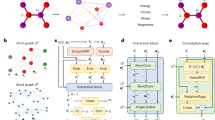
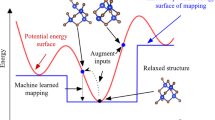
Abstract
Interatomic potentials (IAPs), which describe the potential energy surface of atoms, are a fundamental input for atomistic simulations. However, existing IAPs are either fitted to narrow chemistries or too inaccurate for general applications. Here we report a universal IAP for materials based on graph neural networks with three-body interactions (M3GNet). The M3GNet IAP was trained on the massive database of structural relaxations performed by the Materials Project over the past ten years and has broad applications in structural relaxation, dynamic simulations and property prediction of materials across diverse chemical spaces. About 1.8 million materials from a screening of 31 million hypothetical crystal structures were identified to be potentially stable against existing Materials Project crystals based on M3GNet energies. Of the top 2,000 materials with the lowest energies above the convex hull, 1,578 were verified to be stable using density functional theory calculations. These results demonstrate a machine learning-accelerated pathway to the discovery of synthesizable materials with exceptional properties.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$99.00 per year
only $8.25 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The training data for the universal IAP are available at https://doi.org/10.6084/m9.figshare.19470599 (ref. 61). The phonon dispersion curves of 328 dynamically stable materials are available at https://doi.org/10.6084/m9.figshare.20217212 (ref. 62). The ICSD database used in this study is a commercial product and cannot be shared. All generated hypothetical compounds and their corresponding M3GNet predictions are provided at http://matterverse.ai. Each material can be accessed via a detail page at https://matterverse.ai/details/mv-id, where id ranges from 0 to 31,664,854. Source Data are provided with this paper.
Code availability
The source code for M3GNet is available at https://github.com/materialsvirtuallab/m3gnet and https://doi.org/10.5281/zenodo.7141391 (ref. 63).
References
Weiner, P. K. & Kollman, P. A. AMBER: assisted model building with energy refinement. A general program for modeling molecules and their interactions. J. Comput. Chem. 2, 287–303 (1981).
Case, D. A. et al. The AMBER biomolecular simulation programs. J. Comput. Chem. 26, 1668–1688 (2005).
Rappe, A. K., Casewit, C. J., Colwell, K. S., Goddard, W. A. & Skiff, W. M. UFF, a full periodic table force field for molecular mechanics and molecular dynamics simulations. J. Am. Chem. Soc. 114, 10024–10035 (1992).
Behler, J. & Parrinello, M. Generalized neural-network representation of high-dimensional potential-energy surfaces. Phys. Rev. Lett. 98, 146401 (2007).
Bartók, A. P., Payne, M. C., Kondor, R. & Csányi, G. Gaussian approximation potentials: the accuracy of quantum mechanics, without the electrons. Phys. Rev. Lett. 104, 136403 (2010).
Thompson, A. P., Swiler, L. P., Trott, C. R., Foiles, S. M. & Tucker, G. J. Spectral neighbor analysis method for automated generation of quantum-accurate interatomic potentials. J. Comput. Phys. 285, 316–330 (2015).
Shapeev, A. V. Moment tensor potentials: a class of systematically improvable interatomic potentials. Multiscale Model. Simul. 14, 1153–1173 (2016).
Zhang, L., Han, J., Wang, H., Car, R. & E, W. Deep potential molecular dynamics: a scalable model with the accuracy of quantum mechanics. Phys. Rev. Lett. 120, 143001 (2018).
Zuo, Y. et al. Performance and cost assessment of machine learning interatomic potentials. J. Phys. Chem. A 124, 731–745 (2020).
Schütt, K. et al. SchNet: a continuous-filter convolutional neural network for modeling quantum interactions. In Proc. 31st International Conference on Neural Information Processing Systems Advances in Neural Information Processing Systems 992–1002 (NIPS, 2017).
Klicpera, J., Groß, J. & Günnemann, S. Directional message passing for molecular graphs. Preprint at https://arxiv.org/abs/2003.03123 (2020).
Haghighatlari, M. et al. NewtonNet: a Newtonian message passing network for deep learning of interatomic potentials and forces. Digit. Discov. 1, 333–343 (2022).
