Abstract
A survey of allozyme variation at 17 loci in 14 populations representing four taxonomically problematic Gran Canarian pine forest endemic taxa of Lotus (L. genistoides, L. holosericeus, L. spartioides and some taxonomically uncertain populations collected under the designation Lotus sp.) was conducted to examine their diversification and systematic relationships. All groups exhibited high values of genetic variation, although inbreeding was common within populations. Considerable among-population genetic homogeneity was detected, as inferred from low values of Gst within each of the groups. The high population sizes of these taxa and a lack of evidence for isolation by distance or genetic bottlenecks indicate that diversity has accumulated over a long period of environmental stability. The association of high genetic distances with low linear distances, and the substantial increase in the values of Gst when the taxa considered were merged in different combinations hint at an incipient (yet probably taxonomically insufficient) reproductive isolation. The genetic similarity between L. genistoides, L. holosericeus and L. spartioides, together with the different behaviour of the populations collected under the designation Lotus sp., may have important implications for the restructuring of the taxonomy of this group when the ongoing morphological studies are completed.
Similar content being viewed by others
Introduction
Lotus L. (Fabaceae: Loteae) is a cosmopolitan genus whose Canarian circumscription consists of ca. 24 species, of which 18 are endemic to this oceanic archipelago according to Bramwell (2002), and Acebes Ginovés et al (2001). The Canarian species of Lotus can be ascribed to one of two sections: Heinekenia and Pedrosia. While section Heinekenia is exclusively Canarian, the geographic distribution of section Pedrosia encompasses all Macaronesian islands and the South of Morocco (Sunding, 1979), with L. arenarius Brot. also distributed in the Iberian Peninsula (Talavera et al, 2000).
Gran Canaria hosts 12 species of Lotus. Six of these are endemic to the island, one is endemic to Macaronesia, two are native and three are introduced (Hansen and Sunding, 1993). All six of the exclusively Gran Canarian endemics belong to section Pedrosia, and are listed in the Red List of Spanish Vascular Flora (VV AA, 2000) as critically endangered (L. arinagensis Bramwell, L. kunkelii (Esteve) Bramwell & Davis and L. genistoides Webb ex Pit & Proust), endangered (L. callis-viridis Bramwell and Davis), vulnerable (L. spartioides Webb & Berthel) and minor risk (L. holosericeus Webb & Berthel.). The former five taxa are also listed as prioritary in the Atlas of endangered Spanish Vascular Flora (Bañares et al, 2003).
Although some of these six species are easily recognizable, the taxonomic distinction among L. holosericeus, L. genistoides and L. spartioides is not based on morphologically sound characters and raises considerable controversy. The lack of consensus to define operative biological units in these three Gran Canarian Lotus hinders the study of their diversification, and thus makes it difficult to develop a conservation strategy.
Some of the difficulties stem from the fact that the ecological distributions of these taxa overlap in the pine forests of the Gran Canarian summits (Figure 1). Although the name L. genistoides is used locally to refer to the populations of Lotus occurring in Tirajana (Webb ex Pitard and Proust, 1908), there is no formal description of this taxon, and the material used to obtain its chromosome number was collected in Guayadeque (Aldridge and Ortega, 1976), a 5 km long ravine running from the centre of the island to its eastern coast. However, recent surveys of this ravine have failed to find a single specimen of the taxon. L. spartioides is considered as typical from the Pine forest of Tamadaba (in the North West of Gran Canaria) (Sunding, 1972), but the material of the original description (Webb and Berthelot, 1836–50) was collected in the area of the Caldera de Tirajana (in the South of Gran Canaria), without any further specification. There are also a considerable number of cases where material collected in the same area by the same collector has received different names, confusing L. spartioides and L. genistoides (Ortega, 1976,1979, respectively) and L. spartioides and L. holosericeus (Kunkel, 1969,1977, respectively). The taxonomic uncertainty in these Gran Canarian endemic species of Lotus indirectly suggests that a survey with molecular markers might help improve understanding of their diversification and systematic relationships.
