Abstract
Shox2 plays a vital role in the morphogenesis and physiological function of the sinoatrial node (SAN), the primary cardiac pacemaker, manifested by the formation of a hypoplastic SAN and failed differentiation of pacemaker cells in Shox2 mutants. Shox2 and Nkx2-5 are co-expressed in the developing SAN and regulate the fate of the pacemaker cells through a Shox2-Nkx2-5 antagonistic mechanism. Here we show that simultaneous inactivation of Nkx2-5 in the SAN of Shox2 mutants (dKO) rescued the pacemaking cell fate but not the hypoplastic defects, indicating uncoupling of SAN cell fate determination and morphogenesis. Single-cell RNA-seq revealed that the presumptive SAN cells of Shox2-/- mutants failed to activate pacemaking program but remained in a progenitor state preceding working myocardium, while both wildtype and dKO SAN cells displayed normal pacemaking cell fate with similar cellular state. Shox2 thus acts as a safeguard but not a determinant to ensure the pacemaking cell fate through the Shox2-Nkx2-5 antagonistic mechanism, which is segregated from its morphogenetic regulatory function in SAN development.
Similar content being viewed by others

Introduction
The cardiac rhythm is controlled by the sinoatrial node (SAN), functionally known as the pacemaker1,2,3,4. It has been demonstrated that by embryonic day 7.5 (E7.5) in mice, mesenchymal cardiac progenitor cells (MCPC) possess pacemaking potential, but only a small portion of these cells eventually develop into cardiac conduction system including the SAN, with the majority of MCPC maturating into working myocardium5,6,7,8,9,10,11,12. The SAN primordium appears at E10.5 based on histological structure and becomes further mature and fully functional at E13.59,13. Previous studies have revealed a complex genetic network involving many genes in the regulation of SAN development, including Hcn4, Tbx3, Tbx5, Tbx18, Isl1, Smoc2, Bmp4, and Shox24,14,15,16,17. Among them, Shox2, Tbx3, Isl1, and Hcn4 are specifically expressed in the entire developing SAN, with Hcn4 being regarded as a functional marker of pacemaking capability18,19. Tbx3 deficient mice develop a morphologically normal SAN with compromised function20. In contrast, Tbx18 deficient mice form a hypoplastic yet functional SAN12. These threads of evidence implicate that the molecular mechanisms underlying SAN pacemaking cell fate determination and morphogenesis, a biological process that governs the development of tissue or organ shape by orchestrating the spatial arrangement of cells during embryonic development21, are not identical.
Previous studies showed that Shox2 is indispensable for the physiological function and morphogenesis of the SAN, as Shox2 null mice display hypoplasia of the venous pole structures and failed differentiation of SAN cells, embryonic bradycardia, and upregulation of chamber-program genes in the venous pole structures22,23,24. Nkx2-5 was originally thought to be detrimental to SAN development, and Shox2 was shown to regulate SAN development by preventing ectopic Nkx2-5 activation in the SAN head domain10,22,23,25,26. Further studies showed that Shox2 counterbalances Nkx2-5 in the developing SAN junction domain, an interface between the SAN and atrium that is specialized by Nkx2-5 expression, to regulate the cell fate decisions between pacemaking cells and working myocardial cells27. We have recently found that Nkx2-5 inactivation in the SAN junction cells confers them with the property of the SAN head cells and impairs SAN function, but does not affect SAN morphogenesis28, further supporting the existence of morphogenic genetic programs in the SAN that are separated from pacemaking cell fate determination. However, the molecular mechanisms that distinctly control each process remain unknown.
Results and discussion
Maintenance of pacemaking cell fate in the developing SAN requires a minimal level of Shox2 expression
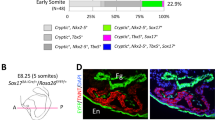
To better understand how Shox2 controls SAN morphogenesis and differentiation, we investigated the physiological consequence of reduced Shox2 expression at different dosages during SAN development. We used a Shox2-Cre knock-in allele that express Cre recombinase instead of Shox2 as Shox2 null allele in our studies29. The Shox2-Cre heterozygous mice appeared indistinguishable from wildtype mice, never exhibited any obvious unusual behaviors, abnormal ECG recordings, and altered gene expression profiles. In the Shox2HA-Neo/+ allele that was used as a hypomorphic allele in this study, the presence of the neomycin cassette interferes with Shox2 expression, resulting in a reduction of Shox2 expression to 65%30 (Supplementary Fig. 1a; Supplementary Data 1). A combination of different Shox2 alleles enabled the manipulation of Shox2 expression at different dosages (Supplementary Table 1). As Hcn4 reflects the pacemaking capability of cells in the SAN, we examined Hcn4 expression in mice carrying different compound Shox2 alleles at various stages. We found that Shox2HA-Neo/Cre mice, which had Shox2 production reduced to 15% and ectopic Nkx2-5 expression (Fig. 1b) compared to wildtype mice (Fig. 1a), displayed Hcn4+ domains in the Shox2+ lineage derived venous pole structures (Fig. 1b). However, such mice had a reduced SAN size with retained head and junction domains and shortened venous valves (Fig. 1b). These results indicate that a 15% level of Shox2 production compared to wildtype was sufficient for the determination and maintenance of pacemaking cell fate, but such residual dosage failed to support normal SAN morphogenesis. As compared to its requirement for normal SAN morphogenesis, the maintenance of pacemaking cell fate requires a lower dose of Shox2. The hypoplastic SAN morphology but with the retaining of the pacemaking cell fate in the Shox2HA-Neo/Cre suggests the existence of distinct Shox2-mediated genetic modules in SAN development dictated by different Shox2 levels.
