Abstract
Histatins are human salivary gland peptides with anti-microbial and anti-inflammatory activities. In this study, we hypothesized that histatin 5 binds to Porphyromonas gingivalis hemagglutinin B (HagB) and attenuates HagB-induced chemokine responses in human myeloid dendritic cells. Histatin 5 bound to immobilized HagB in a surface plasmon resonance (SPR) spectroscopy-based biosensor system. SPR spectroscopy kinetic and equilibrium analyses, protein microarray studies and I-TASSER structural modeling studies all demonstrated two histatin 5 binding sites on HagB. One site had a stronger affinity with a KD1 of 1.9 μM and one site had a weaker affinity with a KD2 of 60.0 μM. Binding has biological implications and predictive modeling studies and exposure of dendritic cells both demonstrated that 20.0 μM histatin 5 attenuated (p < 0.05) 0.02 μM HagB-induced CCL3/MIP-1α, CCL4/MIP-1β and TNFα responses. Thus histatin 5 is capable of attenuating chemokine responses, which may help control oral inflammation.
Similar content being viewed by others
Introduction
Saliva contains an elaborate mixture of peptides, proteins, chemokines, cytokines, growth factors and antimicrobial peptides1,2. Some peptides and proteins play key roles in oral lubrication, mastication, digestion and the demineralization and remineralization of teeth, whereas others play key roles in innate immune defense of oral tissues3,4. Among the latter are the histatins, a group of low molecular-weight histidine-rich peptides and peptide fragments produced by the human parotid, submandibular and sublingual salivary glands5. Although 24 histatin fragments are known6, there are 12 primary histatins arising from HIS1 and HIS2 gene products. Histatins 1 and 3 arise by initial proteolytic events and histatin 5 is a cleavage product of histatin 3.
Histatins bind to specific microorganisms like the periodontal pathogen P. gingivalis7. They also have broad-spectrum antibacterial activity against other oral species of Streptococcus, Actinomyces and Porphyromonas; antifungal activity against species of Candida, Saccharomyces and Cryptococcus; bind copper and zinc; inhibit proteases and toxins; inhibit hemagglutinating activity of some oral bacteria and inhibit co-aggregation of other oral bacteria; inhibit lipopolysaccharide-mediated gelation of Limulus amoebocyte lysate; reverse the anti-complement action of lipopolysaccharide or lipid A; and enhance wound closure8,9,10,11,12. Some histatin fragments also have anti-microbial, DNA-binding and anti-inflammatory activities13,14. Histatin 5 is one of these and pretreatment of outer-membrane protein from P. gingivalis with 10.0 μg/ml (e.g., 3.3 μM) synthetic histatin 5 causes a 37.0% inhibition of IL6 and a 47.0% inhibition of IL8 production in human gingival fibroblasts15.
Hemagglutinin B (HagB) is a major virulence factor of P. gingivalis involved in non-fimbrial adhesion of the microorganism to host cells. It is a 49.0 kDa protein composed of 350 amino acid residues with a pI of 8.42. Recombinant HagB induces a robust chemokine and cytokine response in dendritic cells16. In this study, we hypothesized that histatin 5 binds to recombinant HagB and attenuates HagB-induced chemokine responses in dendritic cells. Our objectives were to i) characterize the interaction of histatin 5 with immobilized HagB by surface plasmon resonance (SPR) spectroscopy using kinetic and equilibrium analyses; ii) identify histatin 5 binding sites on HagB using protein microarrays and I-TASSER molecular modeling; and iii) determine the ability of histatin 5 to attenuate a HagB-induced chemokine and cytokine response using in silico dendritic cell functional simulation modeling of signaling pathways and direct exposure of dendritic cells (Figure 1).
A flow chart on the study design.
SPR spectroscopy kinetic and equilibrium analyses, protein microarray studies and I-TASSER structural modeling studies all were used to assess the ability of histatin 5 to bind to HagB and predictive modeling studies and exposure of dendritic cells in culture were used to assess the ability of histatin 5 to alter HagB-induced chemokine responses.
Results
Binding of histatin 5 to immobilized HagB
Our first objective was to characterize the interaction of histatin 5 with immobilized HagB by kinetic and equilibrium analyses of SPR spectroscopy data (Figure 1). HagB was immobilized at two surface densities to a COOH2 sensor chip using a standard amine coupling method. A third flow channel (FC) was activated and deactivated to serve as a reference in data analysis. Initial binding tests were performed to assess the specificity of histatin 5 for immobilized HagB. In the first test, performed in running buffer without CM-Dextran, notable nonspecific binding of histatin 5 was observed to the reference channel. However, when 0.5 mg/ml CM-Dextran was introduced to the buffer, the non-specific binding became negligible. Subsequent studies on the kinetic binding of histatin 5 to HagB were performed in running buffer with 0.5 mg/ml CM-Dextran. Histatin 5 was observed to fully dissociate from both the high density HagB surface (Figure 2a) and the low density HagB surface (Figure 2b) without the use of regeneration.
The binding responses of histatin 5 demonstrated apparent heterogeneous characteristics, which were thought to be due to the presence of two independent binding sites on immobilized HagB. (Table 1) Equilibrium analysis of a fixed concentration histatin 5 assay (Figure 3a) showed that the dose response plot conformed well to a two-site model (Figure 3b). The maximum binding responses (Rmax) of these data further confirm a multi-site interaction by being larger than the theoretical Rmax. For example, a 1:1 interaction of analyte (MW 3,034.0 Da) and ligand (49.0 kDa) the expected Rmax for FC 2 is 88.0 RU while in Figure 2b responses of >95.0 RU are observed. While individually these results do not conclusively identify a heterogeneous ligand, their agreement suggests this as a valid hypothesis.
Equilibrium and kinetic analysis of SPR spectroscopy data showing histatin 5 binding HagB.
(a,b) Equilibrium responses plotted versus histatin 5 concentration were fit with a one-site Langmuir isotherm (a) and a two-site Langmuir isotherm (b). (c) OneStep™ injection of 16.5 μM histatin 5 over two surface densities of HagB. A two-site kinetic interaction model (blue curves) is shown fit to the data.
