Abstract
Glycosyltransferases typically display acceptor substrate flexibility but more stringent donor specificity. BsGT-1 is a highly effective glycosyltransferase to glycosylate macrolides, including epothilones, promising antitumor compounds. Here, we show that BsGT-1 has three major regions significantly influencing the glycodiversification of epothilone B based on structural molecular docking, “hot spots” alanine scanning, and site saturation mutagenesis. Mutations in the PSPG-like motif region and the C2 loop region are more likely to expand donor preference; mutations of the flexible N3 loop region located at the mouth of the substrate-binding cavity produce novel epothilone oligosaccharides. These “hot spots” also functioned in homologues of BsGT-1. The glycosides showed significantly enhanced water solubility and decreased cytotoxicity, although the glycosyl appendages of epothilone B also reduced drug permeability and attenuated antitumor efficacy. This study laid a foundation for the rational engineering of other GTs to synthesize valuable small molecules.
Similar content being viewed by others
Introduction
Epothilones are a class of 16-membered antitumor macrolides isolated from Sorangium cellulosum strains. As epothilones exhibit potent bioactivities, especially for killing multidrug-resistant tumors1, they have attracted a plethora of research interest ever since discovery2,3. Epothilones A and B1 are the two major members of this family. In comparison to epothilone A, epothilone B (EpoB) contains an additional methyl group at the epoxy ring but shows stronger potency in antitumor activity4,5. Ixabepilone, a semisynthetic analog of EpoB, was approved by the FDA in 2007 for the treatment of unresponsive aggressive metastatic or locally advanced breast cancer6,7. However, the clinical usage of EpoB analogs is still greatly restricted due to severe side effects and poor bioavailability8,9. Therefore, modification(s) appears necessary to overcome the drawbacks of EpoB, whereby glycosylation could potentially come into play, as it is a powerful approach to tune the physicochemical properties and/or biological activities of small molecules10,11. Compared to chemical synthesis, glycosylation reactions catalyzed by enzymes have many advantages, such as high efficiency, regio- and/or stereoselectivity, and environmental compatibility.
Glycosyltransferases (GTs) are natural biocatalysts that transfer glycosyls from activated sugar donors to diverse acceptors12. Topologically, GTs are predominantly classified into GT-A and GT-B folds. The GT-A fold possesses a central β-sheet surrounded by α-helices and two tightly abutting β/α/β domains, with a highly conserved metal-coordinating DxD (ASP-X-ASP) motif. The GT-B fold consists of two separate and flexibly linked Rossmann-like domains, and the catalytic cleft is located between these two domains. The GT-B fold contains a relatively conserved sugar donor binding motif at the C-terminal domain, whereas the N-terminal domain interacting with acceptors is usually variable, exhibiting greater topological plasticity11,13. This peculiarity allows many GT-B folds to show flexible substrate-binding patterns that are known as “enzyme promiscuity”14,15. While the GT superfamily can be classified into 114 families, the members in the GT1 family that typically adopt the typical GT-B fold are invariably promiscuous enzymes16,17. In contrast, the GT1 family has evolved higher specificity for sugar donors by accepting a narrow range of NDP-sugars (nucleoside diphosphate sugars). A highly conserved motif in the C-terminal domain, namely, the “PSPG (or -like) motif”, found in GTs from plants and bacteria18, is known as the major region for the binding of sugar donors. Many studies have focused on the PSPG motif to flexibilize sugar donor specificity; however, site-directed mutagenesis of the PSPG motif often leads to complete loss19 or severe damage20,21 in enzyme activity. More intriguingly, site-directed mutagenesis outside the PSPG motif of the two GTs also broadened the sugar donor scope, indicating that the PSPG motif is probably not the only determinant in sugar donor recognition22,23. With the surge in genome sequencing, a vast number of GTs have been unraveled, but only a very small portion have been characterized confirmatively in function18. In particular, investigations into the underlying structural basis of GTs that determines sugar donor specificity remain insufficient, which limits rational engineering of GTs for glycorandomization of bioactive molecules.