Immunofluorescence shows the expression of Hcn4, Nkx2-5, and Shox2 in the SAN head, SAN junction, and venous valves in E14.5 wildtype control (a) and Shox2HA-Neo/Cre mice (b) (n = 3 for each genotype). Note in the whole SAN (magenta dash-lines), SAN head (red arrowheads), SAN junction (green arrowheads), and venous valves (yellow arrows), compared to controls (a), Shox2HA-Neo/Cre mice (b) exhibited residual Shox2 expression, but visible ectopic Nkx2-5 expression in the SAN head domain and reduced size of these structures. However, Hcn4 expression was retained. RA right atrium, RSVC right superior vena cava. Bar: 50 μm.
Deletion of Nkx2-5 in Shox2 mutant background rescues the pacemaking cell fate and ameliorates the impaired physiological function of the SAN
We have previously reported that Shox2 maintains the pacemaker program in the Nkx2-5+ junction domain of the SAN by antagonizing Nkx2-5 transcriptional output27, we therefore asked whether rebalancing the Shox2-Nkx2-5 antagonism can reset the original pacemaking cell fate in the developing SAN. We took advantage of the compound Shox2Cre/Cre;Nkx2-5F/F alleles to delete Nkx2-5 in the Shox2 null background29,31. In contrast to the embryonic lethality of Shox2Cre/Cre mice at mid-gestation due to cardiac defects, Shox2Cre/Cre;Nkx2-5F/F mice (dKO) survived to postnatal day 0 (P0), indicating amelioration of the impaired physiological function of the SAN. Surface electrocardiogram (ECG) measurements on P0 mice showed regular but slower cardiac rhythm and the normal P waves in the dKO mice as compared to controls (n = dKO: 7/7; WT: 16/16) (Fig. 2a). Additionally, whole-cell patch-clamp recording on cells derived from the Shox2+ population in the SAN of E13.5 dKO mice showed typical AP configurations comparable to controls (Fig. 2d; Supplementary Fig. 2; Supplementary Data 1). Immunohistochemistry and in situ hybridization assays showed that, unlike the lack of Tbx3 and Hcn4 expression in Shox2 null mice22, dKO mice retained the expression of SAN markers, including Tbx3, Hcn4 (Fig. 2e). This observation strongly implies the preservation of the pacemaking cell fate in dKO mice. Morphometric analyses on the SAN of dKO and Shox2 null mice (Supplementary Fig. 1b–d) revealed that the SAN morphology in dKO mice appeared comparable to that in Shox2 null mice22,23. However, upon closer inspection, we discovered that the Hcn4 expression in the SAN head domain (as opposed to the junction domain demarcated by Nkx2-5 expression) of dKO mice was notably restored (Fig. 2b; Supplementary Fig. 1b–d), following the deletion of Nkx2-5 in the Shox2-null background. This result suggests that the dKO mice retain the pacemaking cell fate by developing a hypoplastic SAN lacking the junction domain (Fig. 2c; Supplementary Data 1). Given previous reports indicating that the structural characteristics of the microenvironment surrounding pacemaking cells in the SAN can impact their function through local mechanics32, the sinus bradycardia ECG defects observed in dKO mice (Fig. 2a) may be attributed to the altered SAN morphology (hypoplasia). Above all, these observations demonstrate that rebalancing Shox2-Nkx2-5 antagonism by deletion of Nkx2-5 in Shox2-null background rescues the pacemaking cell fate and ameliorates the impaired SAN physiological function but has no impact on SAN morphogenesis observed in Shox2-/- mice. The exclusive function of Shox2-Nkx2-5 antagonistic mechanism in the regulation of SAN cell fate, but not in SAN morphogenesis, suggests that there are alternative genetic programs utilized by Shox2 to control SAN morphogenesis.