Park, C. W. et al. Accurate and scalable graph neural network force field and molecular dynamics with direct force architecture. npj Comput. Mater. 7, 1–9 (2021).
Cheon, G., Yang, L., McCloskey, K., Reed, E. J. & Cubuk, E. D. Crystal structure search with random relaxations using graph networks. Preprint at https://arxiv.org/abs/2012.02920 (2020).
Lejaeghere, K. et al. Reproducibility in density functional theory calculations of solids. Science 351, aad3000 (2016).
Ong, S. P. et al. Python Materials Genomics (pymatgen): a robust, open-source Python library for materials analysis. Comput. Mater. Sci. 68, 314–319 (2013).
Jain, A. et al. FireWorks: a dynamic workflow system designed for high-throughput applications. Concurr. Comput. 27, 5037–5059 (2015).
Pizzi, G., Cepellotti, A., Sabatini, R., Marzari, N. & Kozinsky, B. AiiDA: automated interactive infrastructure and database for computational science. Comput. Mater. Sci. 111, 218–230 (2016).
Mathew, K. et al. Atomate: a high-level interface to generate, execute, and analyze computational materials science workflows. Comput. Mater. Sci. 139, 140–152 (2017).
Jain, A. et al. Commentary: the materials project: a materials genome approach to accelerating materials innovation. APL Mater. 1, 011002 (2013).
Curtarolo, S. et al. AFLOWLIB.ORG: a distributed materials properties repository from high-throughput ab initio calculations. Comput. Mater. Sci. 58, 227–235 (2012).
Kirklin, S. et al. The Open Quantum Materials Database (OQMD): assessing the accuracy of DFT formation energies. npj Comput. Mater. 1, 1–15 (2015).
Draxl, C. & Scheffler, M. The NOMAD laboratory: from data sharing to artificial intelligence. J. Phys. Mater. 2, 036001 (2019).
Xie, T. & Grossman, J. C. Crystal graph convolutional neural networks for an accurate and interpretable prediction of material properties. Phys. Rev. Lett. 120, 145301 (2018).
Chen, C., Ye, W., Zuo, Y., Zheng, C. & Ong, S. P. Graph networks as a universal machine learning framework for molecules and crystals. Chem. Mater. 31, 3564–3572 (2019).
Chen, C., Zuo, Y., Ye, W., Li, X. & Ong, S. P. Learning properties of ordered and disordered materials from multi-fidelity data. Nat. Comput. Sci. 1, 46–53 (2021).
Choudhary, K. & DeCost, B. Atomistic line graph neural network for improved materials property predictions. npj Comput. Mater. 7, 1–8 (2021).
Tersoff, J. New empirical approach for the structure and energy of covalent systems. Phys. Rev. B 37, 6991–7000 (1988).
Petretto, G. et al. High-throughput density-functional perturbation theory phonons for inorganic materials. Sci. Data 5, 180065 (2018).
Perdew, J. P. et al. Restoring the density-gradient expansion for exchange in solids and surfaces. Phys. Rev. Lett. 100, 136406 (2008).
Kresse, G. & Hafner, J. Ab initio molecular dynamics for liquid metals. Phys. Rev. B 47, 558–561 (1993).
Kresse, G. & Furthmüller, J. Efficiency of ab-initio total energy calculations for metals and semiconductors using a plane-wave basis set. Comput. Mater. Sci. 6, 15–50 (1996).
Dunn, A., Wang, Q., Ganose, A., Dopp, D. & Jain, A. Benchmarking materials property prediction methods: the Matbench test set and Automatminer reference algorithm. npj Comput. Mater. 6, 1–10 (2020).
Sun, W. et al. The thermodynamic scale of inorganic crystalline metastability. Sci. Adv. 2, e160022 (2016).
Qi, J. et al. Bridging the gap between simulated and experimental ionic conductivities in lithium superionic conductors. Mater. Today Phys. 21, 100463 (2021).
Glass, C. W., Oganov, A. R. & Hansen, N. USPEX—evolutionary crystal structure prediction. Comput. Phys. Commun. 175, 713–720 (2006).