We previously characterized the promiscuous enzyme BsGT-1, which displayed highly efficient glycosylation activities toward diverse acceptors24, including the macrolides epothilone A and rapamycin25,26. In this study, we applied BsGT-1 to the glycosylation of EpoB in an attempt to glycodiversify and better this medicinally important small molecule. Directed by molecular docking analysis in tandem with mutagenesis study, we systematically probed functional regions of BsGT-1 and revealed the “hot spots” in three major regions that determine the catalytic efficiency and/or sugar donor specificity. This enabled us to concisely synthesize and characterize seven previously undescribed EpoB glycosides, differentiating in the sugar category and/or size. The glycosyls appended to EpoB significantly improved the water solubility but reduced the antitumor efficacy and drug permeability into cells. The fact that the “hot spots” of BsGT-1 also existed in its orthologues indicated that our work will lay foundations for the future rational engineering of different GTs for the glycodiversification of natural biomolecules with sheer chemical complexity and pronounced bioactivities.
Results
BsGT-1 glycosylated epothilone B using different sugar donors
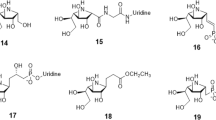
We previously characterized that BsGT-1 could efficiently glycosylate epothilone A at the 7-OH position with UDP-glucose (UDP-Glc, 2a)25. Herein, we further tested the glycosylation ability of BsGT-1 toward epothilone B (EpoB). When using 2a as the sugar donor, HPLC-DAD profiling (249 nm) indicated that EpoB was almost 100% converted by BsGT-1 after 2 h of glycosylation reaction, and the efficiency was congruent with that of epothilone A25. Unexpectedly, three glycosylated products 1a, 1b, and 1c were generated for EpoB (Supplementary Fig. 1), in contrast to the single monoglycoside product for epothilone A. Major product 1a achieved >90% conversion within an hour followed by a gradual decrease, while 1b and 1c increased steadily. After purification, compounds 1a–1c were identified as EpoB 7-O-β-D glucoside, EpoB 7-O-β-D-glucosyl-(1 → 3)-β-D glucoside, and EpoB 7-O-β-D-glucosyl-(1 → 2)-β-D glucoside on the basis of HRMS (high-resolution mass spectrometry) and NMR (nuclear magnetic resonance) (Fig. 1 and Supplementary Table 1, Supplementary Figs. 15–17 and Supplementary Figs. 27–41), respectively. The stepwise tandem assembly of two glucoses by a single GT in a non-regiospecific manner (1 → 3 or 1 → 2 connectivity) is remarkable, which was reminiscent of OleD’s triple mutant (A242V/S132F/P67T) that was able to mediate disaccharide formation through iterative glucosylation27, and AveBI that can consecutively integrate two molecules of l-oleandrose to avermectin28.
a Structures of the 16 tested sugar donors (2a−2p). b Conversion efficiency of wild-type BsGT-1 toward sugar donors 2a−2p after 10 h of incubation. The structures of epothilone B (EpoB) and its glycosides catalyzed by mutants of BsGT-1 are shown, the conversion data (mean ± SD) were obtained from three independent repeat experiments.
We further probed the sugar donor promiscuity of BsGT-1 towards 16 activated forms of monosaccharides (2a–2p, Fig. 1a) in an attempt to glycodiversify EpoB. After 10 h of incubation, the four NDP-glucoses (2a–2d) achieved almost 100% turnover of EpoB, and low conversion efficiency (<20%) was observed for other sugar donors, including 2e, 2f, 2h, 2i, 2l, 2m, 2o, and 2p (Fig. 1b), whereby the corresponding monoglycosylated products 1e, 1f, 1h, 1i, 1l, 1m, 1o, and 1p were confirmed by HRMS/(MS) (Supplementary Figs. 19–26) and/or NMR (Supplementary Table 2 and Supplementary Figs. 48–62). The tolerance of different NDP-sugars by BsGT-1 implied the sugar donor promiscuity of BsGT-1 and its engineering potential for the glycorandomization of EpoB.