a Surface ECG recordings reveal normal P waves and regular rhythms but bradycardia in P0 Shox2Cre/Cre;Nkx2-5F/F;R26RmTmG mice, compared with Shox2Cre/+;R26RmTmG mice. Blue arrowheads point to P waves. b Immunostaining shows the absence of the Nkx2-5+ SAN junction structure and reduced size of the SAN head in P0 Shox2Cre/Cre;Nkx2-5F/F;R26RmTmG mice, compared with Shox2Cre/+;R26RmTmG mice. The SAN region is marked by the magenta dash-lines, and the yellow dash-lines separate the SAN head from the SAN junction (green stars). c 3D reconstructions of the SAN (based on Hcn4 immunostaining) in E13.5 Shox2Cre/+;R26RmTmG and Shox2Cre/Cre;Nkx2-5F/F;R26RmTmG mice (n = 3 for each genotype), respectively, reveal the absence of the SAN junction (green arrowheads) and dramatically reduced size of the entire SAN in the mutants, as confirmed by statistical analysis. d Typical SAN AP configurations were recorded in GFP+ cells from the SAN in E13.5 Shox2Cre/Cre;Nkx2-5F/F;R26RmTmG mice (n = 13), which were comparable to that seen in the SAN cells from E13.5 Shox2Cre/+;R26RmTmG mice (n = 5). e Immunostaining shows colocalization of Shox2, Nkx2-5, Hcn4, Tbx3, and Cx40 in the SAN head (magenta arrowheads) of E13.5 Shox2Cre/+;R26RmTmG and Shox2Cre/Cre;Nkx2-5F/F;R26RmTmG mice. Similar to Shox2Cre/+;R26RmTmG control mice, deletion of Nkx2-5 in the SAN of Shox2 mutants (Shox2Cre/Cre;Nkx2-5F/F;R26RmTmG) retains the expression of the SAN markers. The presented data are mean ± SEM. Statistical analysis was performed with Student’s t-test (two-tailed, **P < 0.01). RA right atrium, dKO Shox2Cre/Cre, Nkx2-5F/F; RSVC right superior vena cava. Bar: 50 μm.
Single-cell transcriptomic analysis confirms the rescued pacemaking cell fate of the dKO SAN
The rescued pacemaking cell fate but not morphology of the SAN in dKO mice as well as the failure of SAN formation in the Shox2-null mice provided a unique model (Supplementary Table 2) for us to investigate how Shox2 interacts with Nkx2-5 to separately safeguard the pacemaker fate and regulate the SAN morphogenesis. To do so, we compounded three genetically modified alleles to generate Shox2Cre/Cre;Nkx2-5F/F;R26RmTmG (dKO), Shox2Cre/Cre;R26RmTmG (Shox2 null), and Shox2Cre/+;R26RmTmG (control) mice, and performed single-cell RNA-seq (scRNA-seq) on Shox2+ cells (labeled by GFP) from the SAN of E13.5 mice with these three different genotypes (control, Shox2-null, and dKO). The datasets obtained were subjected together to unsupervised clustering, resulting in 8 distinct clusters, including Fibroblast, endothelial cells, mesothelial cells (MCs), cardiomyocytes (CM), Macrophages, and epithelial cells (EPIs)28,33,34,35,36,37,38,39 (Fig. 3a). Based on the expression of Tnnt2 and GFP, we were able to identify a Shox2+ cardiomyocyte population (Fig. 3b, c). The Shox2+ cardiomyocytes were then further distinguished into three sub-populations (sP0, sP1, and sP2) (Fig. 3d; Supplementary Fig. 3a). While sP0 and sP2 showed the closest phylogenic distance and expressed pan-SAN marker Hcn440 (Fig. 3e, f), these two sub-populations were defined as pacemaker cells, the sP1 sub-population seemed likely the residual venous valve cells, as they have similar transcriptome with the venous valve we reported previously28. Further unsupervised sub-clustering reclassified the pacemaker cells into two groups, with the control and dKO pacemaker cells falling in group 0 (G0) and Shox2-null pacemaker cells in group 1 (G1) (Fig. 3g; Supplementary Fig. 3b, b’). These results indicate that the control and dKO cells share a high transcriptomic similarity that deviates from that of Shox2-null cells. To infer the cellular states of the pacemaker cells, we used Monocle241 to pseudo-temporally order the pacemaker cells along a developmental trajectory to the differentiated pacemaking cells. Pseudotime trajectory analysis showed that while the control and dKO samples exhibit the most differentiated cellular state, Shox2-null sample displays the most primitive and under-differentiated cellular state (Fig. 3h, i; Supplementary Fig. 3c). This is evidenced by the increased expression of several widely recognized cardiac progenitor cell markers42, including Abcg2. Abcg2, which serves as the molecular marker for identifying side population progenitor cells in multiple adult tissues43, including the adult heart44, is also documented to be transiently expressed during the early stages of heart development45. Thus, the pacemaker cell fate-oriented differentiation process appears unaffected in the dKO cells but is blocked at a progenitor state in the Shox2-null cells, as compared to the control group. These results are in line with the rescued pacemaking cell fate in the dKO mice as shown above, providing a solid foundation for further analyses.
a UMAP visualization of the eight clusters from the SAN and its adjacent atrial cells of E13.5 Shox2Cre/+;R26RmTmG (control), Shox2Cre/Cre;R26RmTmG (Shox2 null), and Shox2Cre/Cre;Nkx2-5F/F;R26RmTmG (dKO) mice. b, c VlnPlot shows the expression of Tnnt2 and GFP probability distributions across eight clusters. d–f Analysis of three sub-populations from Shox2+ (represented by GFP+) cardiomyocytes defines sP0 and sP2 as the SAN cells. g Sankey Plot showing the SAN cells of control, Shox2 null, and dKO reclassified into two groups. h, i Pseudotemporal ordering of SAN cells reveals that Shox2 null cells exhibited the pseudo-start state, while the control and dKO cells displayed the pseudo-end state. Cells are colored by the genotype of samples (i). Cells with dark color represent the pseudo-start, and the bright color represent the pseudo-end (h). CM cardiomyocyte, EC endothelial cells, MC mesothelial cells, EPI epithelial cells, C5, C7 cluster number 5, 7.
