Abstract
α-Enolase 1 (ENO1) is a critical glycolytic enzyme whose aberrant expression drives the pathogenesis of various cancers. ENO1 has been indicated as having additional roles beyond its conventional metabolic activity, but the underlying mechanisms and biological consequences remain elusive. Here, we show that ENO1 suppresses iron regulatory protein 1 (IRP1) expression to regulate iron homeostasis and survival of hepatocellular carcinoma (HCC) cells. Mechanistically, we demonstrate that ENO1, as an RNA-binding protein, recruits CNOT6 to accelerate the messenger RNA decay of IRP1 in cancer cells, leading to inhibition of mitoferrin-1 (Mfrn1) expression and subsequent repression of mitochondrial iron-induced ferroptosis. Moreover, through in vitro and in vivo experiments and clinical sample analysis, we identified IRP1 and Mfrn1 as tumor suppressors by inducing ferroptosis in HCC cells. Taken together, this study establishes an important role for the ENO1–IRP1–Mfrn1 pathway in the pathogenesis of HCC and reveals a previously unknown connection between this pathway and ferroptosis, suggesting a potential innovative cancer therapy.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
RNA-seq data have been deposited in the Gene Expression Omnibus under accession code GSE153989. Source data are provided with this paper. All other data supporting the results of this study can be obtained from the corresponding author upon reasonable request.
References
Faubert, B., Solmonson, A. & DeBerardinis, R. J. Metabolic reprogramming and cancer progression. Science 368, 152–161 (2020).
Hanahan, D. & Weinberg, R. A. Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011).
Pavlova, N. N. & Thompson, C. B. The emerging hallmarks of cancer metabolism. Cell Metab. 23, 27–47 (2016).
Castello, A., Hentze, M. W. & Preiss, T. Metabolic enzymes enjoying new partnerships as RNA-binding proteins. Trends Endocrinol. Metab. 26, 746–757 (2015).
Xu, D. et al. The gluconeogenic enzyme PCK1 phosphorylates INSIG1/2 for lipogenesis. Nature 580, 530–535 (2020).
Chang, C.-H. et al. Posttranscriptional control of T cell effector function by aerobic glycolysis. Cell 153, 1239–1251 (2013).
Lu, Z. & Hunter, T. Metabolic kinases moonlighting as protein kinases. Trends Biochem. Sci. 43, 301–310 (2018).
Simsek, D. et al. The mammalian ribo-interactome reveals ribosome functional diversity and heterogeneity. Cell 169, 1051–1065 (2017).
Castello, A. et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 149, 1393–1406 (2012).
Carpousis, A. J. The RNA degradosome of Escherichia coli: an mRNA-degrading machine assembled on RNase E. Annu. Rev. Microbiol. 61, 71–87 (2007).
Masai, H., Matsumoto, S., You, Z., Yoshizawa-Sugata, N. & Oda, M. Eukaryotic chromosome DNA replication: where, when, and how? Annu. Rev. Biochem. 79, 89–130 (2010).
Hentze, M. W., Muckenthaler, M. U. & Andrews, N. C. Balancing acts: molecular control of mammalian iron metabolism. Cell 117, 285–297 (2004).
Costa da Silva, M. et al. Iron induces anti-tumor activity in tumor-associated macrophages. Front. Immunol. 8, 1479–1479 (2017).
Hassannia, B., Vandenabeele, P. & Vanden Berghe, T. Targeting ferroptosis to iron out cancer. Cancer Cell 35, 830–849 (2019).
Stockwell, B. R. et al. Ferroptosis: a regulated cell death nexus linking metabolism, redox biology, and disease. Cell 171, 273–285 (2017).
Torti, S. V. & Torti, F. M. Iron and cancer: more ore to be mined. Nat. Rev. Cancer 13, 342–355 (2013).
Dixon, S. J. et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell 149, 1060–1072 (2012).
Chen, L., Hambright, W. S., Na, R. & Ran, Q. Ablation of the ferroptosis inhibitor glutathione peroxidase 4 in neurons results in rapid motor neuron degeneration and paralysis. J. Biol. Chem. 290, 28097–28106 (2015).
Chu, B. et al. ALOX12 is required for p53-mediated tumour suppression through a distinct ferroptosis pathway. Nat. Cell Biol. 21, 579–591 (2019).