Structure of BsGT-1 and its docking model with substrates
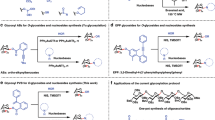
To gain deeper insight into the catalytic mechanism of BsGT-1, we tried to resolve the structure of wild-type BsGT-1. While we made to obtain a crystal structure with low resolution, a higher quality of crystal structure of BsGT-1 (Bs-YjiC) at 2.29 Å was disclosed by another research group (accession number: 7BOV, Fig. 2a)29. BsGT-1 adopts a typical GT-B fold, consisting of two distinct Rossmann-like domains interconnected by a linker region. However, the resolved structure of BsGT-1 (7BOV) is incomplete because two flexible loop regions (residues H57–M71 and A156–F168) with low electron density could not be modeled. To investigate the potential impacts of these two regions on enzyme activity, we deleted residues H57–M71 and A156–F168 respectively from wild-type BsGT-1, and the resulting mutants were both solubly expressed in E. coli (Supplementary Fig. 2). As a result, the deletion of A156-F168 had negligible effects on the glycosylation of EpoB, whereas the ΔH57–M71 mutation caused a severe reduction in catalytic efficiency (Supplementary Fig. 3). In view of this, we used the I-TASSER method and crystal structure 7BOV as a template to restore the complete structure of wild-type BsGT-130 and then subjected it to docking analysis using AutoDock Vina and molecular dynamics simulation of GROMACS (Fig. 2b)31,32. To exploit the catalytic characteristics of BsGT-1 toward substrates EpoB and 2a. The free binding energy (ΔG) was used to characterize the maximum energy of reversible binding of ligands with protein (Supplementary Table 3), and the 35 residues that formed hydrogen bonds or van der waal’s force interactions with ligands of EpoB and 2a were pinpointed (Fig. 2b and Supplementary Table 4). Subsequently, these potential key residues were subjected to alanine scanning mutagenesis to probe their respective functions for EpoB glycosylation towards the 16 different sugar donors 2a–2p displayed in Fig. 1a. The catalytic efficiency of the mutants was classified into five different ranks according to the turnover rate of EpoB (Fig. 3). The mutations at H16 (in Nα1) and S129 (in the N4 loop) that simultaneously interacted with EpoB and 2a (Supplementary Table 4 and Supplementary Fig. 4) caused a dramatic reduction in catalytic efficiency, while the mutation at Y130 (in the N4 loop) that interacted with EpoB absolutely abrogated glycosylating ability. This was consistent with previous results that the residues in the N4 loop of GTs played important roles in substrate catalysis17. Interestingly, many mutations outside the loop N4 region also exhibited evident impacts on either sugar donor recognition or polysaccharification, which compelled us to further investigate the potential “hot spots” in the PSPG-like motif, C2 loop, and N3 loop, as described in the following sections.
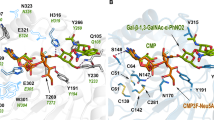
a The incomplete crystal structure of BsGT-1 with resolution at 2.29 Å (PDB ID: 7BOV). b The predicted complete structure of BsGT-1 was docked with the sugar acceptor EpoB and sugar donor UDP-Glc (2a). c The potential hydrogen bond interactions of M296, E317, and Q318 in the PSPG-like motif (surface, yellow) and F231 in loop C2 toward the glucosyl moiety of 2a. d The positional relationship between loop N3 region and EpoB, whereby the distance between the I62 and EpoB was indicated.