With the distinct physiological and morphological traits of the SAN in the three types of mice with distinct genetic background mentioned above, we reasoned that comparative analyses of the transcriptomic profiles between different sample would allow us to segregate the morphogenetic regulatory factors that are downstream of Shox2 from the pacemaking cell fate determination factors (Supplementary Table 2). To identify genes that are potentially involved in SAN morphogenesis, we compared the transcriptomic profiles between the dKO and control mice using Seurat46,47. We identified 1025 genes that are differentially expressed between the dKO and control mice (set 1). Gene ontology (GO) analysis on the set 1 differentially expressed genes (DEGs) revealed the enrichment of morphogenesis-associated GO terms, such as muscle tissue morphogenesis, muscle organ morphogenesis, and cardiac muscle tissue morphogenesis (Fig. 4a). In an attempt to identify genes that are involved in SAN cell fate determination, we then compared the transcriptomes between the dKO and Shox2 null samples and identified 1462 DEGs (set 2). Conversely, the identified set 2 DEGs included genes that are known to participate in pacemaking cell fate regulation, such as Hcn4, Smoc2, and Gata648. These results suggest the feasibility of segregating the SAN morphogenetic regulatory factors from the pacemaking cell fate determination factors by comparative analyses on the transcriptomic profiles of the three samples. To segregate them, we then intersected the DEGs in set 1 and set 2 and found out that except 561 genes that were shared by these two sets, there are 464 genes that are exclusively presented in set 1, and 901 genes that were found exclusively in set 2 (Fig. 4c). These two gene sets are thus potentially involved in the segregation of SAN morphogenesis from cell fate determination. Heatmap analysis confirmed the differential expression of these genes (Fig. 4b). Furthermore, the differential expression of several selected genes including Hcn4, Smoc2, Bmp10, Gata6, Mef2a, and Tbx18 was further confirmed by pseudotime trajectory analysis and by in situ hybridization and immunostaining assays (Fig. 4d, e). Interestingly, although it was shown previously that Pitx2 and Tbx20 act upstream of Nkx2-522,30,49,50, we observed ectopic expression of Pitx2 and Tbx20 in the scRNA-seq profile of the dKO SAN (Fig. 4b), suggesting that the Shox2-Nkx2-5 antagonistic mechanism has a negative feedback effect on the upstream regulator genes, which warrants future investigation. Above all, our unique genetically modified mouse lines allowed us to segregate genes involved in the regulation of morphogenesis from those involved in cell fate determination downstream from Shox2 and Nkx2-5 in SAN development.
a Go analysis on the DEGs between control and dKO reveals GO terms associated with heart development. b Heatmap shows the genes involved in the determination of the pacemaking cell fate and SAN morphogenetic regulation across the three samples. c Venn diagram depicts the number of overlap genes between pacemaking cell fate determination and SAN morphogenetic regulation. d Pseudotime analysis unravels elevated expression of Gata6, Hcn4, Mef2a, and Smoc2 along pseudotime trajectory. e In situ hybridization and immunostaining verified the expression of Smoc2, Tbx18, Bmp10, and Hcn4 in the SAN of E13.5 control, Shox2 null, and dKO mice. RA right atrium, RSVC right superior vena cava. Bar: 50 μm.
ChIP-seq analysis reveals that the segregation of SAN morphogenesis from cell fate determination attributes to a functional interplay between Shox2 and Nkx2-5
Since the two gene sets are identified from Shox2 and Nkx2-5 mutant mice, to verify the identified genes and to further understand how Shox2 interacts with Nkx2-5 to regulate SAN morphogenesis and cell fate determination separately, we re-analyzed our published Shox2 and Nkx2-5 ChIP-seq datasets on the developing heart27. As outlined in our previous report27, Shox2 and Nkx2-5 exhibit genome-wide co-occupancy (Supplementary Fig. 4a). However, characterization of binding peak distribution patterns for Shox2 and Nkx2-5 unveils that Shox2 exhibits a higher affinity for binding to the promoter regions (82.65%) compared to Nkx2-5 (64.81%). Conversely, Nkx2-5 displays a stronger preference for binding to the distal cis-regulatory elements, such as distal intergenic regions (Supplementary Fig. 4b). Intersection of Shox2 and Nkx2-5 binding peaks reveals that the co-binding regions of Shox2-Nkx2-5 are predominantly located within the promoter regions, similar to the distribution of Shox2 peaks (Supplementary Fig. 4c). Interestingly, when functionally categorizing the target genes associated with Shox2 binding, Shox2-Nkx2-5 co-binding, and Nkx2-5 binding peaks, it becomes evident that these genes play distinct roles in cardiac-related GO terms. Specifically, genes bound by Shox2, including those that are co-bound by Shox2-Nkx2-5, were observed to contribute to pacemaker cell differentiation and SAN morphogenesis (Fig. 5a). On the other hand, the genes exclusively bound by Nkx2-5 were linked to the cardiac cell fate commitment (Fig. 5a). This result aligns with Nkx2-5’s binding preference and is consistent with our previous study, where we highlighted its role in defining a subpopulation of pacemaker cells27. More interestingly, only the genes with individual binding of Shox2 and Nkx2-5, rather than co-binding of Shox2-Nkx2-5, are classified within the context of cardiac pacemaker cell development and differentiation (Fig. 5a), implicating that the segregation of Shox2’s morphogenetic regulatory function from its cell fate