Abstract
Translocations occur in a proportion of couples affected by recurrent miscarriages. We describe two such families in which the underlying cause was a cryptic subtelomeric 11p;17p translocation detected only after the birth of an affected child carrying an unbalanced form of the rearrangement. Unbalanced subtelomeric rearrangements are now recognised as a significant cause of mental impairment and we believe that these rearrangements may also be an important cause of recurrent miscarriages. In these two families the translocation is most likely to have arisen from a single ancestral event because all translocation carriers shared almost identical haplotypes around the breakpoints on both chromosomes.
Similar content being viewed by others
Introduction
We present two families with a common subtelomeric 11p;17p translocation detected following the live birth of a child with Miller-Dieker syndrome (MDS). Both families had previously presented with recurrent miscarriages but routine cytogenetic analysis failed to identify the subtle rearrangement. In around 2–5% of couples who have had two or more spontaneous abortions, one member of the couple is a translocation carrier,1,23 the translocation being reciprocal in around two-thirds and Robertsonian in one-third of cases.
Recent reports have suggested that unbalanced submicroscopic telomeric rearrangements might represent a significant cause of idiopathic mental retardation (IMR) either with or without associated phenotypic abnormalities,4,5,6,7,8,9,1011 although this has not been found in all studies.12 In around 50% of reported cases the translocation is familial, the balanced form being carried by a parent and possibly other family members. Subtelomeric translocations have also been identified in couples presenting with recurrent miscarriages and affected live born offspring with mental retardation and a variety of other anomalies13,14,15,16,1718 or with a clinically recognisable syndrome such as cri du chat,19 Wolf-Hirschhorn syndrome20 or MDS.21,22,23,2425 Subtelomeric rearrangements may also be common in such families26 and we present here two additional families in which a cryptic rearrangement is associated with multiple pregnancy losses.
Case reports
Family 1
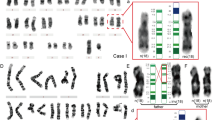
MW was the first child born to healthy, unrelated parents. The parents had previously been investigated cytogenetically, with normal results, because of two spontaneous miscarriages at 8 weeks gestation. Pregnancy was uneventful up to 30 weeks gestation when polyhydramnios was noted. Prenatal cytogenetic analysis following amniocentesis showed an apparently normal female karyotype. At 32 weeks gestation ultrasound examination showed dilated lateral ventricles and probable agenesis of the corpus callosum. The patient was born at 34 weeks gestation, weighing 1520 g, following emergency Caesarean section for intra-uterine growth retardation (IUGR), spontaneous rupture of membranes and polyhydramnios. Postnatal examination showed frontal bossing, a prominent nasal bridge, thin upper lip, micrognathia, tapering fingers with camptodactyly, broad big toes and small toe nails. A head ultrasound scan showed agenesis of the corpus callosum and probable lissencephaly. The patient died aged 1 day. Postnatal directed high resolution cytogenetic analysis, following a clinical suggestion of Miller-Dieker syndrome, showed a subtle terminal deletion of the short arm of one chromosome 17 (ISCN 550 band level). FISH with a cosmid from the locus D17S379 confirmed a deletion of the MDS critical region. Although parental karyotypes had previously been reported as normal following referral for recurrent miscarriages (ISCN 550 band level), high resolution analysis detected an apparently balanced maternal translocation between the distal short arms of chromosomes 11 and 17. FISH with the telomeric probe 2209a2 (11p) and the Miller-Dieker critical region probe D17S379 confirmed this translocation (Figure 1), 46,XX,t(11;17)(p15.5;p13.3).ish t(11;17)(2209a2-, D17S379+; D17S379-, 2209a2+). In addition to the proband's mother three further carriers of this balanced translocation were identified in this family using the telomeric FISH probes (Figure 2). All were male including the maternal grandfather and uncle. The grandmother of MW was noted to have had approximately six first trimester miscarriages over 4 years before the birth of MW's mother and uncles. Since the birth of MW, her parents have had five further 5–10 week miscarriages, of which one was karyotyped and confirmed to carry the same malsegregation pattern seen in MW.