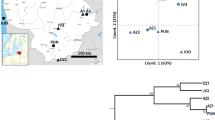
Locations of the 14 populations of Gran Canarian Lotus surveyed. Numerical codes are those presented in Table 1.
Allozyme polymorphisms have been frequently used as neutral molecular markers to study levels and structuring of population genetic variation within species (see Hamrick and Godt (1989) for a comprehensive review), including several species of Lotus (Realson and Grant, 1988,1989; Gauthier et al, 1998). Here, we use them to clarify the relationships among taxa within the complex of Gran Canarian Lotus.
Materials and methods
Plant material
L. spartioides is locally common in the Tamadaba pine forest, North West of Gran Canaria (Bramwell and Bramwell, 2001). It is a small subshrub with yellow flowers, small procumbent or suberect branches and linear leaflets of 5–15 mm in length that are covered with short whitish hairs. It is very frequent between 1000 and 1400 m and shows a high degree of variation in flower size and width, leaf shape and degree of hairiness.
L. holosericeus is widely distributed in the central part of the island of Gran Canaria. It is similar to L. spartioides, but it possesses long silky hairs that cover all of its surface. It inhabits the clearings and the shrubs in the area of Canarian Pine forests in several ravines.
L. genistoides is one of the less known species of Gran Canarian Lotus, with a single locality in the Caldera de Tirajana, a huge volcanic crater in the south of Gran Canaria (Bramwell and Bramwell, 2001). L. genistoides is woody and more robust than the other two taxa, with a smaller flower size.
Available cytogenetic data indicate that all Lotus taxa from Gran Canaria are 2n=14 (Aldridge and Ortega, 1976; Ortega, 1976).
Sampling
We sampled a total of 600 individuals from 14 populations representing the distribution of L. genistoides (one population), L. holosericeus (six populations), L. spartioides (two populations) and five taxonomically undetermined populations that we will refer to as Lotus sp. (Figure 1, Table 1). Since most target populations occupy a large area, each sampling was preceded by a visual survey to provide an idea of population size and area. Based on these surveys, sampling was carried out along transects that subdivided the area of a population into several equally spaced points. Five to 10 individuals (depending on abundance) were sampled from each point. This procedure aimed to provide a representative sample of individuals throughout a population. Sample sizes are strictly related to the size of target populations and ranged from 10 in population LHTE to 100 in population LSTA (Table 1). Leaf samples from individual plants were deposited in numbered zip-lock plastic bags that were kept refrigerated in a portable cooler until they were stored in −80°C freezers at the Jardín Botánico Canario Viera y Clavijo (JBCVC). Samples remained there until used for extract preparation.
Electrophoretic analyses
For each sample, about 0.1 g of frozen leaves per individual were ground in a mortar using an extraction buffer following Shields et al (1983). The resulting protein extract was adsorbed on to a 4 mm × 6 mm Whatman No. 3 chromatography paper wick and stored at −80°C until electrophoresed.
Horizontal starch-gel electrophoresis was carried out in 12% w/v gels using two different gel/electrode buffer systems (Shields et al, 1983) that allowed examination of eight enzyme systems. Histidine pH 7.0 (System E in Shields et al, 1983) was used to examine phosphoglucomutase (PGM, E.C. 5.4.2.2), phosphoglucose isomerase (PGI, E.C. 5.3.1.9), isocitrate dehydrogenase (IDH, E.C.1.1.1.42) and esterase (EST, 3.1.1.1). Morpholine-Citrate pH 6.1 (Clayton and Tretiak, 1972) was used to examine phosphogluconate dehydrogenase (6PGD, E.C.1.1.1.44), shikimate dehydrogenase (SKD, E.C.1.1.1.25), malate dehydrogenase (MDH, E.C.1.1.1.37) and malic enzyme (ME, E.C.1.1.1.40).
For each enzyme, gene loci and alleles were inferred and labelled following numerical and alphabetical sequence, respectively. Intrapopulation, interpopulation and interspecific verifications of enzyme mobilities were determined through side-by-side comparisons of allelic variants on the same gel.