safeguarding role arises due to combinational interactions between Shox2 and Nkx2-5. These results not only reinforce our segregated gene outcomes, but also underscore the significance of delving deeper into the intricate mechanisms of Shox2-Nkx2-5 antagonism. Therefore, we conducted an intersection between the list of 3999 genes displaying exclusive Nkx2-5 binding (Fig. 5b) and the gene set 2 (Fig. 4c), which is potentially involved in cell fate determination. This analysis aimed to pinpoint genes associated with SAN cell fate commitment, yielding a list of 333 genes directly targeted by Nkx2-5. Notably, this list of genes encompasses those that have been previously implicated in cardiac cell fate determination, such as Smoc248 (Fig. 5c) and Taz51 (Supplementary Data 2). To distinguish genes involved in safeguarding cell fate from those associated with morphogenesis, we performed further intersections on the following gene groups: firstly, the group of 1242 genes with both Shox2 and Nkx2-5 binding, but no Shox2-Nkx2-5 co-binding; and the group of 4691 genes with all three binding patterns of Shox2 and Nkx2-5 (Fig. 5b). These intersections were performed with gene set 1, yielding a list of 105 genes possessing both Shox2 and Nkx2-5 binding sites but lacking co-binding sites (Fig. 5d, Supplementary Data 3), and a separate list of 429 genes with all three binding patterns (Fig. 5e, Supplementary Data 4). While the 429 genes with all three binding patterns are potentially involved in SAN morphogenesis, we assumed that these 105 genes with both Shox2 and Nkx2-5 binding but no co-binding sites act as safeguarding factors. Interestingly, among these 105 genes, we found the DEGs that were shown in the regulation of both cell fate determination and morphogenesis through scRNA-seq analysis, including Tbx18, Pitx2 (Supplementary Fig. 4d, e), and Sox11(Fig. 5d). This further substantiates the notion that the segregation during SAN development results from the functional interplay between Shox2 and Nkx2-5. Taken together, our integrative analyses of ChIP-seq and scRNA-seq datasets verified and identified genes that are distinctly involved in the regulation of pacemaking cell fate commitment, safeguarding and SAN morphogenesis, respectively. However, since the ChIP-seq assays were performed on the whole atrium of the heart that contains the SAN, atrial myocardial cells, and non-myocardial cells, while the scRNA-seq was done on the SAN cells, further studies are needed to verify our identified genes.
a Functional categorization of Shox2 and Nkx2-5 binding genes. b Venn diagram depicts the number of overlapped genes between Shox2 binding, Nkx2-5 binding and Shox2-Nkx2-5 co-binding genes. c–e UCSC genome browser track displaying the binding patterns on the representative genes that are potentially involved in the segregation of pacemaking cell fate commitment, safeguarding, and SAN morphogenesis.
In this study, we have uncovered that the essential role of Shox2 in SAN morphogenesis is uncoupled from its function in promoting pacemaking cell fate. We have unraveled that the altered pacemaking cell fate in the SAN of Shox2 mutant mice can be reset by rebalancing Shox2-Nkx2-5 antagonism in Shox2 and Nkx2-5 dKO mice, despite the presence of a hypoplastic SAN. A morphologically integrated SAN is essential for mammals to retain the dominant pacemaker site in a proper position52. Theoretical studies and empirical evidence point toward a morphogenetic program for the SAN that is evolved independently from the pacemaker program12,53,54. In this study, we showed that despite the presence of well-differentiated pacemaking cells in the SAN, the dKO mice nevertheless displayed bradycardia symptom, which was also seen in Shox2 null mutants, indicating the impact of the altered morphology on the physiological function of the SAN. Our scRNA-seq on the SAN cells from those unique mouse lines allow us to establish a Shox2 and Nkx2-5 regulated gene expression profile exclusively implicated in the regulation of SAN morphogenesis along with a gene expression profile specifically involved in pacemaking cell determination and differentiation, which warrants future investigation and verification. These gene expression profiles would also provide candidate genes for the etiology of congenital and acquired abnormalities of the SAN and potential targets for therapeutic intervention of sinus node dysfunction.
Methods
Animal models, embryos, histology, immunofluorescence, and in situ hybridization
The generation and genotyping protocols of Shox2Cre, Shox2HA-Neo, Nkx2.5F/F, and R26RmTmG mice have been described previously29,30,31,55. The animal experiments in this study were approved by the Institutional Animal Care and Use Committee of Tulane University and by the Animal Ethic Committee of Fujian Normal University. We have complied with all relevant ethical regulations for animal use.
Timed pregnant mice were euthanized to collect embryos and embryonic hearts, which were fixed in 4% paraformaldehyde (PFA) at 4 °C overnight, paraffin-embedded, and sectioned at 8 μm for histology and immunofluorescent staining as described previously28. The primary antibodies used in this study were: anti-Shox2 (homemade; 1:500), anti-Nkx2-5 (sc-8697, Santa Cruz; 1:500), anti-Tbx3 (sc-17871, Santa Cruz; 1:500), anti-Hcn4 (ab32675, Abcam; 1:500), anti-Cx40 (sc-20466, Santa Cruz; 1:250), anti-GFP (sc-9996, Santa Cruz; 1:500). The secondary antibodies were all purchased from Life Technologies and used at 1:1000 dilution: donkey-anti rat (A21208, A21209), donkey-anti rabbit (A31573), donkey-anti mouse (A21202, A31571), donkey-anti goat (A32849, A11058, A11055).