Family 2
LT was the first child born to healthy, unrelated parents. The parents had also been investigated cytogenetically with normal results because of four previous spontaneous miscarriages. Prenatal cytogenetic analysis following amniocentesis showed an apparently normal female karyotype. An initial ultrasound scan at 23 weeks gestation showed mild dilation of posterior horns of the cerebral ventricles, but by 33 weeks they were within normal limits. Further scans were performed every 2–3 weeks but showed a gradual decrease in growth velocity. LT was born at 36 weeks by ventouse vaginal delivery. She was noted to have symmetrical IUGR, hypoglycaemia and dysmorphic features – vertical forehead creases, long philtrum, micrognathia, microcephaly, bridged palmar creases and a high pitched cry. The patient was referred for re-investigation at 2 days of age with a suspected diagnosis of either MDS or Pallister Killian syndrome. However, despite a directed high resolution analysis no cytogenetically visible abnormality of 17p was identified (ISCN 500 band level). FISH with a cosmid from the locus D17S379 detected a submicroscopic deletion of the MDS critical region in the proband. Parental karyotypes had previously been reported to be normal following referral for recurrent miscarriages and repeat high resolution examination failed to identify an abnormality (both analyses at ISCN 500 band level). However, FISH analysis of parental blood samples using the same D17S379 cosmid showed that the deletion in LT was the result of unbalanced segregation of a maternal rearrangement involving 11p. FISH with the telomeric probes 2209a2 (11p) and 2111b1 (17p) confirmed the presence of an apparently balanced cryptic translocation between the distal short arms of chromosomes 11 and 17, 46,XX.ish t(11;17)(p15.5;p13.3) mat (2209a2-, 2111b1+; 2111b1-, 2209a2+). LT died at 14 months of age.
Materials and methods
Cytogenetic and subtelomere FISH analysis
Metaphase chromosomes prepared from Fluorodeoxyuridine (FdU)-synchronised phytohaemagglutinin-stimulated cultures27 of peripheral blood lymphocytes were used for Giemsa-trypsin-Leishman's (GTL) banding as well as for fluorescence in situ hybridisation (FISH) analysis. FISH was performed according to the method described by Pinkel et al.28,29 with some modifications. Dual-colour subtelomere FISH was undertaken using 11p and 11q subtelomere specific clones6,30 labelled with digoxigenin and biotin respectively. Sites of hybridisation were subsequently detected using tetramethyl rhodamine isothiocyanate (TRITC) conjugated anti-digoxigenin (Boehringer) and fluorescein isothiocyanate (FITC) conjugated avidin (Vector). Slides were mounted in DAPI-Antifade (Vector) and viewed using a Zeiss Axiophot microscope. Images were captured using MacProbe (PSI) software.
Molecular genetic investigations
DNA was extracted from peripheral blood by a salt precipitation technique as described by Miller et al.31 The translocation breakpoints on chromosomes 11p and 17p were determined by PCR amplification of microsatellite polymorphisms from genomic DNA extracted from MW, LT and their parents. Primer sequences were obtained from the Genome Database (GDB: www.gdb.org) and the relative order of loci inferred from the NCBI (www.ncbi.nlm.nih.gov) and Ensembl (www.ensembl.org) databases.
Results
The translocation breakpoints are shown in Table 1. It should be noted that for both regions the genomic sequence has not been completed and so the locus order has not been unequivocally established. At the level of microsatellite mapping the results are consistent with the breakpoints being identical in both families. On chromosome 17 the breakpoint for LT and MW is between D17S1584 and D17S1810 and on chromosome 11 between D11S1318 and D11S1758 (for the locus D11S4177, between D11S1318 and D11S1758, LT is trisomic but MW is not informative). Thus LT and MW are trisomic for approximately 2 Mb of distal chromosome 11p and, consistent with their MDS phenotypes, are monosomic for approximately 5 Mb of distal chromosome 17p.