Do Van, B. et al. Ferroptosis, a newly characterized form of cell death in Parkinson’s disease that is regulated by PKC. Neurobiol. Dis. 94, 169–178 (2016).
Jiang, L. et al. Ferroptosis as a p53-mediated activity during tumour suppression. Nature 520, 57–62 (2015).
Zhang, Y. et al. BAP1 links metabolic regulation of ferroptosis to tumour suppression. Nat. Cell Biol. 20, 1181–1192 (2018).
Dai, J. et al. Alpha-enolase regulates the malignant phenotype of pulmonary artery smooth muscle cells via the AMPK-Akt pathway. Nat. Commun. 9, 3850 (2018).
Principe, M. et al. Alpha-enolase (ENO1) controls alpha v/beta 3 integrin expression and regulates pancreatic cancer adhesion, invasion, and metastasis. J. Hematol. Oncol. 10, 16 (2017).
Capello, M. et al. Targeting the Warburg effect in cancer cells through ENO1 knockdown rescues oxidative phosphorylation and induces growth arrest. Oncotarget 7, 5598–5612 (2016).
Hudson, W. H. & Ortlund, E. A. The structure, function and evolution of proteins that bind DNA and RNA. Nat. Rev. Mol. Cell Biol. 15, 749–760 (2014).
Wang, W. Y. et al. Identification of alpha-enolase as a nuclear DNA-binding protein in the zona fasciculata but not the zona reticularis of the human adrenal cortex. J. Endocrinol. 184, 85–94 (2005).
Kang, H. J., Jung, S. K., Kim, S. J. & Chung, S. J. Structure of human alpha-enolase (hENO1), a multifunctional glycolytic enzyme. Acta Crystallogr. D Biol. Crystallogr. 64, 651–657 (2008).
Feo, S., Arcuri, D., Piddini, E., Passantino, R. & Giallongo, A. ENO1 gene product binds to the c-myc promoter and acts as a transcriptional repressor: relationship with Myc promoter-binding protein 1 (MBP-1). FEBS Lett. 473, 47–52 (2000).
Sharova, L. V. et al. Database for mRNA Half-Life of 19 977 genes obtained by DNA microarray analysis of pluripotent and differentiating mouse embryonic stem cells. DNA Res. 16, 45–58 (2009).
Lau, N.-C. et al. Human Ccr4-Not complexes contain variable deadenylase subunits. Biochem. J. 422, 443–453 (2009).
Chen, G., Fillebeen, C., Wang, J. & Pantopoulos, K. Overexpression of iron regulatory protein 1 suppresses growth of tumor xenografts. Carcinogenesis 28, 785–791 (2007).
Chapoutot, C. et al. Liver iron excess in patients with hepatocellular carcinoma developed on viral C cirrhosis. Gut 46, 711–714 (2000).
Sorrentino, P. et al. Liver iron excess in patients with hepatocellular carcinoma developed on non-alcoholic steato-hepatitis. J. Hepatol. 50, 351–357 (2009).
Galy, B. et al. Iron regulatory proteins secure mitochondrial iron sufficiency and function. Cell Metab. 12, 194–201 (2010).
Martelli, A. et al. Iron regulatory protein 1 sustains mitochondrial iron loading and function in frataxin deficiency. Cell Metab. 21, 311–322 (2015).
Shaw, G. C. et al. Mitoferrin is essential for erythroid iron assimilation. Nature. 440, 96–100 (2006).
Shen, S. et al. A miR-130a-YAP positive feedback loop promotes organ size and tumorigenesis. Cell Res. 25, 997–1012 (2015).
Troadec, M.-B. et al. Targeted deletion of the mouse Mitoferrin1 gene: from anemia to protoporphyria. Blood 117, 5494–5502 (2011).
Duran, A. et al. p62/SQSTM1 by binding to vitamin D receptor inhibits hepatic stellate cell activity, fibrosis, and liver cancer. Cancer Cell 30, 595–609 (2016).
Ast, T. et al. Hypoxia rescues frataxin loss by restoring iron sulfur cluster biogenesis. Cell 177, 1507–1521 (2019).
Zhang, H. et al. HIF-1 inhibits mitochondrial biogenesis and cellular respiration in VHL-deficient renal cell carcinoma by repression of C-MYC activity. Cancer Cell 11, 407–420 (2007).
