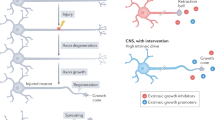
Abstract
Regeneration after injury occurs in axons that lie in the peripheral nervous system but fails in the central nervous system, thereby limiting functional recovery. Differences in axonal signalling in response to injury that might underpin this differential regenerative ability are poorly characterized. Combining axoplasmic proteomics from peripheral sciatic or central projecting dorsal root ganglion (DRG) axons with cell body RNA-seq, we uncover injury-dependent signalling pathways that are uniquely represented in peripheral versus central projecting sciatic DRG axons. We identify AMPK as a crucial regulator of axonal regenerative signalling that is specifically downregulated in injured peripheral, but not central, axons. We find that AMPK in DRG interacts with the 26S proteasome and its CaMKIIα-dependent regulatory subunit PSMC5 to promote AMPKα proteasomal degradation following sciatic axotomy. Conditional deletion of AMPKα1 promotes multiple regenerative signalling pathways after central axonal injury and stimulates robust axonal growth across the spinal cord injury site, suggesting inhibition of AMPK as a therapeutic strategy to enhance regeneration following spinal cord injury.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
The axoplasmic proteomics and AMPK IP–MS data have been deposited in the ProteomeXchange Consortium under accession codes PXD013297 and PXD013318. The pipeline used for the proteomics and AMPK IP analysis is available at: https://github.com/intgenomics/191015.proteomics_analysis. Source data are provided with this paper.
References
Liu, K., Tedeschi, A., Park, K. K. & He, Z. Neuronal intrinsic mechanisms of axon regeneration. Annu. Rev. Neurosci. 34, 131–152 (2011).
Neumann, S. & Woolf, C. J. Regeneration of dorsal column fibers into and beyond the lesion site following adult spinal cord injury. Neuron 23, 83–91 (1999).
Neumann, S., Bradke, F., Tessier-Lavigne, M. & Basbaum, A. I. Regeneration of sensory axons within the injured spinal cord induced by intraganglionic cAMP elevation. Neuron 34, 885–893 (2002).
Saito, A. & Cavalli, V. Signaling over distances. Mol. Cell Proteomics 15, 382–393 (2016).
Yan, D. & Jin, Y. Regulation of DLK-1 kinase activity by calcium-mediated dissociation from an inhibitory isoform. Neuron 76, 534–548 (2012).
Moore, D. L. & Goldberg, J. L. Multiple transcription factor families regulate axon growth and regeneration. Dev. Neurobiol. 71, 1186–1211 (2011).
Gaub, P. et al. HDAC inhibition promotes neuronal outgrowth and counteracts growth cone collapse through CBP/p300 and P/CAF-dependent p53 acetylation. Cell Death Differ. 17, 1392–1408 (2010).
Gaub, P. et al. The histone acetyltransferase p300 promotes intrinsic axonal regeneration. Brain 134, 2134–2148 (2011).
Puttagunta, R. et al. PCAF-dependent epigenetic changes promote axonal regeneration in the central nervous system. Nat. Commun. 5, 3527 (2014).
Hutson, T. H. et al. Cbp-dependent histone acetylation mediates axon regeneration induced by environmental enrichment in rodent spinal cord injury models. Sci. Transl. Med. 11, eaaw2064.
Cho, Y., Sloutsky, R., Naegle, K. M. & Cavalli, V. Injury-induced HDAC5 nuclear export is essential for axon regeneration. Cell 155, 894–908 (2013).
Raivich, G. et al. The AP-1 transcription factor c-Jun is required for efficient axonal regeneration. Neuron 43, 57–67 (2004).
Gao, Y. et al. Activated CREB is sufficient to overcome inhibitors in myelin and promote spinal axon regeneration in vivo. Neuron 44, 609–621 (2004).
Zou, H., Ho, C., Wong, K. & Tessier-Lavigne, M. Axotomy-induced Smad1 activation promotes axonal growth in adult sensory neurons. J. Neurosci. 29, 7116–7123 (2009).
Liu, K. et al. PTEN deletion enhances the regenerative ability of adult corticospinal neurons. Nat. Neurosci. 13, 1075–1081 (2010).