In situ hybridization was performed with the RNAscope 2.5 HD Reagent Kit Brown (Advanced Cell Diagnostics, 322300) according to the manufacturer’s instructions. Probe informations are as below: Bmp10 probe (RNAscope® Probe - Mm-Bmp10, 415921), Smoc2 probe (RNAscope® Probe - Mm-Smoc2, 318541), Tbx18 probe (RNAscope® Probe- Mm-Tbx18-C2, 515221-C2). All experiments were repeated at least three times with consistent results.
Surface ECG, whole-cell patch-clamp recording, and scRNA-seq
Surface ECG recording of P0 mice were conducted as reported previously28. On the morning of the birthday, entire litter from Shox2Cre/+;Nkx2-5F/F female crossed to Shox2Cre/+;Nkx2-5F/F male was collected and each pup was subjected to ECG measurement.
For whole-cell patch-clamp recording, embryonic hearts were collected from E13.5 Shox2Cre/+;R26RmTmG and Shox2Cre/Cre;Nkx2.5F/F;R26RmTmG mice, respectively, and the GFP+ SAN was dissected out under a fluorescent dissecting microscope. Tissues were dissociated into single cells that were plated in culture and subjected to whole-cell patch-clamp recording as previous study28.
For single-cell RNA-seq (scRNA-seq), the GFP+ SAN and residual adjacent atrial tissues from E13.5 Shox2Cre/+;R26RmTmG, Shox2Cre/Cre;R26RmTmG, and Shox2Cre/Cre;Nkx2.5F/F;R26RmTmG mice were isolated under a stereo fluorescent microscope. Tissues were subjected to single-cell dissociation, library preparation, sequencing, and bioinformatic analyses, as described in detail previously28.
RT-qPCR analyses
For quantitative reverse transcription polymerase chain reaction (RT-qPCR), the atrium was isolated from E12.5 WT, Shox2HA-Neo/+, and Shox2HA-Neo/Cre mice, and subjected to RNA extraction (E.Z.N.A.® Total RNA Kit I, R6834-02). The RNAs were subsequently reversely transcribed into complementary DNAs (cDNAs) (PrimeScript™ RT reagent Kit, RR037A). SYBR green and gene-specific primers were used and transcript levels were examined using a Thermo Scientific PikoReal Real-Time PCR System. Primer details are as follows: Shox2 (F, 5′-ACCAATTTTACCCTGGAACAAC-3′; R, 5′-TCGATTTTGAAACCAAACCTG-3′), GAPDH (F, 5′-ATCAAGAAGGTGGTGAAGCAG-3′; R, 5′-GAGTGGGAGTTGCTGTTGAAGT-3′). Differences in the RT-qPCR were analyzed using Student’s t-test in GraphPad Prism7; results are presented as mean ± SEM. P < 0.05 was considered significant.
3D reconstruction
For 3D reconstruction, tissues were processed into consecutive sections at 10 µm, stained by anti-Hcn4 antibodies, imaged, and loaded into Amira 6.0.1, in which subsequent alignment, segmentation, and 3D model generation were performed, as reported previously16,28.
Statistics and Reproducibility
All experiments were repeated at least three times unless specifically indicated. All data were statistically analyzed by GraphPad Prism 7. The data were presented as mean ± standard error of the mean (SEM). Statistical analysis was performed using Student’s t-test. Results were considered statistically significant at P < 0.05.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
ScRNA-Seq data have been deposited to Gene Expression Omnibus (GEO) with access number GSE143997.
References
Boyett, M. R., Honjo, H. & Kodama, I. The sinoatrial node, a heterogeneous pacemaker structure. Cardiovasc. Res. 47, 658–687 (2000).
Liu, J., Dobrzynski, H., Yanni, J., Boyett, M. R. & Lei, M. Organisation of the mouse sinoatrial node: structure and expression of HCN channels. Cardiovasc. Res. 73, 729–738 (2007).
Opthof, T. The mammalian sinoatrial node. Cardiovasc Drugs Ther. 1, 573–597 (1988).
Christoffels, V. M., Smits, G. J., Kispert, A. & Moorman, A. F. Development of the pacemaker tissues of the heart. Circ. Res. 106, 240–254 (2010).
Moorman, A. F. & Christoffels, V. M. Cardiac chamber formation: development, genes, and evolution. Physiol. Rev. 83, 1223–1267 (2003).
Liu, H. et al. Functional redundancy between human SHOX and mouse Shox2 genes in the regulation of sinoatrial node formation and pacemaking function. J. Biol. Chem. 286, 17029–17038 (2011).
Hu, W., Xin, Y., Zhao, Y. & Hu, J. Shox2: the role in differentiation and development of cardiac conduction system. Tohoku J. Exp. Med. 244, 177–186 (2018).
Van Mierop, L. & Gessner, I. H. The morphologic development of the sinoatrial node in the mouse. Am. J. Cardiol. 25, 204–212 (1970).