Badgley, M. A. et al. Cysteine depletion induces pancreatic tumor ferroptosis in mice. Science 368, 85–89 (2020).
Bersuker, K. et al. The CoQ oxidoreductase FSP1 acts parallel to GPX4 to inhibit ferroptosis. Nature 575, 688–692 (2019).
Bock, F. J. & Tait, S. W. G. Mitochondria as multifaceted regulators of cell death. Nat. Rev. Mol. Cell Biol. 21, 85–100 (2020).
Doll, S. et al. FSP1 is a glutathione-independent ferroptosis suppressor. Nature 575, 693–698 (2019).
Gao, M. et al. Role of mitochondria in ferroptosis. Mol. Cell 73, 354–363 (2019).
Cloonan, S. M. et al. Mitochondrial iron chelation ameliorates cigarette smoke-induced bronchitis and emphysema in mice. Nat. Med. 22, 163–174 (2016).
Koppenol, W. H., Bounds, P. L. & Dang, C. V. Otto Warburg’s contributions to current concepts of cancer metabolism. Nat. Rev. Cancer 11, 325–337 (2011).
Muller, F. L. et al. Passenger deletions generate therapeutic vulnerabilities in cancer. Nature 488, 337–342 (2012).
Entelis, N. et al. A glycolytic enzyme, enolase, is recruited as a cofactor of tRNA targeting toward mitochondria in Saccharomyces cerevisiae. Genes Dev. 20, 1609–1620 (2006).
Hernandez-Perez, L. et al. alpha-Enolase binds to RNA. Biochimie 93, 1520–1528 (2011).
Rennekamp, A. J. The ferrous awakens. Cell 171, 1226–1227 (2017).
Angeli, J. P. F., Shah, R., Pratt, D. A. & Conrad, M. Ferroptosis inhibition: mechanisms and opportunities. Trends Pharmacol. Sci. 38, 489–498 (2017).
Reed, J. C. & Pellecchia, M. Ironing out cell death mechanisms. Cell 149, 963–965 (2012).
Zhou, B. et al. Tom20 senses iron-activated ROS signaling to promote melanoma cell pyroptosis. Cell Res. 28, 1171–1185 (2018).
Gao, M. et al. Ferroptosis is an autophagic cell death process. Cell Res. 26, 1021–1032 (2016).
Hassannia, B. et al. Nano-targeted induction of dual ferroptotic mechanisms eradicates high-risk neuroblastoma. J. Clin. Invest. 128, 3341–3355 (2018).
Wang, H. et al. Characterization of ferroptosis in murine models of hemochromatosis. Hepatology 66, 449–465 (2017).
Zhang, Y. et al. Lin28 enhances de novo fatty acid synthesis to promote cancer progression via SREBP-1. EMBO Rep. 20, e48115 (2019).
Chatterjee, A. et al. MOF Acetyl transferase regulates transcription and respiration in mitochondria. Cell. 167, 722–738 (2016).
Salles, F. J. & Strickland, S. Analysis of poly(A) tail lengths by PCR: the PAT assay. Methods Mol. Biol. 118, 441–448 (1999).
Guan, K.-L., Yu, W., Lin, Y., Xiong, Y. & Zhao, S. Generation of acetyllysine antibodies and affinity enrichment of acetylated peptides. Nat. Protoc. 5, 1583–1595 (2010).
Rebouche, C. J., Wilcox, C. L. & Widness, J. A. Microanalysis of non-heme iron in animal tissues. J. Biochem. Biophys. Methods 58, 239–251 (2004).
Birch-Machin, M. A. & Turnbull, D. M. Assaying mitochondrial respiratory complex activity in mitochondria isolated from human cells and tissues. Methods Cell Biol. 65, 97–117 (2001).
Chan, A. C. Y. et al. Evaluation of the seventh edition of the American Joint Committee on Cancer tumournodemetastasis (TNM) staging system for patients undergoing curative resection of hepatocellular carcinoma: implications for the development of a refined staging system. HPB (Oxford) 15, 439–448 (2013).
Li, J. et al. Astrocyte elevated gene-1 is a novel prognostic marker for breast cancer progression and overall patient survival. Clin. Cancer Res. 14, 3319–3326 (2008).
Li, S.-T. et al. Myc-mediated SDHA acetylation triggers epigenetic regulation of gene expression and tumorigenesis. Nat. Metab. 2, 256–269 (2020).