Blackmore, M. G. et al. Kruppel-like factor 7 engineered for transcriptional activation promotes axon regeneration in the adult corticospinal tract. Proc. Natl Acad. Sci. USA 109, 7517–7522 (2012).
Moore, D. L. et al. KLF family members regulate intrinsic axon regeneration ability. Science 326, 298–301 (2009).
Park, K. K. et al. Promoting axon regeneration in the adult CNS by modulation of the PTEN/mTOR pathway. Science 322, 963–966 (2008).
Abe, N., Borson, S. H., Gambello, M. J., Wang, F. & Cavalli, V. Mammalian target of rapamycin (mTOR) activation increases axonal growth capacity of injured peripheral nerves. J. Biol. Chem. 285, 28034–28043 (2010).
Nascimento, A. I., Mar, F. M. & Sousa, M. M. The intriguing nature of dorsal root ganglion neurons: linking structure with polarity and function. Prog. Neurobiol. 168, 86–103.
Hervera, A. et al. Reactive oxygen species regulate axonal regeneration through the release of exosomal NADPH oxidase 2 complexes into injured axons. Nat. Cell Biol. 20, 307–319 (2018).
Usoskin, D. et al. Unbiased classification of sensory neuron types by large-scale single-cell RNA sequencing. Nat. Neurosci. 18, 145–153 (2015).
Clements, M. P. et al. The wound microenvironment reprograms Schwann cells to invasive mesenchymal-like cells to drive peripheral nerve regeneration. Neuron 96, e117 (2017).
Cahoy, J. D. et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J. Neurosci. 28, 264–278 (2008).
Tojkander, S., Gateva, G., Husain, A., Krishnan, R. & Lappalainen, P. Generation of contractile actomyosin bundles depends on mechanosensitive actin filament assembly and disassembly. eLife 4, e06126 (2015).
Xiao, B. et al. Structure of mammalian AMPK and its regulation by ADP. Nature 472, 230 (2011).
Benziane, B. et al. Activation of AMP-activated protein kinase stimulates Na+,K+-ATPase activity in skeletal muscle cells. J. Biol. Chem. 287, 23451–23463 (2012).
Moon, S. et al. Interactome analysis of AMP-activated protein kinase (AMPK)-α1 and -β1 in INS-1 pancreatic beta-cells by affinity purification-mass spectrometry. Sci. Rep. 4, 4376 (2014).
Haider, S. & Pal, R. Integrated analysis of transcriptomic and proteomic data. Curr. Genomics 14, 91–110 (2013).
Kumar, D. et al. Integrating transcriptome and proteome profiling: strategies and applications. Proteomics 16, 2533–2544 (2016).
Rutherford, C. et al. Phosphorylation of Janus kinase 1 (JAK1) by AMP-activated protein kinase (AMPK) links energy sensing to anti-inflammatory signaling. Sci. Signal. 9, ra109 (2016).
Ishizuka, Y. et al. AMP‐activated protein kinase counteracts brain‐derived neurotrophic factor‐induced mammalian target of rapamycin complex 1 signaling in neurons. J. Neurochem. 127, 66–77 (2013).
Huang, B.-P. et al. AMPK activation inhibits expression of proinflammatory mediators through downregulation of PI3K/p38 MAPK and NF-κB signaling in murine macrophages. DNA Cell Biol. 34, 133–141 (2015).
Vazquez-Martin, A., Oliveras-Ferraros, C. & Menendez, J. A. The antidiabetic drug metformin suppresses HER2 (erbB-2) oncoprotein overexpression via inhibition of the mTOR effector p70S6K1 in human breast carcinoma cells. Cell Cycle 8, 88–96 (2009).
Ouchi, N., Shibata, R. & Walsh, K. AMP-activated protein kinase signaling stimulates VEGF expression and angiogenesis in skeletal muscle. Circulation Res. 96, 838–846 (2005).
Huang, W. et al. AMPK plays a dual role in regulation of CREB/BDNF pathway in mouse primary hippocampal cells. J. Mol. Neurosci. 56, 782–788 (2015).
