60%, based on cdw), protein (8.3%, based on cdw) and others21. 58% of carbohydrates in microalgal biomass were soluble starch. We used a mixture of 0.5% (w/v) glucose and 0.5% (w/v) soluble starch as the model carbon source of microalgal biomass for succinate production. These strains and a control strain carrying the vector without an amylase gene (BL-1-pBbEB1c) were tested for their ability to utilize a model carbon source of microalgal biomass. 0.5% glucose was supplemented as an additional carbon source to the CgXII defined medium with 0.5% soluble starch. The control strain BL-1-pBbEB1c in a mixture of glucose and starch grew (maximal growth 1.78 g ± 0.01 cdw/L) where 0.5% glucose was completely depleted and 0.5% starch was not consumed at all. On the other hand, the strains BL-1-pBlAmyS and BL-1-pSbAmyA showed an almost doubled biomass formation (2.64 g ± 0.01 cdw/L and 2.89 ± 0.17 g cdw/L), respectively, in comparison to the control strain (Fig. 3). The BL-1-pSbAmyA and BL-1-pBlAmyS degraded soluble starch in the early stage of cell growth. Then, both BL-1-pBlAmyS and BL-1-pSbAmyA strains completely consumed both glucose and starch within 16 hr.
Profile of glucose, soluble starch, succinate and the growth of C. glutamicum strains.
The succinate-producing C. glutamicum strains BL-1-pBbEB1c (A), BL-1-pSbAmyA (B) and BL-1-pBlAmyS (C) were cultivated with a mixture of 0.5% (w/v) soluble starch and 0.5% (w/v) glucose as an model microalgal biomass. Optical density was measured at 600 nm (closed circle; black). Soluble starch (closed square; blue), glucose (closed triangle, red) and succinate (open circle, green) in the supernatant were quantified. Mean values and standard deviations of triplicate cultures are shown (s.d. less than 1% not shown).
When cultivated with the mixture of 0.5% glucose and 0.5% of starch, strains BL-1-pSbAmyA and BL-1-pBlAmyS produced 1.56 ± 0.01 g/L succinate after 24 hr and 1.44 ± 0.01 g/L succinate after 14 hr, respectively. The yields (g/g) (succinate/total sugars; assuming 100% conversion of starch to glucose) of BL-1-pBlAmyS (0.16 g/g) and BL-1-pSbAmyA (0.14 g/g) were slightly higher compared to the BL-1 strain cultivated on glucose (0.12 g/g)12. As a result, strain BL-1-pBlAmyS showed the fastest cell growth and succinate production due to fast utilization of both glucose and starch. When glucose was supplemented to starch-minimal medium as microalgal biomass, the amylase volume activities of Cg-pSbAmyA or Cg-pBlAmyS were significantly increased by 6.2-folds or 3.1-fold, respectively, compared to the strains with starch as sole carbon source (Table 2) and consequently, the starches were rapidly consumed after 4 hr (lag phase). Initial supply of glucose is necessary for the C. glutamicum-secreting SbAmyA or BlAmyS strains that increase the amylase volume activities and efficiently hydrolyze soluble starch. The increased volume activities of SbAmyA or BlAmyS were also shown in BL-1-pSbAmyA and BL-1-pBlAmyS, which is important for efficient consolidated bioprocessing of microalgal biomass where initial fermentable carbohydrates exist (Fig. 4). For the first time, we successfully constructed starch-degrading and succinate-producing C. glutamicum strains and produced succinate from soluble starch using the engineered strains.
Profile of total sugar, soluble starch, succinate and the growth of C. glutamicum strains.
Succinate-producing C. glutamicum strain BL-1-pBbEB1c (A), BL-1-pSbAmyA (B) and BL-1-pBlAmyS (C) were cultivated with pretreated microalgal biomass as sole carbon source. Total sugar already contains soluble starch in this experiment. Optical density was measured at 600 nm (closed circle; black). Total sugar (open triangle; red), soluble starch (closed square; blue) and succinate (open circle; green) in the supernatant were quantified. Mean values and standard deviations of triplicate cultures are shown (s.d. less than 1% not shown).