Acknowledgements
This work is supported in part by the Chinese Academy of Sciences (no. XDB39020100 to H.Z.); the National Key R&D Program of China (nos. 2018YFA0107103 to H.Z. and 2018YFA0800300 and 2017YFA0205600 to P.G.); the National Natural Science Foundation of China (nos. 81821001 and 81930083 to H.Z., 82130087 and 91957203 to P.G. and 81874060 to L.S.); the Program for Guangdong Introducing Innovative and Entrepreneurial Teams (no. 2017ZT07S054 to P.G.); and the Fundamental Research Funds for the Central Universities (nos. YD2070002008 to H.Z. and 2020ZYGXZR038 to L.S.). All correspondence and requests for materials should be addressed to H.Z. (hzhang22@ustc.edu.cn).
Author information
Authors and Affiliations
Contributions
H.Z. and P.G. conceived the study and supervised experiments. T.Z., L.S., P.G. and H.Z. designed experiments. T.Z., L.S., Y.H., C.S., S.S., H.W., W.M., P.Z., T.W., X.G., R.Y., Y.Z. and Y.C. performed experiments. S.S. analyzed RNA-seq data. W.J. and Z.C. provided clinical specimens. S.-T.L., Y.C., R.Z., W.J. and F.H. provided constructive advice. H.Z., P.G., T.Z. and L.S. wrote the paper. All authors read and approved the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Cancer thanks Marcus Conrad, Michael Kharas and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 ENO1 degrades IRP1 mRNA as an RNA-binding protein.
a, The mRNA expression of the 16 selected genes mentioned in Fig. 1b was measured by qPCR in 33 pairs of clinically matched tumor adjacent noncancerous liver tissues (non-tumor) and human HCC tissues (tumor) (n = 33 patients with HCC). b, qPCR (n = 3 biological replicates) and western blot analysis of IRP1 expression in PLC cells with ENO1 knockdown. c, ANXA1, KANK1, CAT, BIN1, MYO6, RBM25, NCAPD3 and NAB1 mRNA stability was measured in the ENO1-overexpressing PLC cells treated with the transcription inhibitor actinomycin D (5 μM) for 0, 3 or 6 hours (n = 3 biological replicates). Data are presented as the mean ± s.d. of three independent experiments (b,c) or mean ± s.e.m. (a). Statistical significance was determined by two-tailed unpaired Student’s t-test (a-c). Experiments of western blot were repeated three times independently with similar results (b).
Extended Data Fig. 2 ENO1 directly interacts with CNOT6.
a, GST pull−down of His-CNOT3, or His-CNOT10 by GST-ENO1 using proteins purified in E. coli, followed by western blot analysis with anti-CNOT3, anti-CNOT10, and anti-GST antibodies. b, HEK293T cells were transfected with HA-ENO1 plasmids together with Flag-EV or Flag-CNOT6 plasmids. Cell lysates were immunoprecipitated with anti-Flag antibody or IgG, followed by western blot analysis. c, Western blot analysis of IRP1 expression in PLC cells with CNOT6 knockdown (left panel). IRP1 mRNA stability was determined in ENO1-knocking down PLC cells treated with the transcription inhibitor actinomycin D (5 μM) for 0, 3 or 6 hours (right panel) (n = 3 biological replicates). d, HEK293T cells were transfected with HA-CNOT6 plasmids together with Flag-EV or Flag-tagged plasmid expressing ENO1-DBD deletion. Cell lysates were immunoprecipitated with anti-Flag antibody or IgG, followed by western blot analysis. e, HEK293T cells were transfected with HA-ENO1-C terminus plasmids together with Flag-EV or Flag-CNOT6-LR domain plasmids. Cell lysates were immunoprecipitated with anti-Flag antibody or IgG, followed by western blot analysis. f, HEK293T cells were transfected with HA-ENO1 plasmids together with Flag-EV or Flag-CNOT6 plasmids. Cell lysates with or without RNase A treatment were immunoprecipitated with anti-Flag antibody, followed by western blot analysis with anti-HA or anti-Flag antibodies. Data are presented as the mean ± s.d. of three independent experiments (c). Statistical significance was determined by two-tailed unpaired Student’s t-test (c). Experiments of western blot were repeated three times independently with similar results (a-f).