Origin of the translocations
There are at least two simple explanations for the observation of apparently identical translocations in unrelated individuals. First that there is sequence similarity between the breakpoints and the translocation is a recurring one, analogous to the t(11q;22q) (q23;q11), but would go unrecognised in sporadic cases because of its cryptic nature. This hypothesis is supported by our observation that probe 2209a2 which hybridises to the telomeric region of 11p also hybridises weakly to the short arm telomeric region of chromosome 17 (Figure 3).11 This implies that these subtelomeric regions share a degree of sequence similarity. It is possible that during early meiotic prophase, when the telomeres cluster at the nuclear periphery and homology searching and pairing initiates, this sequence similarity promotes a cross-over event resulting in the 11p;17p telomeric rearrangement. However, in each of the previously reported cases of MDS due to a familial cryptic translocation, the cross-over event involved a different chromosome arm: 3q, 8q, 9p, 10q, 19q and 20q.
The second explanation is that the translocation is identical by descent and the patients are related. It is relatively easy to discriminate between these two hypotheses as the first involves two independent events and therefore the regions close to the breakpoints would be expected to have different haplotypes while the second involves a single event and the regions close to the breakpoints would be expected to have identical or very similar haplotypes.
In order to test these hypotheses we undertook haplotyping in the regions surrounding the breakpoints. For MW we had DNA from the maternal grandparents allowing us to determine the phase of alleles present in MW's mother. On chromosome 17, the mothers of LT and MW shared an allele at all nine loci tested between D17S1529 and D17S513, an interval of approximately 5.5 Mb and a genetic distance of 3.97 female cM (fcM) and 24.43 male cM (mcM) (see Table 2). For all seven informative loci a shared allele was transmitted to MW's mother from MW's grandfather, who carries the same translocation. On chromosome 11, the mothers of LT and MW shared at least one allele over an interval exceeding 18 Mb between the loci D11S2071 (the most distal marker tested) and D11S899, a genetic distance of 22.41 fcM and 33.24 mcM (Table 3). With the exception of one locus, D11S4124, a shared allele was demonstrated to be grandpaternal in origin in the mother of MW at all 25 informative loci. Thus all three translocation carriers tested share identical or virtually identical haplotypes for large chromosomal segments that encompass both translocation breakpoints. For chromosome 17 we have also demonstrated that multiple haplotypes exist for this region and could not detect the shared haplotype among any members of five randomly selected nuclear families tested with the same PCR primers (data not shown).
We have utilised sequence based maps of distal 11p and 17p to determine the normal levels of recombination during male and female meiosis. The estimated male genetic distances for the homozygous segments were 24.43 mcM for chromosome 17 and 33.24 mcM for chromosome 11. Given these values, a male recombination event should occur once in 4.1 meioses for chromosome 17 and once in 3.0 meioses for chromosome 11. The shorter female genetic distances of 3.97 fcM for chromosome 17 and 22.41 fcM for chromosome 11, indicate that a female recombination event would occur once in 25.2 meioses and once in 4.5 meioses respectively. Both breakpoints occur in regions with approximately average levels of female recombination but within male recombination hotspots. Thus, by virtue of their extensive sequence homology we consider that these two translocations are identical by descent. Although both families are from Southern England extending the pedigrees back for five generations in family 1 and three generation in family 2 did not identify a common ancestor.
Discussion
An 11p;17p telomeric translocation was found in the mothers of two liveborn probands with MDS. Both families had a history of unexplained recurrent miscarriages. The derived chromosome was visible by high resolution cytogenetic analysis in the proband of family 1 (ISCN 550 band level) but not in the proband of family 2 (ISCN 500 band level). Both maternal translocations were originally reported as normal. On re-examination the translocation was just visible in family 1 (ISCN 550 band level) but remained cryptic in family 2 (ISCN 500 band level). Therefore the rearrangement is very subtle and would only be detectable cytogenetically at an ISCN band level of 550 or above.