The important role of the PSPG-like motif in the sugar donor promiscuity of BsGT-1
The PSPG motif, known to be closely related to sugar binding and recognition, is conserved in most plant GTs, and a PSPG-like motif is also commonly distributed in bacterial GTs18,33. It was reported that mutations of key residues in the PSPG motif usually abolished activity or only caused weak alterations in sugar donor recognition34. We performed alanine-scanning mutagenesis in the PSPG-like motif of BsGT-1, and the results showed that glycosylation with 2a was less affected than the other three NDP-glucoses (2b–2d), since all the mutants of the PSPG-like motif retained conversion unchanged for 2a, or at least better than the other three NDP-glucoses (Fig. 3). Notably, the PSPG-like motif usually begins with the residue Trp and ends with the strictly conserved Glu/Asp-Gln cassette that is critical for hydrogen-bonding interaction with the 2-OH of glucosyl moiety17,35,36. The uracil base of 2a contacts the aromatic side chain of W312 in OleI or W290 in OleD, rendering the preference for 2a rather than other NDP-glucose17,37. Distinctively, BsGT-1 begins with serine in the PSPG-like motif (S277) (Supplementary Fig. 4) and can accept all NDP-glucoses irrespective of nucleotides. The S277A mutation in BsGT-1 caused a considerable decline in the activity (Fig. 3). Moreover, most alanine-scanning mutants of the PSPG-like motif normally decreased the activities toward the other three NDP-glucoses rather than UDP-glucose, while mutants E301A and E317A had weak activities toward all four NDP-glucoses.
Our docking model revealed that E317 had hydrogen bonds with both 2-OH (3.0 Å) and 4-OH (2.9 Å) of glucose. Residue Q318 also formed an interaction (2.7 Å) with the 2-OH of glucose, and M296 potentially formed a hydrogen bond (3.0 Å) with the 4-OH of glucose (Fig. 2c). E317 seemed to be more important than Q318 in capturing the glucosyl moiety because the saturation mutagenesis of E317 almost abolished the conversion activity towards 2a (Supplementary Fig. 5). Notably, although wild-type BsGT-1 gave a relatively low conversion efficiency toward UDP-Gal (2i, 9.3%) and UDP-GlcNAc (2e, 11.2%), their conversion efficiencies were significantly improved by M296A and Q318A mutations, respectively (Fig. 3). Further saturation mutagenesis proved that the M296A mutant was most efficient in 2i catalysis, giving a 10-fold higher catalytic efficiency (kcat/km value, Supplementary Table 5) than WT (Fig. 4a). The Q318C mutation achieved >70% conversion of 2e, in stark contrast to ~10% by wild-type BsGT-1 (Fig. 4b). Taken together, the PSPG-like motif plays an important role in the sugar donor specificity of BsGT-1 and glycodiversification of EpoB.
a Comparison of the conversion efficiency (mean ± SD) of M296 saturated mutants using UDP-Gal and UDP-Glc as sugar donors, and the M296A mutant with the highest efficiency toward UDP-Gal was highlighted. b Comparison of the conversion efficiency of Q318 saturated mutants using UDP-GlcNAc and UDP-Glc as sugar donors, and the Q318C mutant with the highest efficiency toward UDP-GlcNAc was highlighted. c Comparison of the conversion efficiency of F231 saturated mutants using UDP-GlcNAz and UDP-Glc as sugar donors, and the F231S mutant with the highest efficiency toward UDP-GlcNAc was highlighted. d The change in the proportions of EpoB glucosides 1a–1d caused by alanine scanning mutation in the loop N3 region. The I62A mutant that gave the highest portion of polyglycosides (1b–1d) was highlighted, the conversion data were obtained from three independent repeat experiments.