Saha, A. K. et al. Downregulation of AMPK accompanies leucine- and glucose-induced increases in protein synthesis and insulin resistance in rat skeletal muscle. Diabetes 59, 2426–2434 (2010).
Kawaguchi, T., Osatomi, K., Yamashita, H., Kabashima, T. & Uyeda, K. Mechanism for fatty acid ‘sparing’ effect on glucose-induced transcription: regulation of carbohydrate-responsive element-binding protein by AMP-activated protein kinase. J. Biol. Chem. 277, 3829–3835 (2002).
Ning, J. & Clemmons, D. R. AMP-activated protein kinase inhibits IGF-I signaling and protein synthesis in vascular smooth muscle cells via stimulation of insulin receptor substrate 1 S794 and tuberous sclerosis 2 S1345 phosphorylation. Mol. Endocrinol. 24, 1218–1229 (2010).
Lihn, A. S., Jessen, N., Pedersen, S. B., Lund, S. & Richelsen, B. AICAR stimulates adiponectin and inhibits cytokines in adipose tissue. Biochem Biophys. Res Commun. 316, 853–858 (2004).
Sell, H., Dietze-Schroeder, D., Eckardt, K. & Eckel, J. Cytokine secretion by human adipocytes is differentially regulated by adiponectin, AICAR, and troglitazone. Biochem. Biophys. Res. Commun. 343, 700–706 (2006).
Shaw, R. J. LKB1 and AMP‐activated protein kinase control of mTOR signalling and growth. Acta. Physiol. (Oxf.) 196, 65–80 (2009).
Hay, N. & Sonenberg, N. Upstream and downstream of mTOR. Genes Dev. 18, 1926–1945 (2004).
Sokolova, V., Li, F., Polovin, G. & Park, S. Proteasome activation is mediated via a functional switch of the Rpt6 C-terminal tail following chaperone-dependent assembly.Sci. Rep. 5, 14909 (2015).
Djakovic, S. N., Schwarz, L. A., Barylko, B., DeMartino, G. N. & Patrick, G. N. Regulation of the proteasome by neuronal activity and calcium/calmodulin-dependent protein kinase II. J. Biol. Chem. 284, 26655–26665 (2009).
Hasegawa, S., Kohro, Y., Tsuda, M. & Inoue, K. Activation of cytosolic phospholipase A2 in dorsal root ganglion neurons by Ca2+/calmodulin-dependent protein kinase II after peripheral nerve injury. Mol. Pain. 5, 22 (2009).
Schmalbruch, H. Fiber composition of the rat sciatic nerve. Anat. Rec. 215, 71–81 (1986).
Hardie, D. G. AMP-activated/SNF1 protein kinases: conserved guardians of cellular energy. Nat. Rev. Mol. Cell Biol. 8, 774–785 (2007).
Mihaylova, M. M. & Shaw, R. J. The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nat. Cell Biol. 13, 1016–1023 (2011).
Michaelevski, I. et al. Signaling to transcription networks in the neuronal retrograde injury response. Sci. Signal. 3, ra53 (2010).
DeMartino, G. N. & Gillette, T. G. Proteasomes: machines for all reasons. Cell 129, 659–662 (2007).
Jaganathan, S., Malek, E., Vallabhapurapu, S., Vallabhapurapu, S. & Driscoll, J. J. Bortezomib induces AMPK-dependent autophagosome formation uncoupled from apoptosis in drug resistant cells. Oncotarget 5, 12358 (2014).
Kwak, H. J. et al. Bortezomib attenuates palmitic acid-induced ER stress, inflammation and insulin resistance in myotubes via AMPK dependent mechanism. Cell. Signal. 28, 788–797 (2016).
Schmidt, M. & Finley, D. Regulation of proteasome activity in health and disease. Biochim. Biophys. Acta 1843, 13–25 (2014).
Buneeva, O. & Medvedev, A. Ubiquitin-independent degradation of proteins in proteasomes. Biochem. (Mosc.), Suppl. Ser. B: Biomed. Chem. 12, 203–219 (2018).