While it is likely that the sequence similarity between 11p and 17p described in hypothesis 1 contributed to the rearrangement formation, we consider that the translocation arose from a single ancestral event and then segregated in the two families presented here. There were identical haplotypes surrounding 5.5 Mb on the chromosome 17 breakpoint and 18 Mb on the chromosome 11 breakpoint. The single discrepant result at D11S4124 can be explained by microsatellite mutation which, while infrequent (estimates range from 10-2 to 10-4 per generation), is by no means unheard of. The ancestral allele would have been 166 or 170 and we assume the fact that the mothers of LT and MW share an allele present in MW's grandmother is coincidental. Broman and Weber32 have reported that long homozygous segments, up to 77 cM in length, may be relatively common in the human genome. They include the example of 1.6 cM of sequence homology between D11S1794 and D11S4138 which is within the proximal part of our larger homozygous segment on chromosome 11p15.
After transmission of the derived chromosomes from the postulated common ancestor there must have been recombination proximal to the chromosome 11 breakpoint and on both sides of the chromosome 17 breakpoint. The male recombination hotspot on distal 17p is greater than that on distal 11p (4.46 mcM/Mb compared to 1.77 mcM/Mb) and may account for the smaller stretch of shared alleles on chromosome 17. The microsatellite mutation and recombinations suggest that the derived chromosomes were transmitted through a number of generations, although the high rate of pregnancy loss argues against the relationship being very distant.
Irrespective of its original formation, the translocation is responsible for the poor obstetric history in both families presented here. In recent years there has been much emphasis on the search for cryptic telomeric rearrangements as underlying causes in cases of IMR. The possible role of these abnormalities in the etiology of recurrent miscarriages is now being considered. In a study detailing the clinical features associated with submicroscopic subtelomeric rearrangements de Vries et al.33 observed only 2 of 24 probands with a family history of miscarriages. However, the study was not looking specifically for a link with miscarriages and included both sporadic and familial cases. Since the extent of chromosome loss or gain in the unbalanced products from a translocation will determine the clinical consequences, larger imbalances would be more likely to cause spontaneous foetal loss or give rise to severely affected live born children with multiple congenital abnormalities (MCA). Our two patients died aged 1 day and 14 months. The average age of the children studied by de Vries was 8.2 years (range 2 to 20) and a positive family history was common for mental retardation but rare for MCA. Therefore, although miscarriages were rare among the families of the translocation carriers selected, the study population is likely to underestimate the proportion of families with miscarriages. Fan and Zhang34 tried to directly assess the significance of subtelomeric translocations in 80 patients referred with more than three miscarriages. They identified no structural abnormalities amongst their cohort of patients, but the power of the study was halved because the population comprised individuals rather than couples. In contrast, of five couples with five or more miscarriages Yakut et al18 identified two with a balanced subtelomeric rearrangement.
Cryptic subtelomeric rearrangements were first identified in families with recurrent pregnancy losses through direct analysis of individual chromosome arms following a suggested clinical diagnosis.19,20,21,22,23,24,2526 With increased clinical awareness and advances in FISH technology, a growing number of cases are being identified through specific searches of the subtelomeric regions.13,1415,17 The balance of evidence suggests that cryptic translocations are responsible for recurrent miscarriages in a proportion of families and that the proportion is likely to rise in families with five or more miscarriages or where additional relatives are also affected.
In the families reported here, the occurrence of a live birth with a clinically recognisable microdeletion syndrome led to the diagnosis of a cryptic parental balanced translocation, but this is likely to be an uncommon means of ascertainment. Therefore a systematic screen of the subtelomeric regions should be undertaken in a cohort of families to determine the contribution of such rearrangements to reproductive loss.