CO2-derived succinate production from microalgal biomass by engineered C. glutamicum
Finally, we applied our engineered strains to utilize CO2-derived microalgal biomass and to produce CO2-derived succinate. To obtain the microalgal biomass, C. reinhardtii UTEX 90 was grown photoautrophically with 5% (v/v) CO2 and 95% (v/v) air bubbling. Disrupted microalgal biomass was centrifuged and the resulting supernatant was used as only carbon source for succinate production in CgXII medium. It contained 0.2% total sugars, of which 50% were determined to be soluble starch, similar to previous work21. Compared with BL-1-pBbEB1c (0.56 g ± 0.01 cdw/L), BL-1-pSbAmyA and BL-1-pBlAmyS showed doubled biomass formation (1.05 ± 0.01 g cdw/L and 1.02 ± 0.01 g cdw/L) in 24 hr, respectively (Fig. 4). As shown for the cell culture on a mixture of starch and glucose in this study, the BL-1-pSbAmyA and BL-1-pBlAmyS also degraded soluble starch in microalgal biomass from the initial cell growth. Then, the rest of sugars were slowly consumed, but 10% of initial total sugars were not utilized at all, which could be pentose sugars not utilized by the strains, such as xylose or arabinose.
The strains BL-1-pBlAmyS and BL-1-pSbAmyA produced 0.49 ± 0.01 g/L and 0.50 ± 0.01 g/L succinate after 24 hr from 0.2% total sugar including 0.1% starch in pretreated microalgal biomass, respectively. The control strain BL-1-pBbEB1c unable to utilize starch produced only 30% of the succinate (0.15 g/L ± 0.001) found for the amylase-secreting strains. Moreover, the yields (succinate/total sugars used) of BL-1-pBlAmyS (0.28 g/g) and BL-1-pSbAmyA (0.28 g/g) were significantly higher compared to the BL-1-pBbEB1c strain (0.20 g/g) when pretreated microalgal biomass was used. Finally, for the first time, we successfully produced succinate from CO2-derived microalgal biomass using engineered strains capable of degrading starch without a need for additional enzyme treatment.
Discussion
Microalgal biomass of C. reinhardtii is a remarkable carbohydrate feedstock to provide the carbon sources for microbial fermentations. Often separate hydrolysis and fermentation (SHF) or simultaneous saccharification and fermentation (SSF) process have been applied for production for the production of bioethanol22,23. However, additional enzyme loading at either SHF or SSF process could be a crucial bottleneck for economically feasible bioprocess to produce value-added chemicals or biofuels24. Thus, consolidated bioprocessing was suggested as an alternative strategy that microbial strain is capable of producing enzyme for saccharification and producing the target chemicals such as biofuels from lignocellulosic biomass25,26. In this study, we suggested another type of consolidated bioprocessing based on microalgal biomass. A succinate-producing C. glutamicum strain was capable of degrading starch by secreting α-amylase and successfully fermented microalgal biomass and produce succinate without amylase additions.
BL-1-pBlAmyS (0.28 g/g) strain and its fermentation of showed remarkable yield of succinate production due to the utilization of soluble starch that C. glutamicum wild type is not able to consume. This consolidated bioprocessing based on microalgal biomass with the best strain BL-1-pBlAmyS does not require additional costs for loading enzymes but produce the high yield of succinate, compared to the succinate producer BL-1. Furthermore, efficient hydrolysis of soluble starch and co-uptake of other carbohydrates and their cooperative sugar metabolisms could be useful to ensure faster cell growth of C. glutamicum and higher production of succinate. Metabolic engineering by optimizing gene expression of AmyS from B. licheniformis could be possible by tuning translation strengths on ribosomal binding site or changing different signal peptides. Additional sugar transporters and hydrolytic enzymes could be necessary to uptake unused carbohydrates in the total sugars because 10% of total sugars in microalgal biomass were not fermentable. In addition to extensive studies on sugar metabolism27, pentose-sugar fermentations of engineered C. glutamicum have been well investigated28,29. Current synthetic platform (CoryneBrick29) for the gene expression in this study can be easily expanded for additional gene expression of targets. Also, application of cell display system20 (i.e. the B. subtilis PgsA and C. glutamicum PorC protein as anchor) of target amylases in C. glutamicum could be alternative to increase hydrolysis of soluble starch and production of succinate.
Microalgal biomass was shown to serve as an efficient carbon source for the microbial production of succinate, which is considered as a platform chemical, when suitably engineered strains were used which are capable of starch degradation by the secretion of amylases. Ultimately, consolidated bioprocessing based on microalgal biomass offers another options to resolve issues of alternative energy resources, global warming, human health and food security.