Two-site analysis by fitting equilibrium models to the fixed concentration assay indicated the high affinity site to have a KD of 1.9 μM and the weaker affinity site to have a KD of 60.0 μM. Kinetic analysis using the OneStep™ injection, which is modeled by Taylor Dispersion theory, enabled a more accurate estimation of the rate constants for each interaction site17,18. In this injection an internal dispersion line was used to create an analyte gradient of histatin 5, which was then flowed over the FC from low to high concentration (Figure 3c). The gradient binding data were analyzed using a kinetic two-site model, which estimates rate constants for the two individual interactions. (Table 2) The KD1 values determined from the kinetic model fit were 340.0 ± 10.0 and 260.0 ± 10.0 nM from the high and low-density HagB surfaces, respectively. The KD2 values determined were 11.3 ± 0.07 and 15.2 ± 0.2 μM from the high and low-density HagB surfaces, respectively.
A final objective for the SPR spectroscopy experiments was to compare HagB binding of HBDs with histatin 5. HBD1 and HBD3 were observed to bind the sensor chip with more non-specific binding than histatin 5 (data not shown). The concentration of CM-Dextran was therefore increased to 2.0 mg/ml for the HBD assays (Figure 4). HBD1 was observed to have a weak affinity interaction with HagB as expected while HBD3 demonstrated a heterogeneous binding response, which was not able to be fit with a standard interaction model. The NaCl concentration of the buffer was increased to 0.3 M and the assay was repeated for both defensins (Figure 5). HagB binding of HBD1 appeared to be slightly modified by the change in salt concentration. HagB binding of HBD3 however was significantly affected by the change in salt concentration such that an equilibrium model could be fit to the dose response plot (Figure 5d).
SPR spectroscopy data showing binding of HBDs with immobilized HagB.
The assay buffer was modified to contain 2.0 mg/ml CM-Dextran. (a) HBD1 dilution series binding HagB. (b) Dose response plot of the interaction of HBD1 with HagB. (c) HBD3 dilution series binding HagB. (d) Dose response plot of the interaction of HBD3 with HagB.
SPR spectroscopy data showing binding of HBDs with immobilized HagB.
The assay buffer was modified to contain increased NaCl (0.3 M) and 2.0 mg/ml CM-Dextran. (a) HBD1 dilution series binding HagB. (b) Dose response plot of the interaction of HBD1 with HagB. A one-site Langmuir isotherm is shown applied to the data. (c) HBD3 dilution series binding HagB. (d) Dose response plot of the interaction of HBD3 with HagB. A one-site Langmuir isotherm is shown applied to the data.
Identification of binding sites on HagB
Identification of histatin 5, HBD1 and HBD3 binding sites on HagB began with their differential binding to the 350 different, overlapping 15mer peptides in the HagB peptide microarrays. Histatin 5-Cy5-NHS, HBD1-Cy5-NHS and HBD3-Cy5-NHS bound to select 15mer peptides in each array resulting in higher fluorescent intensities than that of adjacent overlapping 15mer peptides on either side. To identify a binding site we assumed that for two overlapping peptides, the significant increase or decrease of fluorescent intensity of one peptide compared with the other adjacent peptide is due to the addition or deletion of a new or old residue. For example, in the histatin 5-HagB microarray, the fluorescent intensity increased from 0.0 to 22.0 from the 15mer peptide (31–45, NIAKLNPKLPELEKA) to peptide (32–46, IAKLNPKLPELEKAI). Based on our assumption, the increase would be due to the addition of the residue I46.
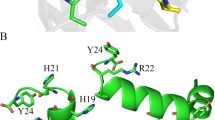
Histatin 5, at 1.0 μg/ml, bound to HagB residues I32, I46, Y76, F88, D117, N154, R165, I242, E256 and Y291. These residues can be mapped to two potential binding sites using the I-TASSER structural model (Figure 6a). The first site contained residues I32, I46, Y76 and F88 (colored in red) and the second site contained residues D117, N154 and R165 (colored in blue); the other three residues (I242, E256 and Y291, colored in green) might belong to the second binding site but they are too far away from the first binding site. This might be caused by the inaccurate structure modeling at the C-terminus (modeled as loop), where the residues I242, E256 and Y291 are located.
The mapped binding sites of peptides to the predicted 3D I-TASSER structural model of HagB.
(a) Histatin 5 binds to residues I32, I46, Y76 and F88 (colored in red) and to residues D117, N154 and R165 (colored in blue). (b) HBD1 binds to residues Y17, L26, K38, Y76, D89, D103 and Y213 (colored in red) likely forming one binding site. (c) HBD3 binds to residues K118, K131, L147, E152 and D160 (colored in red) likely forming one binding site. Histatin 5, HBD1 and HBD3 bind to HagB in different regions: histatin 5 and HBD1 bind more centrally to the HagB molecule whereas HBD3 binds near the end of the loop area. However, one binding site for histatin 5 is near the binding site for HBD3. This may account for the ability of histatin 5 to attenuate HagB-induced chemokine and cytokine responses in dendritic cells similar to that for HBD3, although much higher concentrations of histatin 5 are needed than HagB. For high-resolution images of the binding domains see Supplementary Figures 1a, b & c.
HBD1, at 1.0 μg/ml, bound to HagB residues Y17, L26, K38, Y76, D89, D103, Y213, V261, R273, Y291 and L350. The residues L26, K38, Y76, D89, D103 and Y213 (in red color) likely form one binding site according to the I-TASSER model (Figure 6b). Residues V261, R273, Y291 and L350 are located the loop region of the I-TASSER model, which have low model quality and thus it is difficult to make a conclusion for these residues.
HBD3, at 1.0 μg/ml, bound to HagB residues N31, N54, K118, L147, E152, D199, L212, A281 and G295 and HBD3 at 2.0 μg/ml bound to HagB residues V3, N13, D69, A98, K118, K131, K141, E152, D160, L212, K271, D285 and S338. Using the I-TASSER model and the consensus of the two experiments, we conclude that HBD3 binds to residues K118, K131, L147, E152 and D160 (colored in red in Figure 6c) and this may represent a possible binding site.
Predicted influence of histatin 5 on a HagB-induced dendritic cell response
The in silico dendritic cell functional simulation model predicted that histatin 5 would reduce the HagB-induced IL8 response by 31.8%, the CSF2/GM-CSF response by 55.9%, the CCL3/MIP-1α response by 42.0%, the CCL4/MIP-1β response by 44.4% and the TNFα response by 46.5% (Figure 7a, b).