References
Campana M, Serra A, Neri G . Role of chromosome aberrations in recurrent abortion: a study of 269 balanced translocations Am J Med Genet 1986 24: 341–356
Bourrouillou G, Colombies P, Dastugue N . Chromosome studies in 2136 couples with spontaneous abortions Hum Genet 1986 74: 399–401
Castle D, Bernstein R . Cytogenetic analysis of 688 couples experiencing multiple spontaneous abortions Am J Med Genet 1988 29: 549–556
Flint J, Wilkie AO, Buckle VJ, Winter RM, Holland AJ, Mcdermid HE . The detection of subtelomeric chromosomal rearrangements in idiopathic mental retardation Nature Genet 1995 9: 132–140
Knight SJL, Regan R, Nicod A et al. Subtle chromosomal rearrangements in children with unexplained mental retardation Lancet 1999 354: 1676–1681
Knight SLJ, Lese CM, Precht KS et al. An optimised set of human telomeres clones for studying telomere integrity and architecture Am J Hum Genet 2000 67: 320–332
Reigel M, Baumer A, Jamar M et al. Submicroscopic terminal deletions and duplications in retarded patients with unclassified malformation syndromes Hum Genet 2001 109: 286–294
Rosenberg MJ, Killoran C, Dziadzio L et al. Scanning for telomeric deletions and duplications and uniparental disomy using genetic markers in 120 children with malformations Hum Genet 2001 109: 311–318
Rossi E, Piccini F, Zollino M et al. Cryptic telomeric rearrangements in subjects with mental retardation associated with dysmorphism and congenital malformations J Med Genet 2001 38: 417–420
Slavotinek A, Rosenberg M, Knight S et al. Screening for submicroscopic chromosome rearrangements in children with idiopathic mental retardation using microsatellite markers for the chromosome telomeres J Med Genet 1999 36: 405–411
Fan YS, Zhang Y, Speevak M, Farrell S, Jung JH, Siu VM . Detection of submicroscopic abberration in patients with unexplained mental retardation by fluorescence in situ hybridization Genet Med 2001 3: 416–421
Joyce CA, Dennis NR, Cooper S, Browne CE . Subtelomeric rearrangements: results from a study of selected and unselected probands with idiopathic mental retardation and control individuals by using high-resolution G-banding and FISH Hum Genet 2001 109: 440–451
Bacino CA, Kashork CD, Davino NA, Shaffer LG . Detection of a cryptic translocation in a family with mental retardation using FISH and telomere region-specific probes Am J Med Genet 2000 5: 250–255
Brackley KJ, Kilby MD, Morton J, Whittle MJ, Knight SJ, Flint J . A case of recurrent congenital fetal anomalies associated with a familial subtelomeric translocation Prenat Diagn 1999 19: 570–574
Giardino D, Finelli P, Gottardi G et al. Cryptic subtelomeric translocation t(2;16)(q37;q24) segregating in a family with unexplained stillbirths and a dysmorphic, slightly retarded child Eur J Hum Genet 2001 9: 881–886
Leppig KA, Ball S, Au K, Opheim KE, Norwood T . Familial cryptic (20;21) translocation identified by in situ hybridization technologies Am J Med Genet 2000 93: 273–277
Shaffer LG, Spikes AS, Macha M, Dunn R . Identification of a subtle chromosomal translocation in a family with recurrent miscarriages and a child with multiple congenital anomalies. A case report J Reprod Med 1996 41: 367–371
Wakui K, Tanemure M, Suzumori K et al. Clinical applications of two-color telomeric fluorescence in situ hybridization for prenatal diagnosis: identification of chromosomal translocation in five families with recurrent miscarriages or a child with multiple congenital anomalies J Human Genet 1999 44: 85–90