Virágh, S. & Challice, C. The development of the conduction system in the mouse embryo heart: II. Histogenesis of the atrioventricular node and bundle. Dev. Biol. 56, 397–411 (1977).
Christoffels, V. M. et al. Formation of the venous pole of the heart from an Nkx2–5–negative precursor population requires Tbx18. Circ. Res. 98, 1555–1563 (2006).
Mommersteeg, M. T. et al. Molecular pathway for the localized formation of the sinoatrial node. Circ. Res. 100, 354–362 (2007).
Wiese, C. et al. Formation of the sinus node head and differentiation of sinus node myocardium are independently regulated by Tbx18 and Tbx3. Circ. Res. 104, 388–397 (2009).
Van Mierop, L. Location of pacemaker in chick embryo heart at the time of initiation of heartbeat. Am. J. Physiol. Leg. Content 212, 407–415 (1967).
Munshi, N. V. Gene regulatory networks in cardiac conduction system development. Circ. Res. 110, 1525–1537 (2012).
van Eif, V. W. et al. Transcriptome analysis of mouse and human sinoatrial node cells reveals a conserved genetic program. Development 146, dev173161 (2019).
Sun, C. et al. The short stature homeobox 2 (Shox2)-bone morphogenetic protein (BMP) pathway regulates dorsal mesenchymal protrusion development and its temporary function as a pacemaker during cardiogenesis. J. Biol. Chem. 290, 2007–2023 (2015).
DiFrancesco, D. The role of the funny current in pacemaker activity. Circ. Res. 106, 434–446 (2010).
Moosmang, S. et al. Cellular expression and functional characterization of four hyperpolarization‐activated pacemaker channels in cardiac and neuronal tissues. Eur. J. Biochem. 268, 1646–1652 (2001).
Santoro, B. & Tibbs, G. R. The HCN gene family: molecular basis of the hyperpolarization‐activated pacemaker channels. Ann. N. Y. Acad. Sci. 868, 741–764 (1999).
Frank, D. U. et al. Lethal arrhythmias in Tbx3-deficient mice reveal extreme dosage sensitivity of cardiac conduction system function and homeostasis. Proc. Natl Acad. Sci. 109, E154–E163 (2012).
Tang, W. C. Cell Movement in Health and Disease (eds Michael Schnoor, Lei-Miao Yin, Sean X. Sun) 151–157 (Academic Press, 2022).
Espinoza-Lewis, R. A. et al. Shox2 is essential for the differentiation of cardiac pacemaker cells by repressing Nkx2-5. Dev. Biol. 327, 376–385 (2009).
Blaschke, R. J. et al. Targeted mutation reveals essential functions of the homeodomain transcription factor Shox2 in sinoatrial and pacemaking development. Circulation 115, 1830–1838 (2007).
Vedantham, V. New approaches to biological pacemakers: links to sinoatrial node development. Trends Mol. Med. 21, 749–761 (2015).
Espinoza-Lewis, R. A. et al. Ectopic expression of Nkx2. 5 suppresses the formation of the sinoatrial node in mice. Dev. Biol. 356, 359–369 (2011).
Kasahara, H., Bartunkova, S., Schinke, M., Tanaka, M. & Izumo, S. Cardiac and extracardiac expression of Csx/Nkx2. 5 homeodomain protein. Circ. Res. 82, 936–946 (1998).
Ye, W. et al. A common Shox2–Nkx2-5 antagonistic mechanism primes the pacemaker cell fate in the pulmonary vein myocardium and sinoatrial node. Development 142, 2521–2532 (2015).
Li, H. et al. Nkx2-5 defines a subpopulation of pacemaker cells and is essential for the physiological function of the sinoatrial node in mice. Development 146, dev178145 (2019).
Sun, C., Zhang, T., Liu, C., Gu, S. & Chen, Y. Generation of Shox2‐Cre allele for tissue specific manipulation of genes in the developing heart, palate, and limb. Genesis 51, 515–522 (2013).
Wang, J. et al. Pitx2-microRNA pathway that delimits sinoatrial node development and inhibits predisposition to atrial fibrillation. Proc. Natl Acad. Sci. 111, 9181–9186 (2014).
Pashmforoush, M. et al. Nkx2-5 pathways and congenital heart disease: loss of ventricular myocyte lineage specification leads to progressive cardiomyopathy and complete heart block. Cell 117, 373–386 (2004).
Henley, T. et al. Local tissue mechanics control cardiac pacemaker cell embryonic patterning. Life Sci. Alliance 6, e202201799 (2023).
DeLaughter, D. M. et al. Single-cell resolution of temporal gene expression during heart development. Dev. cell 39, 480–490 (2016).
Khazen, W. et al. Expression of macrophage‐selective markers in human and rodent adipocytes. FEBS Lett. 579, 5631–5634 (2005).
Lee, J. H., Protze, S. I., Laksman, Z., Backx, P. H. & Keller, G. M. Human pluripotent stem cell-derived atrial and ventricular cardiomyocytes develop from distinct mesoderm populations. Cell Stem Cell 21, 179–194.e174 (2017).
Li, G. et al. Transcriptomic profiling maps anatomically patterned subpopulations among single embryonic cardiac cells. Dev. cell 39, 491–507 (2016).