Data analysis
Elementary descriptors of isozyme variation (number of alleles per locus, percentage of polymorphic loci, observed and expected heterozygosity), and genetic distances and identities (Nei, 1978) were calculated using BIOSYS-1 Version 1.7 (Swofford and Selander, 1989). All calculations were made at the species and population levels from genotype data corresponding to each locus. The effective number of alleles (Ae), estimates of interpopulation gene flow and Ewens–Watterson neutrality tests were calculated using Popgene version 1.32 (1997).
Nei's (1973) and Wright's (1951) population-structure statistics were calculated over all loci for L. holosericeus, L. spartioides, Lotus sp. and for all populations (regardless of taxonomic differences) using GeneStat-PC 3.31 (Lewis and Whitkus, 1993) and BIOSYS-1 Version 1.7 (Swofford and Selander, 1989), respectively.
We applied a sign test for heterozygosity excess (Cornuet and Luikart, 1996) to detect whether populations had experienced recent historical bottlenecks. This test compares expected heterozygosity (He) under Hardy–Weinberg equilibrium expectations to the heterozygosity expected at mutation-drift equilibrium (Heq) in a sample that has the same size and the same number of alleles as the sample used to measure He (Luikart and Cornuet, 1998). The rationale for this test is that, since low frequency alleles are lost at a much faster rate than heterozygosity in a bottleneck situation, bottlenecked populations are expected to have a heterozygote excess. Calculations were made based on allele frequency data under the Stepwise Mutation Model (SMM) and the Independent Allele Model (IAM) using the program Bottleneck-PC (Piry et al, 1998).
The relationship between geographic location and genetic makeup of all populations sampled was evaluated by Mantel (1967) tests carried out using the computer program NTSYS-pc version 2.02j (Rohlf, 1998).
Results
Genetic interpretation of the eight enzymes examined allowed the resolution of 17 putative loci, none of which was monomorphic over all populations surveyed. Of the 70 alleles detected, populations LGCJ (L. genistoides), LSTA (L. spartioides) and LSPCM and LSPPN (Lotus sp) possessed one exclusive allele each (Mdh-4c, 6Pgd-4d, Pgm-1g and Mdh-1e, respectively). There were also exclusive allele sharings between the population pairs LGCJ-LSPPN (Est-3e), LSPCM-LSPPA (Idh-1d), LHAR-LHCU (6Pgd-4a), LHAR-LSTA (Pgi-1b), LSPCM-LSPMA (Pgm-2e) and LHRO-LSPCM (Skd-2a). The remaining 60 alleles were shared by different combinations of the 14 populations surveyed. No taxon-diagnostic alleles were found (ie, alleles monomorphic in one taxon and not shared by the other taxa). Also, there were no alleles that were present exclusively in all populations of one taxon (the table of allele frequencies is available upon request).
The basic indicators of polymorphism (Table 2) showed that levels of genetic variation are very similar in each taxon. L. holosericeus contained the maximum number of alleles per locus within a population (Al=2.8 in population LHAR) and the maximum proportion of polymorphic loci (P=76.5 in population LHPI), but also the minimum values of these parameters (Al=1.6 and P=41.2 in LHCU). The values of observed heterozygosity ranged from Ho=0.067 in population LHAR (L. holosericeus) to Ho=0.138 in populations LHTE (L. holosericeus) and LSPMA (Lotus sp.). Expected heterozygosity (He) spanned from He=0.115 (population LHCU) to He=0.204 (population LHPI), both in L. holosericeus. The inbreeding coefficient displayed considerable variation across loci and its average values ranged from Fis=0.294 in L. spartioides to Fis=0.543 in L. holosericeus (Table 3). These values are all much higher than zero (the value that would correspond to a situation of random mating), and indicate a predominance of selfing in the populations surveyed. Consistent with these results, only five out of the 17 loci were in Hardy–Weinberg proportions in L. spartioides, two in Lotus sp. and one in L. holosericeus and L. genistoides (Table 3). None of these populations showed evidence of a recent bottleneck (Table 2), and all loci could be considered neutral according to Ewens–Watterson tests (Table 3).
The proportion of variation that is explained by the differentiation among populations (Table 3) was uniformly low in L. spartioides (average Gst=0.027) and L. holosericeus (average Gst=0.089). By contrast, Lotus sp. exhibited a much higher interpopulation differentiation, with an average value of Gst=0.282.