St-Arnaud, R. Dual functions for transcriptional regulators: myth or reality? J. Cell. Biochem. 75, 32–40 (1999).
Lehr, N. V. D., Johansson, S. & Larsson, L.-G. Implication of the ubiquitin/proteasome system in Myc-regulated transcription. Cell Cycle 2, 402–406 (2003).
Shin, H.-J. R. et al. AMPK–SKP2–CARM1 signalling cascade in transcriptional regulation of autophagy. Nature 534, 553–557 (2016).
Wang, S. et al. AMPKα2 deletion causes aberrant expression and activation of NAD(P)H oxidase and consequent endothelial dysfunction in vivo: role of 26S proteasomes. Circ. Res. 106, 1117 (2010).
Xu, J., Wang, S., Viollet, B. & Zou, M.-H. Regulation of the proteasome by AMPK in endothelial cells: the role of O-GlcNAc transferase (OGT). PloS ONE 7, e36717 (2012).
Tang, Q. et al. Inhibition of integrin-linked kinase expression by emodin through crosstalk of AMPKα and ERK1/2 signaling and reciprocal interplay of Sp1 and c-Jun. Cell. Signal. 27, 1469–1477 (2015).
Vasamsetti, S. B. et al. Metformin inhibits monocyte-to-macrophage differentiation via AMPK-mediated inhibition of STAT3 activation: potential role in atherosclerosis. Diabetes 64, 2028–2041 (2015).
Yang, W. et al. Regulation of transcription by AMP-activated protein kinase: phosphorylation of p300 blocks its interaction with nuclear receptors. J. Biol. Chem. 276, 38341–38344 (2001).
Lee, J. M. et al. AMPK-dependent repression of hepatic gluconeogenesis via disruption of CREB.CRTC2 complex by orphan nuclear receptor small heterodimer partner. J. Biol. Chem. 285, 32182–32191 (2010).
Ramamurthy, S., Chang, E., Cao, Y., Zhu, J. & Ronnett, G. AMPK activation regulates neuronal structure in developing hippocampal neurons. Neuroscience 259, 13–24 (2014).
Amato, S. et al. AMP-activated protein kinase regulates neuronal polarization by interfering with PI 3-kinase localization. Science 332, 247–251 (2011).
Williams, T., Courchet, J., Viollet, B., Brenman, J. E. & Polleux, F. AMP-activated protein kinase (AMPK) activity is not required for neuronal development but regulates axogenesis during metabolic stress. Proc. Natl Acad. Sci. USA 108, 5849–5854 (2011).
Inoki, K. et al. TSC2 integrates Wnt and energy signals via a coordinated phosphorylation by AMPK and GSK3 to regulate cell growth. Cell 126, 955–968 (2006).
Saijilafu et al. PI3K-GSK3 signalling regulates mammalian axon regeneration by inducing the expression of Smad1. Nat. Commun. 4, 2690 (2013).
Joshi, Y. et al. The MDM4/MDM2-p53-IGF1 axis controls axonal regeneration, sprouting and functional recovery after CNS injury. Brain 138, 1843–1862 (2015).
Belin, S. et al. Injury-induced decline of intrinsic regenerative ability revealed by quantitative proteomics. Neuron 86, 1000–1014 (2015).
Tosca, L., Rame, C., Chabrolle, C., Tesseraud, S. & Dupont, J. Metformin decreases IGF1-induced cell proliferation and protein synthesis through AMP-activated protein kinase in cultured bovine granulosa cells. Reproduction 139, 409–418 (2010).
Roe, N. D. et al. Targeted deletion of PTEN in cardiomyocytes renders cardiac contractile dysfunction through interruption of Pink1-AMPK signaling and autophagy. Biochim. Biophys. Acta 1852, 290–298 (2015).
Rogacka, D., Piwkowska, A., Audzeyenka, I., Angielski, S. & Jankowski, M. Involvement of the AMPK-PTEN pathway in insulin resistance induced by high glucose in cultured rat podocytes. Int. J. Biochem. Cell Biol. 51, 120–130 (2014).
Ohtake, Y. et al. Promoting axon regeneration in adult CNS by targeting liver kinase B1. Mol. Ther. 27, 102 (2019).