Methods
Bacterial strains and growth conditions
Strains used in this study are listed in Table 1. For cloning purposes E. coli DH5α was used and grown in lysogeny broth medium (LB) When appropriate, the medium was supplemented with 25 μg/mL chloramphenicol. C. glutamicum ATCC 13032 and its derivatives were cultivated in BHIS medium30 at 30°C and 200 rpm and 7.5 μg/mL chloramphenicol was added when appropriate. For the utilization of soluble starch or microalgal biomass and succinate production, engineered C. glutamicum were pre-cultivated in the BHIS medium overnight and then incubated aerobically in the CgXII defined medium (50 mL in 250 mL baffled Erlenmeyer flasks) containing either 0.5% soluble starch (Sigma-Aldrich) or pretreated microalgal biomass as sole carbon source30 at 30°C on a rotary shaker at 200 rpm with 7.5 μg/mL chloramphenicol. The biomass concentration was calculated from OD600 values using an experimental determined correlation factor of 0.25 g cell dry weight per liter for OD600 = 131.
Microalgal cultivation for biomass preparation
To obtain a large amount of algal biomass containing starch for succinate production by C. glutamicum, a freshwater green alga, C. reinhardtii UTEX 90, was grown photoautotrophically in Tris-acetate-phosphate medium21 without acetic acid (TAP-C). Cultivations were carried out at 23°C in 20 L of the TAP-C medium in a 25 L photobioreactor with 65 mL/min of 5% (v/v) CO2 and 95% (v/v) air aeration and 100 μE/m2/s of illumination with a dark/light cycle (12:12 hr). The cells were harvested by centrifugation at 5,000 × g for 10 min after two weeks cultivation including nitrogen starvation (1.15 g cell dry weight/L). The lyophilized cells were resuspended in distilled water, disrupted by glass bead-beating and centrifuged (10 min at 5,000 × g). The supernatant was used as only carbon source for the cultivation of C. glutamicum.
Construction of α-amylase-secreting C. glutamicum
The amyA (NCBI no. AB000829.1) and amyS (NCBI no. M38570.1) genes from Streptococcus bovis and Bacillus licheniformis, respectively, were chosen since they were well characterized and studied in C. glutamicum19 and E. coli32. Each target gene was synthesized (Genscript, USA) with codon-optimization for C. glutamicum (represented as cg.co). Each gene was assembled using a standard BglBrick cloning method, where the target gene is inserted at the EcoRI and XhoI sites of the CoryneBrick plasmid pBbEB1c29. The Tat-specific TorA signal peptide sequence from C. glutamicum33 was added to the coding sequence of the target gene where the native signal sequences was already deleted. The plasmids used in this study are listed in Table 1. For the transformation of C. glutamicum, competent cell preparation and electroporation were performed with the plasmids according to a previously described protocol34 with some modifications.
Enzyme activity measurement
For the determination of α-amylase volume activity (U/L), the supernatant of the cells were collected and the samples were analyzed using an α-amylase measurement kit (Kikkoman, Tokyo, Japan) using 2-chloro-4-nitrohenyl 65-azido-65-deoxy- β-maltopentaoside (N3-G5-β-CNP) as the substrate in the previous studies35. The assay mixture was incubated at 37°C for 10 min and the enzymatic reaction was terminated by adding 800 μL of a reaction stop solution. α-Amylase activity was determined according to the manufacturer's instruction by measuring the absorbance of the liberated 2-chloro-4-nitrophenol (CNP) at 400 nm. One unit (U) of activity was defined as the amount of enzyme required to release 1 μmol of CNP from N3-G5-β-CNP per minute at 37°C.
Determination of starch, total sugar and succinate
For quantification of starch16, different dilutions of the culture supernatant were assayed with Lugols solution containing iodine (1.5 g/L) and potassium iodide (15 g/L), leading to the formation of a blue complex that was measured in a spectrophotometer at 530 nm. For quantification of total sugars36, a colorimetric method based on the phenol–sulfuric acid reaction was used to determine the amount of total sugars including the starch in pretreated microalgal biomass. Succinate in the supernatant was quantified by HPLC as described previously12.
References
Keasling, J. D. From yeast to alkaloids. Nat. Chem. Biol. 4, 524–525 (2008).
Bokinsky, G. et al. Synthesis of three advanced biofuels from ionic liquid-pretreated switchgrass using engineered Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 108, 19949–19954 (2011).
Vaaje-Kolstad, G. et al. An oxidative enzyme boosting the enzymatic conversion of recalcitrant polysaccharides. Science 330, 219–222 (2010).
Taylor, M. P., Mulako, I., Tuffin, M. & Cowan, D. Understanding physiological responses to pre-treatment inhibitors in ethanologenic fermentations. Biotechnol. J. 7, 1169–1181 (2012).