The values of Gst increased sharply when the assemblages were considered in different combinations (Gst=0.290 for all populations, Gst=0.293 for all populations save for L. genistoides, Gst=0.277 for L. spartioides plus Lotus sp. and Gst=0.159 for L. holosericeus plus Lotus sp.).
Average Nei's (1978) genetic distances were D=0.014 within L. holosericeus, D=0.008 within L. spartioides and D=0.112 within Lotus sp. Lotus sp. was the more genetically heterogeneous assemblage, with a maximum distance value of D=0.203 (Table 4), more than five-fold the maximum genetic distance within L. holosericeus (D=0.033) and within L. spartioides (D=0.008). The highest genetic distance (D=0.245) was detected between one population of L. holosericeus and one of Lotus sp. (LHTE-LSPMA) and the lowest (D=0.002) within L. holosericeus (LHRO-LHPP). Average genetic distances between these assemblages were also relatively low, with a maximum value of D=0.144 between L. genistoides and Lotus sp. and a minimum of D=0.009 between L. holosericeus and L. genistoides.
The UPGMA dendrogram constructed from Nei's (1978) genetic distances (Figure 2) resolved three consistent groups containing: (1) L. holosericeus populations, one population of Lotus sp. (LSPPA) and the population of L. genistoides; (2) L. spartioides populations and one population of Lotus sp. (LSPNU) and (3) the remaining three populations that we assigned to Lotus sp.
UPGMA dendrogram based on Nei's (1978) distance values between pairwise combinations of the 14 populations surveyed. Symbols correspond to Figure 1.
Mantel tests revealed no significant relationship between genetic and geographic distances within L. holosericeus (r=0.187, P=0.241), within Lotus sp. (r=0.539, P=0.102) or in all the populations sampled (r=0.115, P=0.107).
Discussion
General levels of genetic variation
Values for genetic polymorphism in these Gran Canarian endemic Lotus species are about two-fold higher than the average values reported by Hamrick and Godt (1989) for endemic plants (A=1.39, P=0.26, He=0.063). Furthermore, they are only slightly lower than those detected by Gauthier et al (1998) in the mainland diploid species L. alpinus (Schleich ex DC) Ramond (averages A=2.8, Ho=0.219, He=0.279). The degree of genetic variation within these taxa, as measured by the average population diversity (Hs, Table 3), is in all cases much higher than the averages published for plants of Hawaii Islands (Hs=0.064, DeJoode and Wendel, 1992) or the Juan Fernández Islands (Hs=0.042, Crawford et al, 2001) and slightly higher than the average value published for Canarian taxa (Hs=0.137, Francisco-Ortega et al, 2000). It is clear that populations of Gran Canarian endemic Lotus maintain substantial levels of genetic variation, thereby adding to the emerging picture of more variation in Gran Canarian endemics relative to those from other oceanic archipelagos (Francisco-Ortega et al, 2000). The high population sizes of these taxa and lack of evidence for genetic bottlenecks (Table 2) suggest that genetic variation has increased in a context of environmental stability. Lack of continental populations of section Pedrosia in our survey prevent us from knowing whether the levels of variation of these Gran Canarian populations are quantitatively and/or qualitatively different from their closely related mainland congeners.
Since high Fis values are estimated in most populations (Table 3), this variation appears to be maintained despite an overall predominance of inbreeding in the populations surveyed. When the seeds of Lotus have ripened, they detach by their own weight and stay near the parent plant (gravity dispersal or barochory). Barochory fosters small genetic neighbourhoods, where reproduction takes place between related individuals. The Fis values estimated for these Gran Canarian Lotus are much higher than those obtained in the Californian L. scoparius (Nutt.) Ottley, where Montalvo, Clegg and Ellstrand (unpublished data) report low inbreeding in all populations examined based on a study of 14 allozyme loci. The high Fis values in these Canarian Lotus may therefore stem from populations being divided into different mating areas leading to a deficit in heterozygotes in the pooled population due to the Wahlund effect.