Wang, Y., Lv, J., Zhu, L. & Ma, Y. CALYPSO: a method for crystal structure prediction. Comput. Phys. Commun. 183, 2063–2070 (2012).
Xie, T., Fu, X., Ganea, O.-E., Barzilay, R. & Jaakkola, T. Crystal diffusion variational autoencoder for periodic material generation. Preprint at https://arxiv.org/abs/2110.06197 (2021).
Chmiela, S. et al. Machine learning of accurate energy-conserving molecular force fields. Sci. Adv. 3, e1603015 (2017).
Schütt, K. T., Arbabzadah, F., Chmiela, S., Müller, K. R. & Tkatchenko, A. Quantum-chemical insights from deep tensor neural networks. Nat. Commun. 8, 13890 (2017).
Chmiela, S., Sauceda, H. E., Müller, K.-R. & Tkatchenko, A. Towards exact molecular dynamics simulations with machine-learned force fields. Nat. Commun. 9, 3887 (2018).
Zhang, Y., Hu, C. & Jiang, B. Embedded atom neural network potentials: efficient and accurate machine learning with a physically inspired representation. J. Phys. Chem. Lett. 10, 4962–4967 (2019).
Perdew, J. P., Burke, K. & Ernzerhof, M. Generalized gradient approximation made simple. Phys. Rev. Lett. 77, 3865–3868 (1996).
Anisimov, V. I., Zaanen, J. & Andersen, O. K. Band theory and Mott insulators: Hubbard U instead of stoner I. Phys. Rev. B 44, 943–954 (1991).
Hellenbrandt, M. The Inorganic Crystal Structure Database (ICSD)—present and future. Crystallogr. Rev. 10, 17–22 (2004).
Wang, L., Maxisch, T. & Ceder, G. Oxidation energies of transition metal oxides within the GGU+U framework. Phys. Rev. B 73, 195107 (2006).
Singraber, A., Behler, J. & Dellago, C. Library-based LAMMPS implementation of high-dimensional neural network potentials. J. Chem. Theory Comput. 15, 1827–1840 (2019).
Ramachandran, P., Zoph, B. & Le, Q. V. Searching for activation functions. Preprint at https://arxiv.org/abs/1710.05941 (2017).
Kocer, E., Mason, J. K. & Erturk, H. A novel approach to describe chemical environments in high-dimensional neural network potentials. J. Chem. Phys. 150, 154102 (2019).
Kingma, D. P. & Ba, J. Adam: a method for stochastic optimization. Preprint https://arxiv.org/abs/1412.6980 (2017).
Huber, P. J. Robust estimation of a location parameter. Ann. Math. Stat. 35, 73–101 (1964).
Abadi, M. et al. TensorFlow: a system for large-scale machine learning. In 12th Symposium on Operating Systems Design and Implementation 265–283 (OSDI, 2016).
Bitzek, E., Koskinen, P., Gähler, F., Moseler, M. & Gumbsch, P. Structural relaxation made simple. Phys. Rev. Lett. 97, 170201 (2006).
Larsen, A. H. et al. The atomic simulation environment—a Python library for working with atoms. J. Phys. 29, 273002 (2017).
Togo, A. & Tanaka, I. First principles phonon calculations in materials science. Scripta Materialia 108, 1–5 (2015).
Pedregosa, F. et al. Scikit-learn: machine learning in Python. J. Mach. Learn. Res. 12, 2825–2830 (2011).
Seabold, S. & Perktold, J. Statsmodels: econometric and statistical modeling with Python. In Proc. 9th Python in Science Conference 92–96 (SciPy, 2010).
Hunter, J. D. Matplotlib: a 2D graphics environment. Comput. Sci. Eng. 9, 90–95 (2007).
Waskom, M. L. Seaborn: statistical data visualization. J. Open Source Softw. 6, 3021 (2021).
Pandas Development Team Pandas-Dev/Pandas: Pandas (Zenodo, 2020).