The predictive in silico virtual dendritic cell platform generated results showing the percentage change with respect to the control reference baseline.
(a) A high-level view of the HagB and histatin 5 regulation in the dendritic cell platform. HagB provides activating signals to CD14 and activin receptors. On binding to CD14 and activin receptors, it stimulates the release of pro-inflammatory cytokines like CCL3/MIP-1α, CCL4/MIP-1β, CSF2/GM-CSF, IL8 and TNFα via activation of MYD88/TRAF6 and SHC1/ERK pathways, respectively. Both activin and CD14 signals converge at the activation of MAPKs, AKT and PKC to activate transcription factors like NFKB, AP1, SP1, CREB1, NFAT, IRFs, etc., which transcribes different cytokines. Histatin 5 exerts inhibitory effect on dendritic cells. Histatin 5 negatively regulates the pro-inflammatory signal by inhibiting CD14 receptors. The dotted red lines in the figure indicate inhibitory links and dotted fluorescent green lines indicate activation links. The downstream effects and activation links of each transcription factor are shown by uniquely colored solid lines. (b) The illustrated effect of over-expression of histatin 5 along with HagB stimulation on CCL3/MIP-1α, CCL4/MIP-1β, CSF2/GM-CSF, IL8 and TNFα. Compared to HagB stimulation alone on the referenced baseline, the levels of CCL3/MIP-1α, CCL4/MIP-1β, CSF2/GM-CSF, IL8 and TNFα had significantly reduced.
Influence of histatin 5 on a HagB-induced dendritic cell response
None of the solutions, media, or inocula containing 0.2, 2.0, or 20.0 μM histatin 5 or 0.02 or 0.2 μM HagB induced any cytotoxicity in the dendritic cell cultures at 16.0 hours.
0.02 μM HagB induced strong IL8 responses at 2.0 and 4.0 hours that peaked at 8.0 hours and began to decline at 16.0 hours post exposure. Although histatin 5 appeared to be able to attenuate a HagB-induced IL8 response at 8.0 hours in some experimental replications, it was not consistent in all primary dendritic cell cultures and often lost by 16.0 hours (not shown).
0.02 μM HagB also induced strong CCL3/MIP-1α, CCL4/MIP-1β and TNFα responses and a weaker CSF2/GM-CSF response (Figure 8). At 16.0 hours, a 100:1 micromolar ratio of histatin5:HagB, histatin 5 induced a 9.3% decrease in CCL3/MIP-1α, 12.5% decrease in CCL4/MIP-1β, a 25.1% decrease in TNFα and a 42.0% decrease in CSF2/GM-CSF (Figure 8). At 16.0 hours at a 1,000:1 micromolar ratio of histatin5:HagB, histatin 5 significantly (p < 0.05) induced a 49.8% decrease in CCL3/MIP-1α, 39.6% decrease in CCL4/MIP-1β and a 42.4% decrease in TNFα.
Influence of histatin 5 (H5) on a P. gingivalis HagB-induced CCL3/MIP-1α, CCL4/MIP-1β, TNFα and CSF2/GM-CSF dendritic cell response.
In one experiment (top line graph, each panel), dendritic cells were exposed to 0.01 M PBS, pH 7.2; 0.02 μM HagB; 2.0 μM histatin 5; or 2.0 μM histatin 5 + 0.02 μM HagB. Aliquots of cell culture media were removed at 0.0, 2.0, 4.0, 8.0 and 16.0 hours. At 16.0 hours at a 100:1 micromolar ratio of histatin 5:HagB, histatin 5 induced a 9.3% decrease in CCL3/MIP-1α, 12.5% decrease in CCL4/MIP-1β, a 25.1% decrease in TNFα and a 42.0% decrease in CSF2/GM-CSF. In a second experiment increasing the concentration of histatin 5 10-fold (bottom bar graph, each panel), dendritic cells were exposed to 0.01 M PBS, pH 7.2; 0.02 μM HagB; 20.0 μM histatin 5; or 20.0 μM histatin 5 + 0.02 μM HagB. Aliquots of cell culture media were removed at 0.0, 2.0, 4.0, 8.0 and 16.0 hours. At 16.0 hours at a 1000:1 micromolar ratio of histatin 5:HagB, histatin 5 induced a 49.8% decrease in CCL3/MIP-1α, 39.6% decrease in CCL4/MIP-1β, a 42.4% decrease in TNFα and a 55.3% decrease in CSF2/GM-CSF. * = p < 0.05.
Discussion
Histatins are a group of low molecular-weight histidine-rich peptides produced in abundance by the human parotid, submandibular and sublingual salivary glands5,9. Concentrations in parotid saliva range from 7.0 to 28.0 μg/ml for histatin 1, 6.0 to 43.0 μg/ml for histatin 3 and 10.0 to 43.0 μg/ml for histatin 5 and concentrations in submandibular and sublingual saliva range from 28.0 to 122.0 μg/ml for histatin 1, 15.0 to 75.0 μg/ml for histatin 3 and 26.0 to 90.0 μg/ml for histatin 519,20.
Histatins have a variety of innate immune functions. They bind to bacteria, inhibit co-aggregation of oral bacteria and have broad-spectrum antimicrobial activity against bacteria and fungi (e.g., C. albicans). For example, 125I labeled histatin 5 binds to P. gingivalis 3817: binding of histatin 5 is rapid, reversible, saturable and specific; the number of histatin 5-binding sites per cell was estimated to be 3,600; and the dissociation constant (Kd) was calculated to be 1.5 μM. The binding of histatins to virulence agonists alters the physiologic properties of the agonist and neutralizes toxins, inactivates proteases and neutralizes the activities of lipopolysaccharide. Histatin 5 also has anti-inflammatory properties. Histatin 5 is known to suppress the production of IL6 and IL8 to P. gingivalis outer-membrane protein by human gingival fibroblasts15.