The C2 loop is also involved in the sugar donor promiscuity of BsGT-1
In addition to the PSPG-like motif, some C-terminal residues also formed intimate interactions with EpoB and/or UDP-Glc (2a). Alanine-scanning mutagenesis showed that the R207A, K210A, S226A, and S255A mutations had little impact on the catalytic efficiency and sugar donor specificity of BsGT-1 (Fig. 3). However, T229 and F231, located within the loop C2 region (Supplementary Fig. 6), were highly related to sugar donor specificity. The T229A mutation had a detrimental effect on the binding of NDP-glucoses, except TDP-glucose. Although the F231A mutation affected the interactions with NDP-glucoses except 2a, the catalytic activity toward UDP-GlcNAz (2h) of this mutant was surprisingly enhanced (Fig. 3). Notably, the aromatic ring of residue F231 had a close interaction with 2-OH (3.1 Å) of glucose in 2a (Fig. 2c), and the replacement with less bulky alanine might allow the entry of 2h endowed with a large substituent at 2-OH of glucose. Saturation mutagenesis showed that the conversion of 2h was further improved by the F231S mutation, reaching >45% (Fig. 4c). The kcat/Km value of F231S toward 2h was 22.5-fold higher than that of WT (Supplementary Table 5). Nevertheless, most of the saturated mutants of F231 retained potent activity toward 2a, except that the activity of F231P was reduced by half (Fig. 4c). Thus, the disordered C2 loop outside of the PSPG-like motif is also involved in maintaining sugar donor specificity.
The N3 loop of BsGT-1 is related to the polysaccharification of EpoB
It is well known that the N-domain is responsible for the binding of sugar acceptors, and the C-domain holds the sugar donor in GT-B fold glycosyltransferase11,18. We performed alanine-scanning mutagenesis at some residues in the N-domain (from Y4 to I199) that did not exhibit any influence on sugar donor promiscuity (Fig. 3), but the H16A mutation indeed hampered EpoB binding. Interestingly, although the mutants of N61A, I62A, and D63A gave 100% conversion of EpoB as wild type did, the proportion of the generated EpoB glucosides (1a, 1b, 1c) was substantially changed (Fig. 4d), and especially a new compound 1d was produced by these three mutants but not by wild type. Based on the combination of HRMS and intensive 2D NMR interpretation (Supplementary Fig. 18 and Supplementary Figs. 42–47), 1d was identified as an unprecedented linear trisaccharide, epothilone B 7-O-β-D-glucosyl-(1 → 2)-β-D-glucosyl-(1 → 4)-β-D glucoside (1d, Supplementary Fig. 7). The generation of three types of glycosidic bonds (1 → 3 in 1b, 1 → 2 in 1c and 1d, and 1 → 4 in 1d) suggested flexibility in terms of the regioselectivity of BsGT-1.
In BsGT-1, the N3 loop region is located at the entrance of the cleft between the N-domain and C-domain, wherein the I62 residue was 4.2 Å away from C-7 of EpoB in the docking model (Fig. 2d). Because residues N61, I62, and D63 are located in the same N3 loop, we proposed that this region determined polysaccharification. To experimentally confirm this, we carried out systematic alanine scanning of the whole N3 loop region (from Y57 to K73), and each mutant was assessed as a catalyst for glucosylation with 2a as the donor. As a result, polysaccharification peaked at the I62A mutation and then gradually decreased to both sides (Fig. 4d). The I62A mutant dominantly produced diglucosylated derivatives (1b and 1c, accounting for 75%) and a decent amount of 1d (accounting for 10%). Further saturation mutagenesis at site I62 showed that the substitutions of bulky amino acids, such as I62H, I62K, I62F, and I62W, severely affected the enzymatic activity, while the residues with small side chains, such as I62G and I62A, retained both the conversion efficiency and the iterative glucosylation capability; I62A was still the most advantageous mutation for polysaccharification (Supplementary Fig. 8). Notably, the E69A mutation markedly decreased the conversion of epothilone B.
The catalytic efficiency for EpoB→1a conversion by I62A and wild type was comparable, but the kcat/Km values of 1a → 1b and 1a → 1c by I62A were 28.3- and 5.3-fold higher than the wild type, respectively (Supplementary Table 5). The comparatively higher conversion rate of 1a → 1b than 1a → 1c by the I62A mutant could be explanatory to the limited production of 1d, since it was generated on the basis of 1c. Taken together, the dramatic fluctuation in the iterative glycosylation represented by the I62A mutation indicated that the N3 loop is very likely associated with the formation of a catalytic cavity for acceptor EpoB.