Hervera, A. et al. PP4-dependent HDAC3 dephosphorylation discriminates between axonal regeneration and regenerative failure. EMBO J. 38, e101032 (2019).
Rishal, I. et al. Axoplasm isolation from peripheral nerve. Dev. Neurobiol. 70, 126–133 (2010).
Hsu, J. L., Huang, S. Y., Chow, N. H. & Chen, S. H. Stable-isotope dimethyl labeling for quantitative proteomics. Anal. Chem. 75, 6843–6852 (2003).
Cox, J. & Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 (2008).
Benaglia, T., Chauveau, D., Hunter, D. & Young, D. mixtools: an R package for analyzing finite mixture models. J. Stat. Softw. 32, 1–29 (2009).
Kim, S., Carruthers, N., Lee, J., Chinni, S. & Stemmer, P. Classification-based quantitative analysis of stable isotope labeling by amino acids in cell culture (SILAC) data. Comput. Methods Programs Biomed. 137, 137–148 (2016).
Scrucca, L., Fop, M., Murphy, T. B. & Raftery, A. E. mclust 5: clustering, classification and density estimation using Gaussian finite mixture models. R. J. 8, 289–317 (2016).
Sikorski, T. W. et al. Proteomic analysis demonstrates activator-and chromatin-specific recruitment to promoters. J. Biol. Chem. 287, 35397–35408 (2012).
Rauniyar, N., Gupta, V., Balch, W. E. & Yates, J. R. III Quantitative proteomic profiling reveals differentially regulated proteins in cystic fibrosis cells. J. Proteome Res. 13, 4668–4675 (2014).
Edwards, A. W. F. in Landmark Writings in Western Mathematics 1640-1940 856–870 (Elsevier, 2005).
Steward, O., Zheng, B. & Tessier-Lavigne, M. False resurrections: distinguishing regenerated from spared axons in the injured central nervous system. J. Comp. Neurol. 459, 1–8 (2003).
Acknowledgements
We acknowledge start-up funds from the Department of Brain Sciences, Imperial College London (S.D.G.), and the Hertie Foundation for financial support (S.D.G.); the International Spinal Research Trust (Nathalie Rose Barr PhD awards to E.S and E.M.); Wings for Life (S.D.G.); the Deutsche Forschungsgemeinschaft (S.D.G.); the Medical Research Council (S.D.G.); and Rosetrees Trust (S.D.G.). The research was supported by the National Institute for Health Research Imperial Biomedical Research Centre (S.D.G.). The views expressed are those of the author(s) and not necessarily those of the National Health Service, the National Institute for Health Research or the Department of Health. The Q Exactive Plus mass spectrometer was funded by Deutsche Forschungsgemeinschaft INST 247/766-1 FUGG. We would also like to thank Mike Fainzilber for critical discussion of the data and the manuscript.
Author information
Authors and Affiliations
Contributions
G.K. designed and performed experiments, performed data analysis and wrote the paper; L.Z. designed and performed experiments and performed data analysis; E.S. performed experiments and data analysis; I.P. performed data analysis; F.D.V. performed experiments; T.H.H. performed experiments; E.M. performed data analysis; A.F. performed the mass spectrometry experiments and data analysis; P.L.M. performed experiments; K.S. performed data analysis; R.P. provided experimental advice and edited the paper and S.D.G. designed experiments, provided funding and wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Primary Handling Editor: George Caputa.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Venn diagrams show high neuronal enrichment of axoplasm proteins.
a, Area-proportional Venn diagram showing the overlap between the proteins identified in our axoplasm dataset and previously published DRG neuron or Schwann cell specific transcriptomic datasets. Red: peripheral axoplasm; pink: central axoplasm. b, Area-proportional Venn diagram showing the overlap between proteins represented in our axoplasm dataset and previously published DRG neuron or Oligodendrocyte specific transcriptomic datasets.