In this study, we show that histatin 5 binds to P. gingivalis HagB. The interaction of histatin 5 with immobilized HagB was characterized by kinetic and equilibrium analysis of SPR spectroscopy data. Evidence for a two-site interaction was observed and considered when choosing the appropriate models. A higher affinity binding site was identified with a KD1 of 340.0 or 260.0 nM when measured at two HagB surface densities. A weaker affinity site was calculated to have a KD2 of 11.3 or 15.2 μM when measured at two HagB surface densities. We therefore conclude that histatin 5 has a moderate affinity for HagB that includes a weaker affinity interaction that could be due to the peptide's basic isoelectric point. Analysis with OneStep™ provided additional confidence in the kinetic and affinity constants obtained for both interaction events.
We also showed that histatin 5 attenuates HagB-induced CCL3/MIP-1α, CCL4/MIP-1β and TNFα responses at 16.0 hours post-inoculation of dendritic cells. Attenuation more readily occurs at 100:1 and 1,000:1 micromolar ratios of histatin 5:HagB than at 1:1 and 10:1 micromolar ratios of histatin 5:HagB (Figure 8). It is worth noting that 2.0 μM (6.1 μg/ml) and 20.0 μM (60.7 μg/ml) of histatin 5 are well within the physiologic concentration of histatin 5 reported above in the oral cavity19,20.
HBD1 demonstrated a very weak affinity for HagB compared with histatin 5 and HBD3. HBD3 demonstrated a strong affinity with HagB that was highly charge based. After the non-specific binding of HBD3 was reduced by increasing the NaCl concentration, the resulting interaction affinity was estimated with a KD value of 35.0 μM. Histatin 5 therefore has a more favorable affinity for HagB than the defensin peptides and which is apparently more specific than just electrostatic attraction.
The exact mechanism for histatin 5 attenuation of the HagB-induced response is not yet fully known. It is easy to speculate that histatin 5 simply binds to HagB and inhibits its attachment to dendritic cell surfaces. If that were the case, we would expect to see attenuation of all of the HagB-induced chemokine and cytokine responses. More likely, histatin 5 may be first altering the binding of HagB to the surface of dendritic cells like that seen by HBD321 and then selectively altering HagB-induced signal transduction and transcription factors (Figure 7a), also like that seen by HBD316,22. The MAPK pathways are important in controlling the type and magnitude of the inflammatory response to P. gingivalis and its extracellular products including HagB and histatin 5 may work via a similar mechanism.
We believe that histatin 5 may now be grouped with a family of anti-inflammatory components present in the oral cavity. These include salivary gland derived peptides23, fibronectin24, LL-3725,26,27, lactoferrin28,29, human neutrophil peptide (HNP) α defensins30,31 and HBDs16,22. Therefore, we believe that, histatin 5, in the physiologic range of human saliva, has the ability to significantly attenuate select chemokines and cytokines produced in dendritic cells exposed to HagB, a virulence factor of the periodontal pathogen P. gingivalis. Together with this family of anti-inflammatory peptides in saliva, histatin 5 may complement existing mechanisms to maintain oral homeostasis and control and contain oral infection and inflammation by down regulating chemokine and pro-inflammatory cytokine responses.
In summary, histatin 5 behaves like HBD3. Both histatin 5 and HBD3 bind to HagB and both attenuate HagB-induced chemokine and cytokine responses in dendritic cells when co-incubated together at 37°C for 30 minutes with HagB. Although the trends of histatin 5 with HagB are reproducible, they are much more subtle than the trends seen with HBD3 and HagB and much higher concentrations of histatin 5 are needed. One explanation may be the difference in binding domains of histatin 5 and HBD3 on HagB (Figure 6): HBD3 binding occurred near the end of the loop whereas histatin 5 and HBD1 binding occurred more centrally to the HagB molecule. Future work will focus on identifying the chemokine and cytokine active domain on HagB and the binding domain of antimicrobial peptides on HagB.
Methods
Reagents
All solutions, media and cell culture inocula were prepared as previously described16. 0.01 M sodium phosphate with 0.14 M NaCl, pH 7.2 (0.01 M PBS, pH 7.2) contained 4.0 ± 0.7 (±Std Err, n = 3) pg/ml endotoxin; and Lymphocyte Growth Media-3 (LGM-3, Lonza Walkersville, Inc.) contained 53.0 ± 1.0 (±Std Err, n = 3 tests) pg/ml endotoxin.
For SPR spectroscopy studies, histatin 5 (Sigma-Aldrich, St. Louis, MO) was suspended in 0.01 M HEPES pH 7.4, 0.15 M NaCl, 3.4 mM ethylenediaminetetraacetic acid (EDTA), 0.005% (w/w) Tween-20 and 0.5 mg/ml CM-Dextran to 16.5 μM. For cell culture studies, histatin 5 was suspended in 0.01 M PBS, pH 7.2, filtered and diluted to 20× stock solutions containing 400.0, 40.0 and 4.0 μM. These preparations contained 0.07 (n = 3 tests) ng/ml endotoxin.
Human β defensins HBD1 and HBD3 (PeproTech, Inc., Rocky Hill, NJ) were used as weak and strong HagB binding controls in SPR spectroscopy studies, respectively, for comparison to histatin 5 responses32. HBD1 and HBD3 each contained 0.07 (n = 3 tests) pg/ml endotoxin. HBD1 and HBD3 were suspended in 0.01 M HEPES pH 7.4, 0.15 M NaCl (or 0.3 M NaCl), 3.4 mM EDTA, 0.005% (w/w) Tween-20 and 2.0 mg/ml CM-Dextran.
HagB was prepared by cloning hagb of P. gingivalis (1.4 kb) into the vector pQE31 (QIAGEN Inc., Valencia, CA); expressing HagB in E. coli M15(pREP4)pQE-31-TX1; and isolating HagB from E. coli lysate supernatants with a Ni-NTA spin column (HisPur, Pierce Biotechnology, Rockford, IL)16,22,33. Bound HagB was eluted with 0.25 M imidazole in 0.01 M Tris, 0.5 M NaCl, 20% glycerol, pH 7.4 and dialyzed against 0.01 M Tris with 0.5 M NaCl, pH 7.4 at 4°C and diluted with 0.01 M PBS, pH 7.2 to give 20× stock solutions containing 4.0 and 0.4 μM. The purity of HagB was verified by SDS-PAGE. HagB contained 0.07 (n = 3 tests) pg endotoxin/mg HagB.
