Abstract
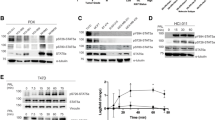
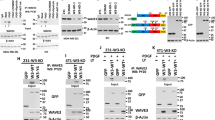
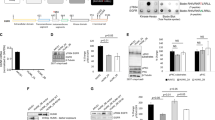
Breast tumor kinase (Brk) is a non-receptor tyrosine kinase distantly related to the Src family kinase. It is expressed in more than 60% of breast tumors, but the biological role of this kinase remains to be determined. Only a limited number of substates have been identified for Brk, and the link of Brk to tumorigenesis remains largely unknown. In this study, we provide evidence that the signal transducer and activator of transcription 3, STAT3, is a physiological target of Brk. Activation of STAT3 previously has been linked to oncogenesis, and results in this study demonstrate that STAT3 is tyrosine phosphorylated and transcriptionally activated in cells expressing endogenous Brk. Signal transducer and activator of transcription 3 is specifically targeted since other STAT members are not responsive to Brk expression. Signal transducer and activator of transcription 3 activation requires the catalytic activity of Brk, and expression of both STAT3 and Brk stimulate cellular proliferation. In addition, we have identified a negative regulator of Brk, the suppressor of cytokine signaling, SOCS3. The SOCS3 protein is known to block signaling mediated by cytokine receptors, and here we find that SOCS3 is able to repress the activity of the Brk non-receptor tyrosine kinase.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Akira S . (1999). Stem Cell 17: 138–146.
Banninger G, Reich NC . (2004). J Biol Chem 279: 39199–39206.
Barker KT, Jackson LE, Crompton MR . (1997). Oncogene 15: 799–805.
Benekli M, Baer MR, Baumann H, Wetzler M . (2003). Blood 101: 2940–2954.
Bowman T, Garcia R, Turkson J, Jove R . (2000). Oncogene 19: 2474–2488.
Bromberg J . (2002). J Clin Invest 109: 1139–1142.
Bromberg JF, Wrzeszczynska MH, Devgan G, Zhao Y, Pestell RG, Albanese C et al. (1999). Cell 98: 295–303.
Chen HY, Shen CH, Tsai YT, Lin FC, Huang YP, Chen RH . (2004). Mol Cell Biol 24: 10558–10572.
Chiarle R, Simmons WJ, Cai H, Dhall G, Zamo A, Raz R et al. (2005). Nat Med 11: 623–629.
Coppo P, Dusanter-Fourt I, Millot G, Nogueira MM, Dugray A, Bonnet ML et al. (2003). Oncogene 22: 4102–4110.
Coyle JH, Guzik BW, Bor YC, Jin L, Eisner-Smerage L, Taylor SJ et al. (2003). Mol Cell Biol 23: 92–103.
Darnell Jr JE . (1997). Science 277: 1630–1635.
Darnell Jr JE, Kerr IM, Stark GR . (1994). Science 264: 1415–1421.
Decker T . (1999). Cell Mol Life Sci 55: 1505–1508.
Derry JJ, Prins GS, Ray V, Tyner AL . (2003). Oncogene 22: 4212–4220.
Derry JJ, Richard S, Valderrama Carvajal H, Ye X, Vasioukhin V, Cochrane AW et al. (2000). Mol Cell Biol 20: 6114–6126.
Easty DJ, Mitchell PJ, Patel K, Florenes VA, Spritz RA, Bennett DC . (1997). Int J Cancer 71: 1061–1065.
Endo TA, Masuhara M, Yokouchi M, Suzuki R, Sakamoto H, Mitsui K et al. (1997). Nature 387: 921–924.
Faruqi TR, Gomez D, Bustelo XR, Bar-Sagi D, Reich NC . (2001). Proc Natl Acad Sci USA 98: 9014–9019.
Garcia R, Bowman TL, Niu G, Yu H, Minton S, Muro-Cacho CA et al. (2001). Oncogene 20: 2499–2513.
Haegebarth A, Heap D, Bie W, Derry JJ, Richard S, Tyner AL . (2004). J Biol Chem 279: 54398–54404.
Harvey AJ, Crompton MR . (2003). Oncogene 22: 5006–5010.
Hennipman A, van Oirschot BA, Smits J, Rijksen G, Staal GE . (1989). Cancer Res 49: 516–521.
Ihle JN . (2001). Curr Opin Cell Biol 13: 211–217.
Kamalati T, Jolin HE, Fry MJ, Crompton MR . (2000). Oncogene 19: 5471–5476.
Kamalati T, Jolin HE, Mitchell PJ, Barker KT, Jackson LE, Dean CJ et al. (1996). J Biol Chem 271: 30956–30963.
Kang KN, Kim M, Pae KM, Lee ST . (2002). Biochim Biophys Acta 1574: 365–369.
Kim H, Lee ST . (2005). J Biol Chem 280: 28973–28980.
Krebs DL, Hilton DJ . (2000). J Cell Sci 113 (Part 16): 2813–2819.