Extended Data Fig. 2 Injury to the DRG peripheral and central axons elicit differential protein expression profiles.
a, Heat map of the log2 ratio of differentially expressed proteins (FDR < 0.05) identified by mass spectrometry in the axoplasmic extract from peripheral and central DRG axons. Comparisons include peripheral nerve after sciatic nerve axotomy (SNA) vs sham (control injury); central branches after dorsal column axotomy (DCA) vs Lam (control injury). Red and Blue indicates up- and down-regulated proteins, respectively. b, Venn diagram shows the number of differentially expressed proteins following SNA and DCA (FDR < 0.05, absolute log2 ratio > 0.58) and how many proteins are overlapped between these two compartments after injury. c–f, Heatmap graphs show Gene ontology (GO) analysis of differentially expressed proteins following SNA and DCA. Differentially expressed proteins were selected with cut off FDR < 0.05, log2 ratio > 0.58 (Red) or log2 ratio < -0.58 (Blue). Gene ontology was performed by DAVID. Only enriched GO items with Fisher’s exact P value < 0.05 were selected and categories that share the same protein groups were combined in one category. Categories in orange of (c, e, f) are known to be regulated by or to regulate AMPK.
Extended Data Fig. 3 Regeneration associated genes and immunoblotting validation of axoplasmic protein expression identified by mass spectrometry.
a, Bar graphs show axoplasmic proteins belonging to RAGs with FDR < 0.05, log2 ratio > 0.58 (SNA vs Sham) that are plotted in a log2 ratio scale. b, d, Immunoblots show validation of axoplasmic proteins identified by mass spectrometry after SNA vs Sham or DCA vs Lam (SN: sciatic nerve; SC: spinal cord). c, e, Bar graphs show quantification of immunoblots in (b) and (d) respectively. n = 3 independent experiments. The expression level of each protein was quantified after normalization to GAPDH. Values represent means ± SEM. Two-tailed unpaired Welch’s t-test.
Extended Data Fig. 4 AMPKα1 expression in NF200, PARV, CGRP and IB4 positive DRG neurons.
a, Representative fluorescence images of immunostaining for AMPKα1 and parvalbumin (PARV) or CGRP, or IB4+ in DRG neurons. n = 3. Scale bar, 100 μm. b, Representative fluorescence images of immunostaining for AMPKα1 and parvalbumin (PARV) in DRG neurons following Sham and SNA. n = 3. Scale bar, 100 μm. c, Percentage of NF200, PARV, CGRP or IB4 positive neurons expressing AMPKα1. d, Quantification of immunostaining for AMPKα1 expression level of (b). n = 3 mice each group. Each mouse represents an independent replicate. The relative AMPKα1 expression level was quantified after normalization to the background (secondary antibody only). Values represent means ± SEM. Two-tailed unpaired Welch’s t-test.
Extended Data Fig. 5 AMPKα1 expression in DRG neurons following AMPKα1 conditional deletion or overexpression.
a, Representative fluorescence images of immunostaining for GFP; AMPKα1 and DAPI in cultured DRG cells dissected from AMPKα1 floxed mice 48 h after transfection with AAV-GFP or AAV-Cre-GFP. n = 3. Scale bar, 100 μm. b, Representative fluorescence images of immunostaining for GFP; AMPKα1 and DAPI in cultured DRG cells after electroporation with GFP plasmid or AMPKα1 plasmid at 48 h. Arrows show non-pycnotic DAPI positive nuclei. Scale bar, 50 μm. c, Quantification of the percentage of cells with non-pycnotic nuclei (b). n = 3 independent experiments. Values represent means ± SEM. Two-tailed unpaired Welch’s t-test.
Extended Data Fig. 6 AMPKα1 and AMPKα2 interaction with PSMC5.
a, b, Immunoblots for PSMC5, AMPKα1 and AMPKα2 after AMPKα1 and AMPKα2 IP from DRG. Repeated twice with similar results.