Extended Data Fig. 3 IRP1 inhibits cell proliferation.
a, b, Cell growth curves were measured in PLC, HepG2 and Huh7 cells overexpressing IRP1 (a) or in PLC-shIRP1cells (b) in the presence of 200 µM ferrous iron (n = 3 biological replicates). c, Cell viability was measured in PLC cells treated with ferrous iron (n = 5 biological replicates). d, Mouse tumor masses in Fig. 3a were measured at the end of the experiment (n = 5 male mice per group). e, Western blot analysis of IRP1 protein from tumor tissues in Fig. 3a. f, Mfrn1 mRNA (n = 3 biological replicates) and protein expression in PLC-shIRP1 cells. g, Mfrn1 protein in HepG2 and Huh7 cells with IRP1 knockdown in the presence of ferrous iron. h, Mfrn1 protein and cell growth curves (n = 3 biological replicates) were measured in PLC cells expressing wild-type IRP1 or the IRP1C437S in the presence of 200 µM ferrous iron. i, j, HepG2 cells stably expressing EV or IRP1C437S were injected subcutaneously into nude mice (n = 5 male mice per group). Tumor masses (i) and protein of IRP1 and Mfrn1 in tumor tissues (j) were measured at the end of the experiment. Data are presented as the mean ± s.d. of three (a,b,f,h) or five (c) independent experiments or mean ± s.e.m. (d,i). Statistical significance was determined by two-tailed unpaired Student’s t-test (c,d,f,i), one-way analysis of variance (ANOVA) (b,h), or two-way analysis of variance (ANOVA) (a). Experiments of western blot were repeated three times independently with similar results (f-h).
Extended Data Fig. 4 ENO1 suppresses Mfrn1 expression by inhibiting IRP1.
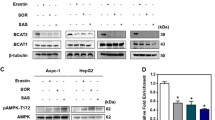
a, The total and phosphorylated CREB protein in PLC cells expressing IRP1 in the presence of ferrous iron (left). Mfrn1 mRNA (n = 3 biological replicates) and protein in PLC-shCREB cells (right). b, ChIP experiments were performed in PLC cells using IgG or anti-CREB antibody (n = 3 biological replicates). CRE: cAMP response element. c, Cell growth curves were measured in PLC, HepG2 and Huh7 cells expressing Mfrn1 in the absence or presence of 200 µM ferrous iron (n = 3 biological replicates). d, mRNA (n = 3 biological replicates) and protein expression of IRP1 and Mfrn1 in PLC-shENO1 cells in the presence of ferrous iron. e,f, Western blots analysis in PLC cells stably expressing shENO1 with further knockdown of IRP1 (e), or in PLC cells expressing wide-type ENO1, ENO1S40A or ENO1D245R (f) in the presence of ferrous iron. Data are presented as the mean ± s.d. of three independent experiments (a-d). Statistical significance was determined by two-tailed unpaired Student’s t-test (a,b,d), or two-way analysis of variance (ANOVA) (c). Experiments of western blot were repeated three times independently with similar results (a,d-f).
Extended Data Fig. 5 ENO1 promotes tumor growth by inhibiting the IRP1/Mfrn1 axis.
a, b, PLC cells overexpressing ENO1 were infected with viruses expressing IRP1(a) or Mfrn1(b) in the presence of ferrous iron. Protein levels and cell growth (n = 3 biological replicates) were measured, respectively. c, Tumors were extracted and tumor masses were measured at the end of the experiment as shown in Fig. 4a (n = 5 male mice per group). d, PLC cells with IRP1 knockdown were infected with viruses expressing Mfrn1 in the presence of ferrous iron. Protein levels and cell growth (n = 3 biological replicates) were measured, respectively. e, Tumors were extracted and tumor masses were measured at the end of the experiment as shown in Fig. 4b (n = 5 male mice per group). Data are presented as the mean ± s.d. of three independent experiments (a,b,d) or mean ± s.e.m. (c,e). Statistical significance was determined by two-tailed unpaired Student’s t-test (c,e), or two-way analysis of variance (ANOVA) (a,b,d). Experiments of western blot were repeated three times independently with similar results (a,b,d).