The “hot spots” of BsGT-1 also exist in other homologous GTs
Sequence alignment of the regions of the PSPG-like motif, N3 loop, and C2 loop in ten related GTs (Supplementary Table 6) revealed that YjiC, BaGT, BgGT, and BpGT were highly conserved at the “hot spots” characterized in BsGT-1 (Supplementary Fig. 6 and Supplementary Fig. 9). These four macrolide GTs were further subjected to TM alignment (https://zhanglab.ccmb.med.umich.edu/TM-align/)38 to compare their 3D architectures. The topologies of these enzymes, especially for the PSPG-like motif, C2 loop and N3 loop regions, showed a high degree of similarity to BsGT-1 (Supplementary Fig. 10), probably rendering them similar capabilities for glycosylation of EpoB.25 We subsequently performed site-directed mutagenesis of BaGT, YjiC, BpGT, and BgGT at the sites corresponding to the “hot spots” of BsGT-1 to probe their putative roles in EpoB glycosylation. As a result, the conclusions drawn for the four “hot spots” (I62, F231, M296, and Q318) of BsGT-1 were actually applicable to the four enzymes YjiC, BaGT, BgGT, and BpGT (Fig. 5a). Specifically, mutagenesis at the two “hot spots” in the PSPG-like motif of these four enzymes corresponding to M296 and Q318 in BsGT-1 significantly increased the catalytic activities toward UDP-Gal (2i) and UDP-GlcNAc (2e) by ~2.5- and 5-fold, respectively. Likewise, the site mutagenesis of the four GTs corresponding to F231S in BsGT-1 was favorable for the enhanced acceptance of UDP-GlcNAz (2h) as a sugar donor (Fig. 5b). The mutations at position 62 of the four GTs gave rise to an elevated ratio in the polysaccharification compared to the wild type, as represented by the new generation of 1d (Fig. 5c). Collectively, similar improvements in the promiscuity of sugar donors after mutagenesis at the “hot spots” of the four macrolide GTs implied that the structural knowledge revealed for BsGT-1 in this study has somewhat generality, and would provide guidance for the rational engineering of other GTs to glycodiversify a broad range of pharmaceutically important small molecules.
a The alignment of BsGT-1 with its four homologs YjiC, BaGT, BgGT, and BpGT, wherein the “hot spots” of I62, F231, M296, and Q318 characterized in BsGT-1 were highlighted. b The elevated conversion efficiency towards the sugar donors UDP-GlcNAz (2h), UDP-Gal (2i), and GlcNAc (2e) achieved by the mutations of F231S, M296A, and Q318C, respectively. The displayed values (fold change) are the conversion efficiency of mutants relative to the respective wild type. c Comparison of the conversion rate (mean ± SD) of glycosides 1a–1d by the mutants (I62A or V62A) and wild type (WT). The 62nd site in the five GTs was uniformly related to polysaccharification because the proportion of monoglycoside 1a was reduced and the polyglycosides (1b–1d) in the mutant were accordingly increased, the conversion data were obtained from three independent repeat experiments, the displayed fold changes were calculated based on the averages of the conversion data.