SPR spectroscopy
SensíQ Pioneer is an SPR-based biosensor that was used to characterize the interactions of histatin 5 and defensins with immobilized HagB. The assay running buffer was 0.01 M HEPES pH 7.4, 0.15 M NaCl, 3.4 mM EDTA, 0.005% (w/w) Tween-20 and 0.5 mg/ml CM-Dextran (Fluka 86524) unless otherwise noted and was filtered (0.22 μm filter) before use. The SensíQ Pioneer analysis chamber was set at a controlled temperature of 25°C and the sample racks were cooled to 18°C. SPR spectroscopy data for histatin experiments were recorded at 4 Hz and HBD experiments were recorded at 2 Hz.
A COOH2 sensor chip was used and was preconditioned by two one minute injections, each of 10.0 mM HCl, 50.0 mM NaOH and 0.1% SDS and then primed in running buffer without CM-Dextran. A 30.0 μg/ml HagB solution was prepared in 10.0 mM sodium acetate buffer pH 5.0. Activation solution containing 10.0 mM EDC and 2.5 mM NHS was injected over flow channels (FC) 1 and 2 for 4.5 minutes. The HagB solution was then injected over FC 1 and 2 for approximately 1.5 minutes. FC 3 was then activated similarly with the EDC/NHS solution. All three FC's were then capped by injecting 1.0 M ethanolamine, pH 7.5 for 5.0 minutes. Approximately 3,470 response units (RU) of HagB was immobilized on FC 1 (e.g., high density HagB surface) and 1,430 RU of HagB was immobilized on FC 2 (e.g., low density HagB surface).
A solution of 16.5 μM histatin 5 was prepared in running buffer without CM-Dextran and was injected over all three FC for 3.0 minutes. Running buffer was then modified to contain 0.5 mg/ml CM-Dextran and was primed into the SensíQ Pioneer. Another 16.5 μM solution of histatin 5 was prepared in running buffer and was injected for 3.0 minutes. Considerably less nonspecific binding was seen when histatin 5 was injected in the FC 3 reference channel with CM-Dextran in the running buffer than without CM-Dextran in the running buffer (data not shown).
Two-fold dilution series of histatin 5, HBD1 and HBD3 were prepared in running buffer from 33.0 to 0.1 μM. The HBD samples were prepared in the running buffer with 2.0 mg/ml CM-Dextran. Each sample and several buffer blanks were injected for one minute (histatin 5) at a flow rate of 20.0 μl/minute or two minutes (HBDs) at a flow rate of 10.0 μl/minute. Histatin 5 samples were tested in triplicate and HBDs were tested once in order from the lowest to the highest concentration. HBDs were then prepared in an additional two-fold dilution series in running buffer with 0.3 M NaCl and 2.0 mg/ml CM-Dextran from 25.4 to 0.4 μM (HBD1) and from 19.4 to 0.3 μM (HBD3). HBD samples were injected for three minutes at a flow rate of 15.0 μl/minute. All HBD assays included two one-minute surface regeneration injections of 2.0 M NaCl. SPR spectroscopy data were recorded at a sampling rate of 4 Hz. The assay data were zeroed (Y-normalized) and the response of the FC 3 reference channel was subtracted from the FC 1 and 2 specific channel responses. The average response of the buffer blanks was then subtracted from the sample injections.
Equilibrium responses of histatin 5, HBD1 and HBD3 were plotted versus peptide concentration and the resulting dose response was fitted with a Langmuir isotherm. In these analyses, the Rmax term(s) were fitted individually for each channel data set while the KD term(s) were fitted as shared values for the two data sets. Histatin 5 data were fitted with a two-site isotherm and HBD data were fitted with a one-site isotherm.
A OneStep™ injection assay was performed to resolve the heterogeneous histatin 5 binding in the time domain. 3% (w/v) sucrose was also prepared in running buffer as a diffusion standard. Running buffer, sucrose solution and 16.5 μM histatin 5 were each injected in triplicate over all three FC as a sigmoidal OneStep™ at a flow rate of 20.0 μl/minute. Kinetic analysis of the histatin 5 binding response was performed by fitting a two-site interaction model to the referenced data.
HagB peptide microarray
To assess putative binding sites of histatin 5 on HagB, we developed a HagB peptide microarray on glass slides (PEPperPRINT GmbH, Heidelberg, Germany). For this, the protein sequence of HagB was translated into 350 different 15mer peptides as triplicates with a peptide-peptide overlap of 14 amino acids giving 1,050 peptide spots per array copy. The C-and N-terminus of HagB was elongated by neutral GSGSGSG linkers to avoid truncated peptides. Each microarray contained five array copies with the same peptide content. Each array copy was framed by FLAG tag (e.g., DYKDDDDKGG; 86 spots) and HA tag (e.g., YPYDVPDYAG; 86 spots) as control peptides.
Slides were incubated with 0.01 M PBS, pH 7.4, 0.05% Tween 20 and 1% bovine serum albumin (BSA) with slight shaking for 1.0 hour at room temperature to reduce non-specific interactions with histatin 5, HBD1, or HBD3. 1.0 μg/ml histatin 5-Cy5-NHS; 1.0 μg/ml HBD1-Cy5-NHS; and 1.0 and 2.0 μg/ml HBD3-Cy5-NHS in 0.01 M PBS, pH 7.4, 0.05% Tween 20 with 0.1% BSA were added to the slide chambers. 1.0 μg/ml control peptide-Cy-NHS; mouse monoclonal anti-HA antibody-Cy5; and mouse monoclonal anti-FLAG (M2) antibody-Cy3-NHS in 0.01 M PBS, pH 7.4, 0.05% Tween 20, with 0.1% BSA were added to control slide arrays. All slides were incubated with slight shaking overnight at 4°C. After 16.0 hours, the arrays were washed three times for one minute each with 0.01 M PBS, pH 7.4, 0.05% Tween 20.
The slides were scanned in a Typhoon 9410 Variable Mode Imager and the extent of histatin 5, HBD1 and HBD3 binding was determined from the fluorescence images. Spot intensities were quantified with PepSlide® Analyzer. Based on averaged median foreground intensities, an intensity map was generated and binders in the peptide map highlighted by an intensity color code with red for high and white for low spot intensities. Peptides fragments on the array containing arginine and lysine-rich regions like PKKKKK (e.g., HagB amino acid residues 309–314) tended to interact with labeled samples due to ion-exchange effects with the charged dye molecules and thus were regarded as non-specific.