Souders, C. A., Bowers, S. L. & Baudino, T. A. Cardiac fibroblast: the renaissance cell. Circ. Res. 105, 1164–1176 (2009).
Tarnawski, L. et al. Integrin based isolation enables purification of murine lineage committed cardiomyocytes. PloS One 10, e0135880 (2015).
Vanlandewijck, M. et al. A molecular atlas of cell types and zonation in the brain vasculature. Nature 554, 475 (2018).
Zappia, L. & Oshlack, A. Clustering trees: a visualization for evaluating clusterings at multiple resolutions. GigaScience 7, giy083 (2018).
Trapnell, C. et al. Pseudo-temporal ordering of individual cells reveals dynamics and regulators of cell fate decisions. Nat. Biotechnol. 32, 381 (2014).
Iancu, C. B. et al. Molecular signatures of cardiac stem cells. Rom. J. Morphol. Embryol. 56, 1255–1262 (2015).
Bunting, K. D. ABC transporters as phenotypic markers and functional regulators of stem cells. Stem Cells 20, 11–20 (2002).
Pfister, O. et al. Role of the ATP-binding cassette transporter Abcg2 in the phenotype and function of cardiac side population cells. Circ. Res 103, 825–835 (2008).
Martin, C. M. et al. Persistent expression of the ATP-binding cassette transporter, Abcg2, identifies cardiac SP cells in the developing and adult heart. Dev. Biol. 265, 262–275 (2004).
Butler, A., Hoffman, P., Smibert, P., Papalexi, E. & Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411 (2018).
Stuart, T. et al. Comprehensive integration of single-cell data. Cell https://doi.org/10.1016/j.cell.2019.05.031 (2019).
van der Maarel, L. E., Postma, A. V. & Christoffels, V. M. Genetics of sinoatrial node function and heart rate disorders. Dis. Model. Mech. 16, https://doi.org/10.1242/dmm.050101 (2023).
Stennard, F. A. et al. Cardiac T-box factor Tbx20 directly interacts with Nkx2-5, GATA4, and GATA5 in regulation of gene expression in the developing heart. Dev. Biol. 262, 206–224 (2003).
Takeuchi, J. K. et al. Tbx20 dose-dependently regulates transcription factor networks required for mouse heart and motoneuron development. Development 132, 2463–2474 (2005).
Zheng, M. et al. Hippo-Yap Signaling Maintains Sinoatrial Node Homeostasis. Circulation 146, 1694–1711 (2022)
Burkhard, S., Van Eif, V., Garric, L., Christoffels, V. M. & Bakkers, J. On the evolution of the cardiac pacemaker. J. Cardiovasc. Dev. Dis. 4, 4 (2017).
Jensen, B., Boukens, B. J., Wang, T., Moorman, A. F. & Christoffels, V. M. Evolution of the sinus venosus from fish to human. J. Cardiovasc. Dev. Dis. 1, 14–28 (2014).
Ye, W., Song, Y., Huang, Z., Zhang, Y. & Chen, Y. Genetic regulation of sinoatrial node development and pacemaker program in the venous pole. J. Cardiovasc. Dev. Dis. 2, 282–298 (2015).
Muzumdar, M. D., Tasic, B., Miyamichi, K., Li, L. & Luo, L. A global double‐fluorescent Cre reporter mouse. Genesis 45, 593–605 (2007).
Acknowledgements
We thank Kejing Song and Alanna Wanek at the NextGen Sequencing Core in the Tulane Center for Translational Research in Infection and Inflammation for technical assistance for scRNA-seq and initial bioinformatic analyses. This research was supported in part using high performance computing (HPC) resources and services provided by Technology Services at Tulane University. We acknowledge financial support by a grant (R01HL136326) from the National Institutes of Health (to Y.C.). T.Y. is supported by a Predoctoral Fellowship (20PRE35040002) from the American Heart Association (AHA). H.H. is supported by NIH grant R01DC016324. H.L. was supported in part by a fellowship from Fujian Normal University and by a grant (2021J01681) from the Natural Science Foundation of Fujian Province, P.R. China. L.W. received a fellowship from the China Scholarship Council during the initial phase of the studies.
Author information
Authors and Affiliations
Contributions
H.L. and Y.C. conceived the project. H.L. performed most experiments, collected, and analyzed data, prepared figures. H.L. and T.Y. prepared the first draft of the manuscript. D.L. and H.H. conducted whole-cell patch-clamp recordings and data processing and explanation. T.Y., Z.W., L.W. L.L., Yaoyi C., and Y.Z. helped with histology, immunohistochemistry, RT-qPCR, ECG, and 3D reconstruction. Q.T., and H.L., with the help of T.Y., L.W. performed bioinformatic analyses. Q.T., H.H., and Y.Z. provided insights during the studies and helped in manuscript preparation and editing. Y.C. conducted the final revision and editing of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks Vincent Christoffels, Gudrun Rappold, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: Madhumita Basu and Joao Valente.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Li, H., Tang, Q., Yang, T. et al. Segregation of morphogenetic regulatory function of Shox2 from its cell fate guardian role in sinoatrial node development. Commun Biol 7, 385 (2024). https://doi.org/10.1038/s42003-024-06039-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-024-06039-2
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.