However, L. spartioides and L. holosericeus exhibit a considerable genetic homogeneity over populations, which hints at an abundant interpopulation gene flow and manifests qualitatively in the fact that exclusive alleles are very infrequent and scattered throughout the populations. Quantitatively, levels of interpopulation differentiation as estimated by Gst and Fst (Table 3) are quite low in these taxa. Likewise, Gauthier et al (1998) detect a remarkable genetic cohesion in diploid populations of L. alpinus (Gst=0.030).
By contrast, the average value of Gst=0.282 in Lotus sp. suggests a high degree of genetic heterogeneity in this assemblage. Mantel tests for isolation by distance between populations were not significant. In fact, some of the highest genetic distance values (Table 4) were detected between closely located populations. Therefore, sharp geographic discontinuities (eg, deep ravines and cliffs) may be important causes of genetic differentiation between some populations, especially in Lotus sp.
Except for some populations of Lotus sp, the average estimated number of migrants per generation is high within the three assemblages (data not shown), ranking far above the theoretical threshold of one migrant per generation assumed to prevent independent population evolution according to Slatkin (1985),(1987),(1994). These population assemblages could therefore be regarded as single reproductive units that have maintained high levels of variation through abundant genetic interchange. However, the greater genetic distances detected within Lotus sp. (see Table 4) suggest that this assemblage may either contain taxonomically distinct entities or is in a more advanced stage of differentiation.
Taxonomic implications
Since the relationship between phenotype and genotype is in general simpler for isozyme evidence than for morphological characters (Gottlieb, 1977), the genetic differences detected might be useful in assessing the taxonomic congruence in these Gran Canarian Lotus.
The existence of different taxa is in the first place a question of reproductive isolation. Therefore, if the population assemblages considered for these Gran Canarian endemic Lotus are indeed different taxonomic entities, then we should expect reduced levels of gene flow among them. In agreement with this prediction, the low values of Gst within L. holosericeus and L. spartioides are substantially increased when assemblages are compared. This finding indicates incipient reproductive isolation. Thus, the pertinent question to address is whether this degree of genetic heterogeneity is enough to sustain the ascription of these population groups to different taxonomic categories.
The average genetic identities within L. holosericeus (I=0.986), L. spartioides (I=0.992) and Lotus sp. (I=0.896) are within the range reported by Gottlieb (1977) for con-specific populations (I=0.95±0.2), as are those between pairwise combinations of L. holosericeus, L. genistoides and L. spartioides (Table 5). Remarkably, populations LSPPA and LSPNU have higher genetic identities with populations of L. spartioides or L. holosericeus than with other populations of Lotus sp. Removing these two populations from the calculations results in a further decrease of the values (Table 5) to a region between Gottlieb's (1977) averages for con-specific populations (I=0.95±0.2) and congeneric species (I=0.67±0.07).
Overall, our results suggest that L. genistoides L. holosericeus and L. spartioides are very close genetically, and that Lotus sp. is a heterogeneous assemblage that contains two populations (LSPNU and LSPPA) much closer to L. holosericeus and L. spartioides than to other populations designated as Lotus sp. A joint consideration of these data with ongoing morphological analyses (Oliva et al, in prep.) will allow us to determine whether the triad L. holosericeus, L. spartioides and L. genistoides should be considered a single taxonomic entity and whether populations LSPCM, LSPMA, LSPPN within the assemblage that we designated Lotus sp. are taxonomically distinct.
References
Acebes Ginovés JR, Del Arco Aguilar M, García Gallo A, León Arencibia MC, Pérez De Paz PL, Rodríguez Delgado O et al (2001). Pteridophyta & spermatophyta. In: Izquierdo I, Martín JL, Zurita N, Arechavaleta M (eds) Lista de especies silvestres de Canarias (hongos, plantas y animales terrestres). Consejería de Política Territorial y Medio Ambiente: Gobierno de Canarias, pp 98–140.
Aldridge A, Ortega J (1976). Estudios en la flora macaronésica. Bot Macaronésica 2: 9–18.
Bañares A, Blanca G, Güemes J, Moreno JC, Ortiz S (Eds.) (2003). Atlas y Libro Rojo De La Flora Vascular Amenazada De España. Dirección General de Conservación de la Naturaleza: Madrid.