Parmar, A., Singh, N. K., Pandey, A., Gnansounou, E. & Madamwar, D. Cyanobacteria and microalgae: a positive prospect for biofuels. Bioresour. Technol. 102, 10163–10172 (2011).
Yen, H. -W., Hu, I. -C., Chen, C. -Y., Ho, S. -H., Lee, D. -J. & Chang, J. -S. Microalgae-based biorefinery - From biofuels to natural products. Bioresour. Technol. 135, 166–174 (2012).
John, R. P., Anisha, G. S., Nampoothiri, K. M. & Pandey, A. Micro and macroalgal biomass: a renewable source for bioethanol. Bioresour. Technol. 102, 186–193 (2011).
Aikawa, S. et al. Direct conversion of Spirulina to ethanol without pretreatment or enzymatic hydrolysis processes. Energy Environ. Sci. 6, 1844–1849 (2013).
Kalinowski, J. et al. The complete Corynebacterium glutamicum ATCC 13032 genome sequence and its impact on the production of l-aspartate-derived amino acids and vitamins. J. Biotechnol. 104, 5–25 (2003).
Xu, J., Zhang, J., Guo, Y., Zai, Y. & Zhang, W. Improvement of cell growth and L-lysine production by genetically modified Corynebacterium glutamicum during growth on molasses. J. Ind. Microbiol. Biotechnol. 40, 1423–1432 (2013).
Becker, J. & Wittmann, C. Bio-based production of chemicals, materials and fuels -Corynebacterium glutamicum as versatile cell factory. Curr. Opin. Biotechnol. 23, 631–640 (2012).
Litsanov, B., Kabus, A., Brocker, M. & Bott, M. Efficient aerobic succinate production from glucose in minimal medium with Corynebacterium glutamicum. Microb. Biotechnol. 5, 116–128 (2012).
Woo, H. M. & Park, J. B. Recent progress in development of synthetic biology platforms and metabolic engineering of Corynebacterium glutamicum. J. Biotechnol. 180, 43–51 (2014).
Uhde, A. et al. Glucosamine as carbon source for amino acid-producing Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 97, 1679–1687 (2013).
Adachi, N., Takahashi, C., Ono-Murota, N., Yamaguchi, R., Tanaka, T. & Kondo, A. Direct L-lysine production from cellobiose by Corynebacterium glutamicum displaying beta-glucosidase on its cell surface. Appl. Microbiol. Biotechnol. 97, 7165–7172 (2013).
Seibold, G., Auchter, M., Berens, S., Kalinowski, J. & Eikmanns, B. J. Utilization of soluble starch by a recombinant Corynebacterium glutamicum strain: growth and lysine production. J. Biotechnol. 124, 381–391 (2006).
Tateno, T., Okada, Y., Tsuchidate, T., Tanaka, T., Fukuda, H. & Kondo, A. Direct production of cadaverine from soluble starch using Corynebacterium glutamicum coexpressing alpha-amylase and lysine decarboxylase. Appl. Microbiol. Biotechnol. 82, 115–121 (2009).
Yao, W. et al. Display of alpha-amylase on the surface of Corynebacterium glutamicum cells by using NCgl1221 as the anchoring protein and production of glutamate from starch. Arch. Microbiol. 191, 751–759 (2009).
Tateno, T., Fukuda, H. & Kondo, A. Direct production of L-lysine from raw corn starch by Corynebacterium glutamicum secreting Streptococcus bovis alpha-amylase using cspB promoter and signal sequence. Appl. Microbiol. Biotechnol. 77, 533–541 (2007).
Tsuge, Y., Tateno, T., Sasaki, K., Hasunuma, T., Tanaka, T. & Kondo, A. Direct production of organic acids from starch by cell surface-engineered Corynebacterium glutamicum in anaerobic conditions. AMB Express 3, 72 (2013).
Nguyen, M. T., Choi, S. P., Lee, J., Lee, J. H. & Sim, S. J. Hydrothermal acid pretreatment of Chlamydomonas reinhardtii biomass for ethanol production. J. Microbiol. Biotechnol. 19, 161–166 (2009).
Choi, S. P., Nguyen, M. T. & Sim, S. J. Enzymatic pretreatment of Chlamydomonas reinhardtii biomass for ethanol production. Bioresour. Technol. 101, 5330–5336 (2010).
Ho, S. H., Huang, S. W., Chen, C. Y., Hasunuma, T., Kondo, A. & Chang, J. S. Bioethanol production, using carbohydrate-rich microalgae biomass as feedstock. Bioresour. Technol. 135, 191–198 (2013).