Extended Data Fig. 6 IRP1 and Mfrn1 serve as tumor suppressors in HCC.
a, H&E staining of the livers shown in Fig. 4c were presented. N, normal liver tissue. T, liver tumor area. b-d, NRAS/shp53-induced liver tumorigenesis was analyzed 60 days after injection of corresponding plasmids. Plasmids expressing NRAS/shp53 alone or NRAS/shp53 plus mouse-IRP1 together with plasmids expressing SB and PB transposase were delivered into mice by hydrodynamic injection (n = 5 male mice per group). RFP served as a control. Liver/body weight ratios (b), total number of tumors, number of tumors bigger than 3 mm (diameter), and maximal tumor diameters were measured (c). H&E staining of livers were performed (d). e, The Mfrn1 protein was measured by western blot in wide type or Mfrn1+/− mice. f-i, YAP-5SA-induced liver tumors in wide-type and Mfrn1+/− mice were analyzed 100 days after injection of corresponding plasmids, respectively (n = 5 male mice per group). RFP served as a control. Liver/body weight ratios (f), total number of tumors, number of tumors bigger than 3 mm (diameter), and maximal tumor diameters were measured (g). H&E staining of livers were performed (h). Mfrn1 protein in tissues was measured (i). j,k, H&E staining (j) and Mfrn1 protein in liver tissues in Fig. 4e were presented (k). l, Mfrn1 protein in liver tissues shown in Fig. 4g were measured. Data are presented as the mean ± s.e.m. (b,c,f,g). Statistical significance was determined by two-tailed unpaired Student’s t-test (b,c,f,g). Experiments of micrographs were repeated three times independently with similar results (a,d,h,j).
Extended Data Fig. 7 ENO1 inhibits mitochondrial respiration by inhibiting the IRP1/Mfrn1 axis.
a, Mitochondrial complex I and II activities were detected in PLC cells overexpressing IRP1 or Mfrn1 (the upper panel), or in PLC cells expressing shRNAs targeting IRP1 or Mfrn1 (the lower panel) in the presence of ferrous iron (n = 3 biological replicates). b, The O2 consumption rates (OCR) of PLC cells stably overexpressing IRP1 or Mfrn1 were detected in the presence of ferrous iron (n = 3 biological replicates). c,d, Mitochondrial complex I and II activities (c) and OCR (d) in PLC cells stably overexpressing ENO1 were measured in the presence of ferrous iron (n = 3 biological replicates). e, PLC cells overexpressing IRP1 were further infected with viruses expressing NTC or shMfrn1 in the presence of ferrous iron, followed by detection of mitochondrial complex I and II activities (n = 3 biological replicates). f,g, PLC cells overexpressing ENO1 were further infected with viruses overexpressing IRP1 (f) or Mfrn1 (g) in the presence of ferrous iron, followed by detection of mitochondrial complex I and II activities (n = 3 biological replicates). h,i, Mitochondrial complex I and II activities (h) (n = 3 biological replicates) and FECH and POLD1 protein levels (i) were detected using xenograft tumor samples in Fig. 3a generated from Huh7 cells overexpressing IRP1. Data are presented as the mean ± s.d. of three independent experiments (a-h). Statistical significance was determined by two-tailed unpaired Student’s t-test (a-h). Experiments of western blot were repeated three times independently with similar results (i).
Extended Data Fig. 8 Roles of IRP1 and Mfrn1 in ferroptosis.
a, Mitochondrial ROS levels were measured by flow cytometry using MitoSOX staining in PLC cells overexpressing IRP1 or Mfrn1 in the presence of ferrous iron. b, PLC cells overexpressing IRP1 were further infected with viruses expressing NTC or shMfrn1 in the presence of ferrous iron, followed by mitochondrial ROS detection. c, Western blot analysis of the expression of Caspase-3, cleaved Caspase-3, PARP1, cleaved PARP1 and LC3-I/II in PLC cells overexpressing IRP1 or Mfrn1 in the absence or presence of ferrous iron. d,e, Cell growth curves were determined in PLC cells overexpressing IRP1 (d) or Mfrn1 (e) treated with 200 µM ferrous iron combined with 2 µM Ferrostatin-1 or 5 µM deferiprone (n = 3 biological replicates). f, LDH release was detected in PLC cells overexpressing IRP1 or Mfrn1 in the absence or presence of ferrous iron (n = 3 biological replicates). g,h, Lipid peroxidation was assessed by flow cytometry after C11-BODIPY staining in PLC and Huh7 cells overexpressing IRP1 (g) or Mfrn1 (h) in the presence of ferroptosis inhibitors Ferr-1 or Lip-1. (i) Western blot analysis of the expression of iron-related proteins and canonical ferroptosis regulators in PLC, HepG2 and Huh7 cells overexpressing IRP1 in the presence of ferrous iron. Data are presented as the mean ± s.d. of three independent experiments (d-f). Statistical significance was determined by two-tailed unpaired Student’s t-test (f), or two-way analysis of variance (ANOVA) (d,e). Experiments of western blot were repeated three times independently with similar results (c,i).