Extended Data Fig. 7 In vivo AMPKα1 conditional deletion in DRG neurons.
a, Representative images of AMPKα1 and GFP immunostaining in DRG sections 5 weeks following AAV-GFP or AAV-Cre-GFP sciatic nerve injection. Scale bar, 50 μm. b, Representative images of AMPKα2 and GFP immunostaining in DRG sections 5 weeks following AAV-GFP or AAV-Cre-GFP sciatic nerve injection. Arrowheads mark GFP positive neurons showing the presence or loss of AMPKα2 staining. Scale bar, 50 μm. c, Quantification of AMPKα1 level of (a). n = 3 mice. Values represent means ± SEM. Two-tailed unpaired Welch’s t-test. d, Quantification of AMPKα2 level of (b). n = 3 mice. Values represent means ± SEM. Two-tailed unpaired Welch’s t-test.
Extended Data Fig. 8 GFP and dextran co-localization in DRG neurons.
a, Representative images of DRG sections from AAV-Cre-GFP sciatic nerve injected mice co-immunostained with antibodies against GFP and Dextran. Scale bar, 50 μm. b, c, Quantification of the percentage of GFP+ and Dextran+ / GFP+ cells following AAV-GFP or AAV-Cre-GFP. AAV-GFP, n = 13 mice; AAV-Cre-GFP, n = 10 mice. Percentage of GFP positive cells was calculated as the ratio of GFP+ versus TUJ1+ cells; percentage of Dextran+/GFP+ was calculated as the ratio of Dextran+/GFP+ versus TUJ1+ cells. Values represent means ± SEM. Two-tailed unpaired Welch’s t-test. d, Longitudinal spinal cord section 5 weeks after SCI showing axonal labelling across the injured dorsal columns following deletion of AMPKα1. Dorsal column axons are labelled by sciatic nerve injected Dextran. Asterisk indicates the lesion epicentre. D; dorsal; V; ventral, C; caudal; R; rostral. Scale bar; 250 μm. Similar results were found in eight AMPKα1 conditionally deleted mice. The quantification source data is provided in statistical source data, Fig. 8b.
Supplementary information
Supplementary Information
Supplementary Tables 3, 6 and 7
Supplementary Table 1
Proteomic dataset
Supplementary Table 2
GO KEGG SNAvsSham and DCAvsLam
Supplementary Table 4
Combined KEGG pathways
Supplementary Table 5
AMPK IP–MS
Source data
Source Data Fig. 2
Unprocessed Western Blots
Source Data Fig. 2
Statistical Source Data
Source Data Fig. 3
Unprocessed Western Blots
Source Data Fig. 3
Statistical Source Data
Source Data Fig. 4
Unprocessed Western Blots
Source Data Fig. 4
Statistical Source Data
Source Data Fig. 5
Unprocessed Western Blots
Source Data Fig. 5
Statistical Source Data
Source Data Fig. 6
Unprocessed Western Blots
Source Data Fig. 6
Statistical Source Data
Source Data Fig. 7
Statistical Source Data
Source Data Fig. 8
Statistical Source Data
Source Data Extended Data Fig. 3
Unprocessed Western Blots
Source Data Extended Data Fig. 3
Statistical Source Data
Source Data Extended Data Fig. 4
Statistical Source Data
Source Data Extended Data Fig. 5
Statistical Source Data
Source Data Extended Data Fig. 6
Unprocessed Western Blots
Source Data Extended Data Fig. 7
Statistical Source Data
Source Data Extended Data Fig. 8
Statistical Source Data
Rights and permissions
About this article
Cite this article
Kong, G., Zhou, L., Serger, E. et al. AMPK controls the axonal regenerative ability of dorsal root ganglia sensory neurons after spinal cord injury. Nat Metab 2, 918–933 (2020). https://doi.org/10.1038/s42255-020-0252-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s42255-020-0252-3
This article is cited by
-
Unleashing Axonal Regeneration Capacities: Neuronal and Non-neuronal Changes After Injuries to Dorsal Root Ganglion Neuron Central and Peripheral Axonal Branches
Molecular Neurobiology (2024)
-
Circular RNAs in organ injury: recent development
Journal of Translational Medicine (2022)
-
NPC transplantation rescues sci-driven cAMP/EPAC2 alterations, leading to neuroprotection and microglial modulation
Cellular and Molecular Life Sciences (2022)
-
Differential hippocampal protein expression between normal mice and mice with the perioperative neurocognitive disorder: a proteomic analysis
European Journal of Medical Research (2021)