Lacronique V, Boureux A, Monni R, Dumon S, Mauchauffe M, Mayeux P et al. (2000). Blood 95: 2076–2083.
Leong PL, Andrews GA, Johnson DE, Dyer KF, Xi S, Mai JC et al. (2003). Proc Natl Acad Sci USA 100: 4138–4143.
Levy DE, Darnell Jr JE . (2002). Nat Rev Mol Cell Biol 3: 651–662.
Liu L, McBride KM, Reich NC . (2005). Proc Natl Acad Sci USA 102: 8150–8155.
Llor X, Serfas MS, Bie W, Vasioukhin V, Polonskaia M, Derry J et al. (1999). Clin Cancer Res 5: 1767–1777.
McBride KM, McDonald C, Reich NC . (2000). EMBO J 19: 6196–6206.
Minoguchi M, Minoguchi S, Aki D, Joo A, Yamamoto T, Yumioka T et al. (2003). J Biol Chem 278: 11182–11189.
Mitchell PJ, Barker KT, Martindale JE, Kamalati T, Lowe PN, Page MJ et al. (1994). Oncogene 9: 2383–2390.
Mitchell PJ, Barker KT, Shipley J, Crompton MR . (1997). Oncogene 15: 1497–1502.
Mitchell PJ, Sara EA, Crompton MR . (2000). Oncogene 19: 4273–4282.
Mossman T . (1983). J Immunol Methods 65: 55–63.
Naka T, Narazaki M, Hirata M, Matsumoto T, Minamoto S, Aono A et al. (1997). Nature 387: 924–929.
Nicholson SE, Willson TA, Farley A, Starr R, Zhang JG, Baca M et al. (1999). EMBO J 18: 375–385.
Niu G, Bowman T, Huang M, Shivers S, Reintgen D, Daud A et al. (2002). Oncogene 21: 7001–7010.
Olayioye MA, Beuvink I, Horsch K, Daly JM, Hynes NE . (1999). J Biol Chem 274: 17209–17218.
Qiu H, Miller WT . (2002). J Biol Chem 277: 34634–34641.
Qiu H, Miller WT . (2004). Oncogene 23: 2216–2223.
Qiu H, Zappacosta F, Su W, Annan RS, Miller WT . (2005). Oncogene 24: 5656–5664.
Sasaki A, Yasukawa H, Shouda T, Kitamura T, Dikic I, Yoshimura A . (2000). J Biol Chem 275: 29338–29347.
Serfas MS, Tyner AL . (2003). Oncol Res 13: 409–419.
Siyanova EY, Serfas MS, Mazo IA, Tyner AL . (1994). Oncogene 9: 2053–2057.
Starr R, Willson TA, Viney EM, Murray LJ, Rayner JR, Jenkins BJ et al. (1997). Nature 387: 917–921.
Takeda K, Noguchi K, Shi W, Tanaka T, Matsumoto M, Yoshida N et al. (1997). Proc Natl Acad Sci USA 94: 3801–3804.
van den Brule FA, Engel J, Stetler-Stevenson WG, Liu FT, Sobel ME, Castronovo V . (1992). Int J Cancer 52: 653–657.
Vasioukhin V, Serfas MS, Siyanova EY, Polonskaia M, Costigan VJ, Liu B et al. (1995). Oncogene 10: 349–357.
Yu CL, Meyer DJ, Campbell GS, Larner AC, Carter-Su C, Schwartz J et al. (1995). Science 269: 81–83.
Acknowledgements
We thank all the members of the NCR laboratory, especially the assistance of Sarah Van Scoy. We thank our colleagues Dafna Bar-Sagi, Michael Hayman, and Patrick Hearing for their advice along the way. These studies were supported by NIH grants (RO1CA50773) (PPGCA28146), and in part by grants from the Carol Baldwin Breast Cancer Research Program to NCR, and the Italian Cancer Research Association to VP.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Liu, L., Gao, Y., Qiu, H. et al. Identification of STAT3 as a specific substrate of breast tumor kinase. Oncogene 25, 4904–4912 (2006). https://doi.org/10.1038/sj.onc.1209501
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.onc.1209501
Keywords
This article is cited by
-
PTK6 inhibits autophagy to promote uveal melanoma tumorigenesis by binding to SOCS3 and regulating mTOR phosphorylation
Cell Death & Disease (2023)
-
Gene prioritization, communality analysis, networking and metabolic integrated pathway to better understand breast cancer pathogenesis
Scientific Reports (2018)
-
Protein tyrosine kinase 6 promotes ERBB2-induced mammary gland tumorigenesis in the mouse
Cell Death & Disease (2015)
-
Protein Tyrosine Kinase 6 Regulates UVB-Induced Signaling and Tumorigenesis in Mouse Skin
Journal of Investigative Dermatology (2015)
-
The impact of Cysteine-Rich Intestinal Protein 1 (CRIP1) in human breast cancer
Molecular Cancer (2013)