Chen, C. & Ong, S. P. MPF.2021.2.8. (FigShare, 2022); https://doi.org/10.6084/m9.figshare.19470599.v3
Chen, C. m3gnet Phonon Dispersion Curve of 328 Materials (FigShare, 2022); https://doi.org/10.6084/m9.figshare.20217212.v1
Ong, S. P. et al. materialsvirtuallab/m3gnet: v0.1.0. 2022 (FigShare, 2022); https://doi.org/10.5281/zenodo.7141391
de Jong, M. et al. Charting the complete elastic properties of inorganic crystalline compounds. Sci. Data 2, 150009 (2015).
Acknowledgements
This work was primarily supported by the Materials Project, funded by the US Department of Energy, Office of Science, Office of Basic Energy Sciences, Materials Sciences and Engineering Division under contract no. DE-AC02-05-CH11231: Materials Project program KC23MP. The lithium superionic conductor analysis portion of the work was funded by the LG Energy Solution through the Frontier Research Laboratory (FRL) Program. This work used the Extreme Science and Engineering Discovery Environment (XSEDE), which is supported by National Science Foundation (grant no ACI-1548562). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
C.C. and S.P.O. conceived the idea and designed the work. C.C. implemented the models and performed the analysis. C.C. and S.P.O. wrote the manuscript and contributed to the discussion and revision.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no financial interests.
Peer review
Peer review information
Nature Computational Science thanks Ekin Cubuk and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Handling editor: Jie Pan, in collaboration with the Nature Computational Science team. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 M3GNet-calculated phonon dispersion curves of four new materials predicted to be thermodynamically and dynamically stable.
a, Sr6Sc2Al4O15; b, K2Li3AlO4; c, KMN4V2O12; d, MnCd(GAO2)4.
Supplementary information
Supplementary Information
Supplementary Figs. 1–13, Sections 1–7 and Tables 1–5.
Source data
Source Data Fig. 2
Sheet Figure_2a: 2D energy-force distribution counts Sheet Figure_2b: 2D energy-stress distribution counts Sheet Figure_2c: Radial distribution Sheet Figure_2d: Element counts in the dataset.
Source Data Fig. 3
Sheet Figure_3a: DFT energy vs M3GNet energy Sheet Figure_3b: DFT force vs M3GNet force Sheet Figure_3c: DFT stress vs M3GNet stress Sheet Figure_3g: DFT DOS center vs M3GNet DOS center Sheet Figure_3h: DFT Debye temperature vs M3GNet Debye temperature.
Source Data Fig. 4
Sheet Figure_4a: Volume change with model relax vs no model relax Sheet Figure_4b: Energy difference between M3GNet energies and DFT ground-state energies using DFT initial structures, M3GNet-relaxed structures and DFT-relaxed structures.
Source Data Fig. 5
Sheet Figure_5a: Signed Ehull of M3GNet and DFT for all cases and oxide cases Sheet Figure_5b: Stable materials ratio for all and oxide cases as a function of DFT ehull thresholds. Sheet Figure_5c: M3GNet energy vs DFT energy for the all cases. Sheet Figure_5d: M3GNet energy vs DFT energy for the oxide cases.
Source Data Extended Data Fig. 1
Sheet a-special_points: special high symmetry points of phonon dispersion curve and their high symmetry labels Sheet a-high_symmetry_line-{d}: distance and the frequency for different vibration modes. d goes from 0 to the max number of symmetry lines considered for this phonon dispersion curve. Panels b, c, d are stored in Sheet {x}-special points, Sheet {x}-high_symmetry_line-{d}, where x = b, c, or d.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chen, C., Ong, S.P. A universal graph deep learning interatomic potential for the periodic table. Nat Comput Sci 2, 718–728 (2022). https://doi.org/10.1038/s43588-022-00349-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s43588-022-00349-3
This article is cited by
-
The rise of high-entropy battery materials
Nature Communications (2024)
-
Robust training of machine learning interatomic potentials with dimensionality reduction and stratified sampling
npj Computational Materials (2024)
-
Exploring the frontiers of condensed-phase chemistry with a general reactive machine learning potential
Nature Chemistry (2024)
-
MLMD: a programming-free AI platform to predict and design materials
npj Computational Materials (2024)
-
Towards atom-level understanding of metal oxide catalysts for the oxygen evolution reaction with machine learning
npj Computational Materials (2024)