Bramwell D (2002). Lotus: pico de paloma, corazoncillos. Aguayro 228: 42–45.
Bramwell D, Bramwell ZI (2001). Flores Silvestres de las Islas Canarias, 4th edn. Editorial Rueda: Madrid.
Clayton JW, Tretiak DN (1972). Amine citrate buffer for pH control in starch gel electrophoresis. J Fish Res Board Can 29: 1169–1172.
Cornuet JM, Luikart G (1996). Description and evaluation of two tests for detecting recent bottlenecks. Genetics 144: 2001–2014.
Crawford DJ, Ruiz E, Stuessy TF, Tepe E, Aqeveque P, Gonzalez F et al (2001). Allozyme diversity in endemic flowering plant species of the Juan Fernandez Archipelago, Chile: ecological and historical factors with implications for conservation. Am J Bot 88: 2195–2203.
DeJoode DR, Wendel JF (1992). Genetic diversity and origin of the Hawaiian Island cotton, Gossypium tomentosum. Am J Bot 79: 1311–1319.
Francisco-Ortega J, Santos-Guerra A, Kim S-C, Crawford DJ (2000). Plant genetic diversity in Canary Islands: a conservation perspective. Am J Bot 87: 909–919.
Gauthier P, Lumaret R, Bédécarrats A (1998). Genetic variation and gene flow in alpine diploid and tetraploid populations of Lotus (L. alpinus (D.C.) Schleicher/L. corniculatus L.): I. Insights from morphological and allozyme markers. Heredity 80: 683–693.
Gottlieb LD (1977). Electrophoresis evidence and plant systematics. Ann Missouri Bot Gard 64: 161–180.
Hamrick JL, Godt MJW (1989). Allozyme diversity in plant species. In: Brown AHD, Clegg MT, Kahler AL, Weir BS (eds) Plant Population Genetics, Breeding and Germplasm Resources. Sinauer Assosiates Inc.: Sunderland, MA. pp 43–63.
Hansen A, Sunding P (1993). Flora of Macaronesia. Checklist of vascular plants. 4th rev. edn. Sommerfeltia 17: 1–295.
Kunkel G (1969). Aliens to the Canary Flora. Cuadernos Bot Canaria 5: 27–44.
Kunkel G (1977). Endemismos canarios. Inventerio de las plantas vasculares endémicas en la provincia de Las Palmas. ICONA Monografías 15, 436pp.
Lewis PO, Whitkus R (1993). GeneStat-PC version 3.3. North Carolina State University: Raleigh, North Carolina.
Luikart G, Cornuet JM (1998). Empirical evaluation of a test for identifying recently bottlenecked populations from allele frequency data. Conserv Biol 12: 228–237.
Mantel N (1967). The detection of disease clustering and a generalized regression approach. Cancer Res 27: 209–220.
Nei M (1973). Analysis of gene diversity in subdivided populations. Proc Natl Acad Sci USA 70: 3321–3323.
Nei M (1978). Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89: 583–590.
Ortega J (1976). Citogenética del género Lotus en Macaronesia. Bot Macaronésica 1: 17–24.
Ortega J (1979). Citogenética del Género Lotus en Macaronesia III. Variación en el contenido de glucósidos cianogénicos en Lotus de las Islas Canarias y en Macaronesia. Bot Macaronésica 5: 9–19.
Piry S, Luikart G, Cornuet JM (1998). Bottleneck, a program for detecting recent effective population size reductions from allele frequency data. INRA, URLB, Laboratoire de Modélisation et Biologie Evolutive. Montpellier, France.
Pitard J, Proust L (1908). Les Iles Canaries. Flore de ĹArchipiel: Paris.
Popgene Version 1.32 (1997). Ag/For Molecular Biology and Biotechnology Centre (University of Alberta) and Center for International Forestry Research. Alberta, Canada.
Realson J, Grant WF (1988). Evaluation of hypotheses concerning the origin of Lotus corniculatus (Fabaceae) using isozyme data. Theor Appl Genet 76: 267–276.