Modeling the structure of HagB
The state-of-the-art protein structure prediction method I-TASSER34,35,36 was used to predict the structure and function of HagB. First, the HagB sequence was threaded against the PDB library37 by the meta-threading algorithm LOMETS38 to identify homologues templates. The fragments excised from the threading templates were then reassembled into full-length models by replica-exchange Monte Carlo simulations with the threading unaligned regions (mainly loops) built by ab initio folding. The lowest free-energy conformation was selected by clustering the Monte Carlo simulation structures using SPICKER39. Next, fragment assembly simulation was performed again starting from the SPICKER cluster centroids, where the spatial restraints collected from both the LOMETS templates and the analogy PDB structures by TM-align40 were used to guide the reassembly simulations. Finally, the models were refined in the atomic-level by the fragment-guided molecular dynamics (FG-MD) simulations41. To examine the biological function of the HagB sequence, the COFACTOR program36,42 was used to match the I-TASSER models to the protein function library BioLiP43 by local and global structural comparisons.
The overall topology of the HagB structure model was a three-helix bundle attached by a long loop at the C-terminus (Figure 6). The confidence score of the I-TASSER structure prediction was −2.52 with the estimated TM-score = 0.42 ± 0.14, which corresponds to an approximately correct topology. The major threading templates used by I-TASSER include human APPL2BARPH domain (PDBID: 4h8sA), nucleotide exchange factor Son of Sevenless (1xd4A) and BAR-PH domain of APPL1 (2q13A).
Modeling of the dendritic cell response of histatin 5 on HagB
An in silico dendritic cell functional simulation model was used to predict the effect of histatin 5 on the HagB-induced IL8, CSF2/GM-CSF, CCL3/MIP-1α, CCL4/MIP-1β and TNFα responses of dendritic cells16. This model provides a dynamic and transparent view of dendritic cell physiology at the functional proteomics abstraction level and predicts outcomes for different experimental scenarios in an automated high-throughput system. The model was manually aggregated and is currently updated, from published research and collaborative data on signaling pathways, intermediates, transcription factors, enzyme kinetics and gene regulations comprising 3,860 biological species with over 15,433 cross-talk interactions16. The technology has been recently used to predict a) elastin triggered transient pro-inflammatory responses by human dermal fibroblasts44; b) anti-aggregation of alpha-synuclein in M17 dopaminergic cells45; ATPase activity of heat shock protein 90 in tumor cells, endothelial cells and stromal cells46; c) effects of progesterone and Cox2 inhibitors on progesterone receptor isoforms A and B in myometrial cells47; d) ability of butein to inhibit STAT3 expression in HepG2, SNU-387 and PLC/PRF5 human hepatocellular carcinoma cells48; and e) the potential effects of isorhamnetin on the peroxisome proliferator-activated receptor gamma signaling cascade49.
For this study, HagB was thought to activate dendritic cells through TLR and activin receptor signaling16. Histatin 5 was thought to inhibit the activation of dendritic cells by TLR signaling. HagB was introduced with a concentration of 8.0 × 10−4 μM along with a 5-fold over-expression of histatin 5 and the effect on downstream markers were analyzed compared to HagB stimulation alone on the referenced baseline. The first simulation protocol response consisted of control simulation for 15,000 seconds, stimulation with 8.0 × 10−4 μM HagB and trigger simulation for 50,000 seconds. This response was compared with a second simulation protocol response consisting of control simulation for 15,000 seconds, stimulation with 8.0 × 10−4 μM HagB, trigger simulation for 50,000 seconds, histatin 5 inhibition on CD14 activation flux and drug simulation for 75,000 seconds.
Dendritic cell response of histatin 5 on HagB
Mononuclear cell-derived human myeloid dendritic cells (Lonza Walkersville, Inc., Walkersville, MD and StemCell Technologies, Inc., Vancouver, BC Canada) were used and contained cells staining 31.5% CD1c+/79.1% CD11c+ and 54.4% CD141+/79.1% CD11c+ (Flow cytometry kit FMC016, R&D Systems, Inc., Minneapolis, MN). Adhered dendritic cells in 24 well plates (4.1 × 104 cells/cm2; Corning, Tewksbury MA) were treated with 0.01 M PBS, pH 7.2 in LGM-3 (diluent control); 2.0, or 20.0 μM histatin 5 in LGM-3 (histatin 5 controls), or 0.02 μM HagB in LGM-3 (HagB control). Dendritic cells were also treated with 0.02 μM HagB + 2.0 μM histatin 5, previously mixed together at 37°C for 30 minutes or 0.02 μM HagB + 20.0 μM histatin 5, previously mixed together at 37°C for 30 minutes. After treatment, cells were incubated at 37°C in 5% CO2. At 0.0, 2.0, 4.0, 8.0 and 16.0 hours, 0.1 ml was removed from each well and 0.1 ml of LGM-3 was added back. Samples were centrifuged at 13,200 × g (16,100 rpm) at 25°C for 5.0 minutes to pellet non-adherent cells and the supernatants were removed and frozen at −80°C.
Cytotoxicity
After collection of the last timed sample, LGM-3 was removed and fresh LGM-3 containing Alamar blue (AlamarBlue, Invitrogen Corp., Frederick, MD), was added and the cells were incubated for 16 hours. The fluorescence intensity of the resorufin was determined using an excitation wavelength of 544 nm and an emission wavelength of 590 nm (SpectraMax M2e Multi-Mode Microplate Reader, Molecular Devices, LLC, Sunnyvale, CA).
Determination of chemokines and cytokines
Briefly, 25.0 μl of dendritic cell supernatant was added to anti-human multi-cytokine magnetic beads (Milliplex immunoassay, Millipore, Billerica, MA USA) and incubated at 4°C for 18.0 hours. Unbound material was removed by aspiration (ELx405TS magnetic plate washer, BioTek, Winooski, VT USA); anti-human multi-cytokine biotin reporter was added; and the reactions were incubated at room temperature for 1.5 hours in the dark. Streptavidin–phycoerythrin was then added and the plates were incubated at room temperature for an additional 30 minutes. Stop solution was added and the plates were read (Luminex model 100 IS, Austin, TX). Standard curves for each cytokine were prepared from 2.3 to 10,000.0 pg/ml and concentrations of chemokines and cytokines in each sample were interpolated from standard curves (xPonent v3.1, Luminex, Austin, TX USA; MILLIPLEX Analyst v5.1, Millipore, Billerica, MA USA).