Lynd, L. R. et al. How biotech can transform biofuels. Nat. Biotechnol. 26, 169–172 (2008).
Steen, E. J. et al. Microbial production of fatty-acid-derived fuels and chemicals from plant biomass. Nature 463, 559–562 (2010).
Bokinsky, G. et al. Synthesis of three advanced biofuels from ionic liquid-pretreated switchgrass using engineered Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 108, 19949–19954 (2011).
Ikeda, M. Sugar transport systems in Corynebacterium glutamicum: features and applications to strain development. Appl. Microbiol. Biotechnol. 96, 1191–1200 (2012).
Sasaki, M., Jojima, T., Kawaguchi, H., Inui, M. & Yukawa, H. Engineering of pentose transport in Corynebacterium glutamicum to improve simultaneous utilization of mixed sugars. Appl. Microbiol. Biotechnol. 85, 105–115 (2009).
Kang, M. -K. et al. Synthetic biology platform of CoryneBrick vectors for gene expression in Corynebacterium glutamicum and its application to xylose utilization. Appl. Microbiol. Biotechnol. 98, 5991–6002 (2014).
Eggeling, L. & Bott, M. [23. Experiments]. .In: Handbook of Corynebacterium glutamicum [Eggeling, L. and Bott, M. (ed.)] [537–568]. (CRC Press, Boca Raton, Florida, 2005).
Kabus, A., Niebisch, A. & Bott, M. Role of cytochrome bd oxidase from Corynebacterium glutamicum in growth and lysine production. Appl. Environ. Microbiol. 73, 861–868 (2007).
Sibakov, M. & Palva, I. Isolation and the 5′-end nucleotide sequence of Bacillus licheniformis alpha-amylase gene. Eur. J. Biochem. 145, 567–572 (1984).
Scheele, S. et al. Secretory production of an FAD cofactor-containing cytosolic enzyme (sorbitol-xylitol oxidase from Streptomyces coelicolor) using the twin-arginine translocation (Tat) pathway of Corynebacterium glutamicum. Microb. Biotechnol. 6, 202–206 (2013).
Van der Rest, M., Lange, C. & Molenaar, D. A heat shock following electroporation induces highly efficient transformation of Corynebacterium glutamicum with xenogeneic plasmid DNA. Appl. Microbiol. Biotechnol. 52, 541–545 (1999).
Narita, J. et al. Display of active enzymes on the cell surface of Escherichia coli using PgsA anchor protein and their application to bioconversion. Appl. Microbiol. Biotechnol. 70, 564–572 (2006).
DuBois, M., Gilles, K. A., Hamilton, J. K., Rebers, P. A. & Smith, F. Colorimetric method for determination of sugars and related substances. Anal. Chem. 28, 350–356 (1956).
Acknowledgements
Authors thank Dr. Kyoungseon Min and Mr. Min Eui Hong for technical support. This work was supported by Korea CCS R&D Center (KCRC) (no. 2014M1A8A1049277) and by the National Research Foundation of Korea grant-funded by the Korean Government (Ministry of Science, ICT & Future Planning) (2014, University-Institute cooperation program).
Author information
Authors and Affiliations
Contributions
H.M.W. conceived and supervised the project. J.L. and H.M.W. designed experiments. J.L. and H.M.W. performed experiments. Y.U., S.J.S., M.B. and M.-K.O. oversaw the project. J.L., M.B. and H.M.W. wrote and revised the manuscript. All authors analyzed data and discussed the results and reviewed the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License. The images or other third party material in this article are included in the article's Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder in order to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Lee, J., Sim, S., Bott, M. et al. Succinate production from CO2-grown microalgal biomass as carbon source using engineered Corynebacterium glutamicum through consolidated bioprocessing. Sci Rep 4, 5819 (2014). https://doi.org/10.1038/srep05819
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep05819
This article is cited by
-
Application of Corynebacterium glutamicum engineering display system in three generations of biorefinery
Microbial Cell Factories (2022)
-
Metabolic engineering of Corynebacterium glutamicum for fermentative production of chemicals in biorefinery
Applied Microbiology and Biotechnology (2018)
-
Characterization of a heat-tolerant Chlorella sp. GD mutant with enhanced photosynthetic CO2 fixation efficiency and its implication as lactic acid fermentation feedstock
Biotechnology for Biofuels (2017)
-
Comparative analysis of Corynebacterium glutamicum genomes: a new perspective for the industrial production of amino acids
BMC Genomics (2017)
-
Magnetophoretic sorting of microdroplets with different microalgal cell densities for rapid isolation of fast growing strains
Scientific Reports (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.