Yakut S, Berker-Karauzum S, Simsek M, Zorlu G, Trak B, Luleci G . Telomere-specific fluorescence in situ hybridization analysis of couples with five or more recurrent miscarriages Clin Genet 2002 61: 26–31
Overhauser J, Bengtsson U, McMahon J et al. Prenatal diagnosis and carrier detection of a cryptic translocation by using DNA markers from the short arm of chromosome 5 Am J Hum Genet 1989 45: 296–303
Altherr MR, Bengtsson U, Elder FFB et al. Molecular confirmation of Wolf-Hirschhorn syndrome with a subtle translocation of chromosome 4 Am J Hum Genet 1991 49: 1235–1242
Alvarado M, Bass HN, Caldwell S, Jamehdor M, Miller AA, Jacob P . Miller-Dieker syndrome. Detection of a cryptic chromosome translocation using in situ hybridization in a family with multiple affected offspring Am J Dis Child 1993 147: 1291–1294
Kohler A, Hain J, Muller U . Familial half-cryptic translocation t(9;17) J Med Genet 1994 31: 712–714
Kuwano A, Ledbetter SA, Dobyns WB, Emanual BS, Ledbetter DH . Detection of deletions and cryptic translocations in Miller-Dieker syndrome by in situ hybridisation Am J Hum Genet 1991 49: 707–714
Masuno M, Imaizumi K, Nakamura M, Matsui K, Goto A, Kuroki Y . Miller-Dieker syndrome due to maternal cryptic translocation t(10;17)(q26.3;p13.1) Am J Med Genet 1995 4: 441–443
van Zelderen-Bhola SL, Breslau-Siderius EJ, Beverstock GC et al. Prenatal and postnatal investigation of a case with Miller-Dieker syndrome due to a familial cryptic translocation t(17;20)(p13.3;q13.3) detected by fluorescence in situ hybridization Prenat Diagn 1997 17: 173–179
Weber LM, Garson OM . Fluorodeoxyuridine synchronization of bone marrow cultures Cancer Genet Cyogenet 1983 8: 123–132
Pinkel D, Straume T, Gray JW . Cytogenetic analysis using quantitative, high sensitivity, fluorescence hybridization Proc Natl Acad Sci USA 1986 83: 2934–2938
Pinkel D, Landegent J, Collins C et al. Fluorescence in situ hybridization with human chromosome specific libraries: detection of trisomy 21 and translocations of chromosome 4 Proc Natl Acad Sci USA 1988 85: 9138–9142
National Institutes of Health and Institute of Molecular Medicine Collaboration. A complete set of human telomeric probes and their clinical application Nature Genet 1996 14: 86–89
Miller SA, Dykes DD, Polesky HF . A simple salting out procedure for extracting DNA from human nucleated cells Nucleic Acids Res 1988 16: 1215
Broman KW, Weber JL . Long homozygous chromosomal segments in reference families from the Centre d'Etude du Polymorphisme Humain Am J Hum Genet 1999 65: 1493–1500
de Vries BBA, White SM, Knight SJL et al. Clinical studies on submicroscopic subtelomeric rearrangements: a checklist J Med Genet 2001 38: 145–150
Fan YS, Zhang Y . Subtelomeric translocations are not a frequent cause of recurrent miscarriages Am J Med Genet 2002 109: 154
Acknowledgements
We are indebted to the families of MW and LT, without whom this study could not have taken place. We thank Dr J Flint and Dr L Kearney for kindly donating the panel of subtelomere clones used in this study and Will Tapper for his expert database analysis. We are particularly grateful to Prof PA Jacobs and Dr JA Crolla for their advice and encouragement. We also wish to thank Mary Hart and Rebecca Protheroe for maintaining the library of FISH probes.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Joyce, C., Dennis, N., Howard, F. et al. An 11p;17p telomeric translocation in two families associated with recurrent miscarriages andMiller-Dieker syndrome. Eur J Hum Genet 10, 707–714 (2002). https://doi.org/10.1038/sj.ejhg.5200882
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.ejhg.5200882