Statistical analysis
A log10-transformation was applied to IL8, CSF2/GM-CSF, CCL3/MIP-1α, CCL4/MIP-1β and TNFα concentrations detected in the tissue culture supernatants, as recently described16. The log transformation attenuates the positive skew in the distributions of the chemokine concentrations and makes the normality assumption more defensible. One-way fixed-effects ANOVA models were fit to the log-transformed concentrations. Pairwise group comparisons were conducted using the method of Tukey's Honest Significant Differences (HSD). A 0.05 level was used to determine statistically significant differences. All analyses were conducted using JMP (Version 10.0, SAS, Cary, NC).
References
Gorr, S. U. Antimicrobial peptides of the oral cavity. Periodontol. 2000 51, 152–180 (2009).
Khan, A. Detection and quantitation of forty eight cytokines, chemokines, growth factors and nine acute phase proteins in healthy human plasma, saliva and urine. J. Proteomics 75, 4802–4819 (2012).
Schenkels, L. C., Veerman, E. C. & Nieuw Amerongen, A. V. Biochemical composition of human saliva in relation to other mucosal fluids. Crit. Rev. Oral Biol. Med. 6, 161–175 (1995).
Lamkin, M. S. & Oppenheim, F. G. Structural features of salivary function. Crit. Rev. Oral Biol. Med. 4, 251–259 (1993).
Oppenheim, F. G. et al. Histatins, a novel family of histidine-rich proteins in human parotid secretion. Isolation, characterization, primary structure and fungistatic effects on Candida albicans. J. Biol. Chem. 263, 7472–7477 (1988).
Castagnola, M. et al. A cascade of 24 histatins (histatin 3 fragments) in human saliva. Suggestions for a pre-secretory sequential cleavage pathway. J. Biol. Chem. 279, 41436–41443 (2004).
Murakami, Y. et al. Binding of a histidine-rich peptide to Porphyromonas gingivalis. FEMS Microbiol. Lett. 66, 253–256 (1991).
Sugiyama, K. Anti-lipopolysaccharide activity of histatins, peptides from human saliva. Experientia 49, 1095–1097 (1993).
Tsai, H. & Bobek, L. A. Human salivary histatins: promising anti-fungal therapeutic agents. Crit. Rev. Oral Biol. Med. 9, 480–497 (1998).
Gyurko, C., Lendenmann, U., Helmerhorst, E. J., Troxler, R. F. & Oppenheim, F. G. Killing of Candida albicans by histatin 5: cellular uptake and energy requirement. Antonie Van Leeuwenhoek 79, 297–309 (2001).
Grogan, J., McKnight, C. J., Troxler, R. F. & Oppenheim, F. G. Zinc and copper bind to unique sites of histatin 5. FEBS Lett. 491, 76–80 (2001).
Oudhoff, M. J. et al. Histatins enhance wound closure with oral and non-oral cells. J. Dent. Res. 88, 846–850 (2009).
Rothstein, D. M. et al. Anticandida activity is retained in p-113, a 12-amino-acid fragment of histatin 5. Antimicrob. Agents Chemother. 45, 1367–1373 (2001).
Huo, L. et al. Antimicrobial and DNA-binding activities of the peptide fragments of human lactoferrin and histatin 5 against Streptococcus mutans. Arch. Oral Biol. 56, 869–876 (2011).
Imatani, T., Kato, T., Minaguchi, K. & Okuda, K. Histatin 5 inhibits inflammatory cytokine induction from human gingival fibroblasts by Porphyromonas gingivalis. Oral Microbiol. Immunol. 15, 378–382 (2000).
Harvey, L. E. et al. Defensin DEFB103 bidirectionally regulates chemokine and cytokine responses to a pro-inflammatory stimulus. Sci. Rep. 3, 1232 (2013).
Quinn, J. G. Evaluation of Taylor dispersion injections: determining kinetic/affinity interaction constants and diffusion coefficients in label-free biosensing. Anal. Biochem. 421, 401–410 (2012).
Quinn, J. G. Modeling Taylor dispersion injections: determination of kinetic/affinity interaction constants and diffusion coefficients in label-free biosensing. Anal. Biochem. 421, 391–400 (2012).
Gusman, H. et al. Human salivary gland-specific daily variations in histatin concentrations determined by a novel quantitation technique. Arch. Oral Biol. 49, 11–22 (2004).
Campese, M., Sun, X., Bosch, J. A., Oppenheim, F. G. & Helmerhorst, E. J. Concentration and fate of histatins and acidic proline-rich proteins in the oral environment. Arch. Oral Biol. 54, 345–353 (2009).
Van Hemert, J. R. et al. Human beta-defensin-3 alters, but does not inhibit, the binding of Porphyromonas gingivalis haemagglutinin B to the surface of human dendritic cells. Int. J. Antimicrob. Agents 40, 75–79 (2012).
Pingel, L. C. et al. Human beta-defensin 3 binds to hemagglutinin B (rHagB), a non-fimbrial adhesin from Porphyromonas gingivalis and attenuates a pro-inflammatory cytokine response. Immunol. Cell Biol. 86, 643–649 (2008).
Mathison, R. D., Davison, J. S., Befus, A. D. & Gingerich, D. A. Salivary gland derived peptides as a new class of anti-inflammatory agents: review of preclinical pharmacology of C-terminal peptides of SMR1 protein. J. Inflamm. (Lond). 7, 49 (2010).
Murakami, Y. et al. Fibronectin in saliva inhibits Porphyromonas gingivalis fimbria-induced expression of inflammatory cytokine gene in mouse macrophages. FEMS Immunol. Med. Microbiol. 22, 257–262 (1998).
Scott, M. G., Davidson, D. J., Gold, M. R., Bowdish, D. & Hancock, R. E. The human antimicrobial peptide LL-37 is a multifunctional modulator of innate immune responses. J. Immunol. 169, 3883–3891 (2002).
Mookherjee, N. et al. Modulation of the TLR-mediated inflammatory response by the endogenous human host defense peptide LL-37. J. Immunol. 176, 2455–2464 (2006).