Extended Data Fig. 9 IRP1 and Mfrn1 promote ferroptosis in vivo.
a,b, Tumor masses were measured at the end of the experiment shown in Fig. 6a (a) and western blot analysis of IRP1 and Mfrn1 protein levels from tumor tissues as shown in Fig. 6a (b) (n = 3 male mice per group). c,d, Tumor masses were measured at the end of the experiment shown in Fig. 6c (c) and western blot analysis of IRP1 and Mfrn1 protein levels from tumor tissues as shown in Fig. 6c (d) (n = 3 male mice per group). Data are presented as the mean ± s.e.m. (a,c). Statistical significance was determined by two-tailed unpaired Student’s t-test (a,c).
Extended Data Fig. 10 IRP1 and Mfrn1 deficiency correlates with human HCC.
a,b, The mRNA levels of Mfrn1 (a) and ENO1 (b) were measured by qPCR in 33 pairs of clinically matched tumor adjacent noncancerous liver tissues (non-tumor) and human HCC tissues (tumor) (n = 33 patients with HCC) (Right panel: n = 3 technical replicates). c, Statistical quantification of the mean optical density (MOD) values of ENO1 staining in IHC assay between normal liver tissues and HCC specimens of different clinical stages (I–IV) (Healthy donors: n = 9, patients with HCC: I (n = 14), II (n = 75), III (n = 30), IV (n = 16)). d, Kaplan-Meier curves with univariate analyses of patients with low versus high ENO1 expression (high ENO1: n = 65 patients, low ENO1: n = 70 patients). e,f, Representative IHC images of IRP1 (e) and Mfrn1 (f) staining were shown from the HCC specimens that included cancerous tissue and adjacent tissue. Data are presented as the mean ± s.e.m. (a,b,c). Statistical significance was determined by two-tailed unpaired Student’s t-test (a,b,c) or Log-rank test (d).
Supplementary information
Supplementary Information
Supplementary Tables 1–12.
Source data
Source Data Fig. 1
Unprocessed immunoblots.
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Unprocessed immunoblots and gels.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Unprocessed immunoblots.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Statistical source data.
Source Data Fig. 7
Unprocessed immunoblots.
Source Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 1
Unprocessed immunoblots.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 2
Unprocessed immunoblots.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 3
Unprocessed immunoblots.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 4
Unprocessed immunoblots.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 5
Unprocessed immunoblots.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 6
Unprocessed immunoblots.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 7
Unprocessed immunoblots.
Source Data Extended Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 8
Unprocessed immunoblots.
Source Data Extended Data Fig. 8
Statistical source data.
Source Data Extended Data Fig. 9
Unprocessed immunoblots.
Source Data Extended Data Fig. 9
Statistical Source Data
Source Data Extended Data Fig. 10
Statistical source data.
Rights and permissions
About this article
Cite this article
Zhang, T., Sun, L., Hao, Y. et al. ENO1 suppresses cancer cell ferroptosis by degrading the mRNA of iron regulatory protein 1. Nat Cancer 3, 75–89 (2022). https://doi.org/10.1038/s43018-021-00299-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s43018-021-00299-1
This article is cited by
-
Multi-omics reveals the role of ENO1 in bladder cancer and constructs an epithelial-related prognostic model to predict prognosis and efficacy
Scientific Reports (2024)
-
Mechanisms controlling cellular and systemic iron homeostasis
Nature Reviews Molecular Cell Biology (2024)
-
Carnosine regulation of intracellular pH homeostasis promotes lysosome-dependent tumor immunoevasion
Nature Immunology (2024)
-
The cell biology of ferroptosis
Nature Reviews Molecular Cell Biology (2024)
-
Targeting ferroptosis in hepatocellular carcinoma
Hepatology International (2024)