Realson J, Grant WF (1989). An isozyme study in the genus Lotus (Fabaceae). Experimental protocols and genetic basis of the electrophoretic phenotype. Theor Appl Genet 77: 595–607.
Rohlf FJ (1998). NTSYS-pc. Numerical Taxonomy and Multivariate Analysis Version 2.02j. Applied Biostatistics Inc.: Setauket, NY.
Shields CR, Orton TJ, Stuber CW (1983). An outline of general resource needs and procedures for the electrophoretic separation of active enzymes from plant tissue. In: Tanskley SD, Orton TJ (eds) Isozymes in Plant Genetics Breeding, Part A. Elsevier Science Publishing Company Inc.: New York, USA. pp 443–468.
Slatkin M (1985). Gene flow in natural populations. Annu Rev Ecol Systemat 16: 393–430.
Slatkin M (1987). Gene flow and the geographic structure of natural populations. Science 236: 787–792.
Slatkin M (1994). Gene flow and population structure. In: Real LA (ed) Ecological Genetics. Princeton University Press: Princeton, NJ. pp 3–17.
Sunding P (1972). The Vegetation of Gran Canaria – Oslo. I. Mat -Naturv Klasse. Serie. No. 29, 186pp.
Sunding P (1979). Origins of the Macaronesian flora. In: Bramwell D (ed) Plants and Islands. Academic Press: London. pp 13–40.
Swofford DL, Selander RB (1989). BYOSYS-1: a fortran program for the comprehensive analysis of electrophoretic data in population genetics and systematics. J Hered 72: 281–283.
Talavera S, Aedo C, Castroviejo S, Herrero A, Romero-Zarco C, Salgueiro FJ et al (2000). Flora Peninsula Iberica VII (II). Leguminosae (Partim). Real Jardín Botánico: Madrid.
VV AA (2000). Lista Roja de Flora Vascular Española (valoración según categorías UICN). Conservación Vegetal 6(Extra): 11–38.
Webb PB, Berthelot S (1836–50). Histoire Naturelle des Iles Canaries 3. Phytographia Canariensis (2/2):1–496. lam 37–136 B.
Wright S (1951). The genetical structure of populations. Ann Eugen 15: 323–354.
Acknowledgements
We thank the Cabildo Insular de Gran Canaria (CIGC) for a pre-doctoral research grant to Felicia Oliva and for supporting this investigation. The biologists at the Jardín Botánico Canario ‘Viera y Clavijo’ provided helpful suggestions and field assistance, especially Bernardo Navarro, Isabel Santana, Blas Vilches, Olga Fernández-Palacios and Carolina Suárez. Roque M López, environment official at the Gobierno de Canarias, helped us locate some of the populations. This paper was written while Juli Caujapé-Castells (JCC) was contracted by the Ministerio de Ciencia y Tecnología (MCYT) and the CIGC. JCC thanks the MCYT and the CIGC for the cofunding of his Ramón y Cajal Project.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Oliva-Tejera, F., Caujapé-Castells, J., Naranjo-Suárez, J. et al. Population genetic differentiation in taxa of Lotus (Fabaceae: Loteae) endemic to the Gran Canarian pine forest. Heredity 94, 199–206 (2005). https://doi.org/10.1038/sj.hdy.6800598
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.hdy.6800598
Keywords
This article is cited by
-
Population differentiation in relation to conservation: nuclear microsatellite variation in the Canary Island endemic Lotus sessilifolius (Fabaceae)
Conservation Genetics Resources (2018)
-
The intersection of conservation and horticulture: bird-pollinated Lotus species from the Canary Islands (Leguminosae)
Biodiversity and Conservation (2011)
-
General G ST and θ inflation due to biased intra-population sampling, and its consequences for the conservation of the Canarian Flora
Conservation Genetics (2010)
-
Genetic diversity and floral dimorphism in Limonium dendroides (Plumbaginaceae), a woody Canarian species on the way of extinction
Plant Systematics and Evolution (2009)
-
Isozyme diversity in some Canarian woody endemisms of the genus Echium L. (Boraginaceae)
Plant Systematics and Evolution (2009)