Molhoek, E. M. et al. Structure-function relationship of the human antimicrobial peptide LL-37 and LL-37 fragments in the modulation of TLR responses. Biol. Chem. 390, 295–303 (2009).
Baveye, S., Elass, E., Mazurier, J., Spik, G. & Legrand, D. Lactoferrin: a multifunctional glycoprotein involved in the modulation of the inflammatory process. Clin. Chem. Lab. Med. 37, 281–286 (1999).
Caccavo, D. et al. Antimicrobial and immunoregulatory functions of lactoferrin and its potential therapeutic application. J. Endotoxin Res. 8, 403–417 (2002).
Shi, J. et al. A novel role for defensins in intestinal homeostasis: regulation of IL-1beta secretion. J. Immunol. 179, 1245–1253 (2007).
Miles, K. et al. Dying and necrotic neutrophils are anti-inflammatory secondary to the release of alpha-defensins. J. Immunol. 183, 2122–2132 (2009).
Dietrich, D. E. et al. Human alpha- and beta-defensins bind to immobilized adhesins from Porphyromonas gingivalis. Infect. Immun. 76, 5714–5720 (2008).
Song, H., Belanger, M., Whitlock, J., Kozarov, E. & Progulske-Fox, A. Hemagglutinin B is involved in the adherence of Porphyromonas gingivalis to human coronary artery endothelial cells. Infect. Immun. 73, 7267–7273 (2005).
Roy, A., Kucukural, A. & Zhang, Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 5, 725–738 (2010).
Zhang, Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9, 40 (2008).
Roy, A., Yang, J. & Zhang, Y. COFACTOR: an accurate comparative algorithm for structure-based protein function annotation. Nucleic Acids Res. 40, W471–477 (2012).
Rose, P. W. et al. The RCSB Protein Data Bank: redesigned web site and web services. Nucleic Acids Res. 39, D392–401 (2011).
Wu, S. & Zhang, Y. LOMETS: a local meta-threading-server for protein structure prediction. Nucleic Acids Res. 35, 3375–3382 (2007).
Zhang, Y. & Skolnick, J. SPICKER: a clustering approach to identify near-native protein folds. J. Comput. Chem. 25, 865–871 (2004).
Zhang, Y. & Skolnick, J. TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 33, 2302–2309 (2005).
Zhang, J., Liang, Y. & Zhang, Y. Atomic-level protein structure refinement using fragment-guided molecular dynamics conformation sampling. Structure 19, 1784–1795 (2011).
Roy, A. & Zhang, Y. Recognizing protein-ligand binding sites by global structural alignment and local geometry refinement. Structure 20, 987–997 (2012).
Yang, J., Roy, A. & Zhang, Y. BioLiP: a semi-manually curated database for biologically relevant ligand-protein interactions. Nucleic Acids Res. 41, D1096–1103 (2013).
Almine, J. F. et al. Elastin sequences trigger transient proinflammatory responses by human dermal fibroblasts. FASEB J. 27, 3455–3465 (2013).
Sultana, Z. et al. Dynamic modeling of alpha-synuclein aggregation in dopaminergic neuronal system indicates points of neuroprotective intervention: experimental validation with implications for Parkinson's therapy. Neuroscience 199, 303–317 (2011).
Vali, S., Pallavi, R., Kapoor, S. & Tatu, U. Virtual prototyping study shows increased ATPase activity of Hsp90 to be the key determinant of cancer phenotype. Syst. Synth. Biol. 4, 25–33 (2010).
Equils, O. et al. A computer simulation of progesterone and Cox2 inhibitor treatment for preterm labor. PLoS ONE 5, e8502 (2010).
Rajendran, P. et al. Suppression of signal transducer and activator of transcription 3 activation by butein inhibits growth of human hepatocellular carcinoma in vivo. Clin. Cancer Res. 17, 1425–1439 (2011).
Ramachandran, L. et al. Isorhamnetin inhibits proliferation and invasion and induces apoptosis through the modulation of peroxisome proliferator-activated receptor gamma activation pathway in gastric cancer. J. Biol. Chem. 287, 38028–38040 (2012).
Acknowledgements
This work was supported by NIH NIDCR grant R01 DE014390.
Author information
Authors and Affiliations
Contributions
In this study, we assessed the interaction of histatin 5 with immobilized HagB (SPR spectroscopy-based biosensor, A.M.); determined the histatin 5, HBD1 and HBD3 binding sites on HagB (HagB protein microarray, J.V.H., E.N.R. and K.A.B.); modeled the preliminary histatin 5, HBD1 and HBD3 binding domains on HagB (TASSER imaging, J.Y. and Y.Z.); assessed the histatin 5 attenuation of HagB-induced chemokine and cytokine response in human dendritic cells (Luminex 100 IS, D.S.B., C.L.F., E.N.R., A.P.F., D.D. and K.A.B.); and predicted the pathways or transcription factors altered by histatin 5 (P.R.N., R.V., S.C. and S.V.). J.E.C. oversaw the statistical analysis and D.S.B. and K.A.B. wrote the main manuscript.
Ethics declarations
Competing interests
D.S.B., J.R.V.H., J.Y., C.L.F., E.N.R., A.P.F., D.D., J.E.C., Y.Z. and K.A.B. declare no competing financial interests in the findings of this study or with Cellworks Group, Inc., Saratoga, CA; Cellworks Research India Pvt Ltd.; or with Sensı´Q Technologies Inc., 800 Research Parkway, Suite 100, Oklahoma City,OK 73104, USA. A.M. works for Sensı´Q Technologies Inc., 800 Research Parkway, Suite 100, Oklahoma City, OK 73104, USA. P.R.N., R.V. and S.C. work for Cellworks Research India Pvt Ltd., Bangalore, India and S.V. works for Cellworks Group, Inc., Saratoga, CA.
Electronic supplementary material
Supplementary Information
Supplemental Figure 1
Supplementary Information
Supplemental Fig 1a
Supplementary Information
Supplemental Fig 1b
Supplementary Information
Supplemental Fig 1c
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Borgwardt, D., Martin, A., Van Hemert, J. et al. Histatin 5 binds to Porphyromonas gingivalis hemagglutinin B (HagB) and alters HagB-induced chemokine responses. Sci Rep 4, 3904 (2014). https://doi.org/10.1038/srep03904
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep03904
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.