Biological properties of epothilone B glycosides
We first tested the water solubility of epothilone B (EpoB) glycosides. Not unexpectedly, the more glycosyl groups attached to EpoB, the better solubility was observed, whereby mono-, di-, and tri-glycosylation evidently improved the water solubility of EpoB to 40-, 400-, 1000-fold, respectively (Table 1). Next, the isolated glycosides were subjected to antitumor and cytotoxicity assays using the liver carcinoma cell line HepG2 and hepatocyte cell line HL-7702. Consistent with previous studies1,2,39, EpoB exhibited potent antitumor activity at the nanomolar level toward HepG2 cells (IC50 < 0.002 μM) but also caused severe cytotoxicity (IC50 < 0.003 μM, Table 1). Both the inhibitory effects and cytotoxicities of all the glycosides decreased >1000-fold, despite different extensions depending on the properties of sugar moieties. In general, the size of the sugar moiety negatively correlated with the biological activity, so the IC50 values of EpoB with substituents of mono-Glc (1a), GlcNAc (1e), di-Glc (1c), and tri-Glc (1d) showed an ascending trend. Notably, the galactose derivative (1i) showed the best antitumor activity (IC50, 1.09 μM) among the EpoB glycosides. To understand the reason why 7-O-glycosylation had a detrimental effect on the bioactivity of EpoB, we conducted molecular docking analysis of the representative monoglycoside 1a with tubulin (target protein), whereby the cocrystal structure of β-tubulin subunit-EpoB deposited in the PDB database (7DAE) was used as a template model (Supplementary Fig. 11). In comparison, EpoB intimately interacted with the β-tubulin subunit through hydrogen bonding (T276, Q281, and D226) and hydrophobic interactions (L230, F272, and L275), which were disrupted in the docking model for 1a. However, although the hydrogen bonding between the key residue D226 and the 7-OH group of 1a was blocked by the appended glycosyl, a new hydrogen bond was formed with the 6-OH group of sugar, which could be explanatory for the appreciable antitumor activity of 1a (IC50, 9.84 μM).
As compound penetration into cells plays an important role in their antitumor effects40, we further carried out immunofluorescence experiments to compare the cellular intake of EpoB and glycosides 1a and 1i. As shown in Figs. 1a, 6 could penetrate into the tumor cells and interact with tubulin, although the amount was less than that of EpoB. Further quantitative analysis showed that the cellular intake rate of glycosides 1a and 1i was 12- and 8-fold lower than that of EpoB, which also contributed to the reduced activities of EpoB glycosides (Supplementary Fig. 12).
Discussion
Glycosylation is an effective way to tune the physicochemical properties and/or bioactivities of various natural products41, and glycosyltransferases (GTs) are powerful enzymatic toolkits for this purpose42. Wild-type GTs often have limitations in terms of low catalytic efficiency and/or unwanted substrate specificity/selectivity; thus, rational engineering is always needed43. Protein structure-guided mutagenesis has been proven to be a rewarding approach to enhance the catalytic activity and/or alter the product specificity of GTs. For instance, the solution of the crystal structure of triterpene glycosyltransferase UGT74AC1 from Siraitia grosvenorii facilitated directed evolution to improve enzymatic activity and substrate promiscuity, leading to one variant that exhibited up to 4.17 × 104-fold increase in catalytic efficiency toward the terpenoid mogrol and 1.53 × 104-fold increase for sugar donor UDP-glucose, respectively44; γ-Cyclodextrin (γ-CD) is produced from starch by using Bacillus clarkii γ‐cyclodextrin glycosyltransferase (CGTase), but this enzymatic process also gives rise to the interfering side products α-CD and β-CD45. The site-directed mutagenesis of γ-CGTase significantly improved the product preference toward γ-CD, whereby the mutant Y186W displayed a high selectivity rate of 94.6% versus 77.1% given by the wild type46. In recent years, the Ye lab has been productive in solving the 3D structures of plant O- and C-glycosyltransferases (i.e., TcCGT121, GgCGT47, TcOGT448, and Sb3GT149) and accordingly characterized the structural basis that determines the catalytic efficiency and/or substrate specificity, which has greatly promoted the glycodiversification of important flavonoid-class natural products. In this study, we likewise employed structure-guided engineering of the versatile glycosyltransferase BsGT-126,29 to improve its catalytic efficiency towards different sugar donors and guaranteed sufficient yields for the chromatographic purification of seven previously undescribed EpoB glycosides. Hence, the docking-guided engineering of GTs is an effective and economical strategy for the glycosylation of important molecules.
In general, wild-type GTs have a relatively wide spectrum of acceptors but a narrow window of sugar donor selectivity18. As diverse sugar moieties can positively fine-tune the physicochemical properties and/or alter the bioactivities of drug acceptors42,46, it appears important to understand the structural basis shaping sugar donor preference. The PSPG motif has been regarded as the key region for sugar donor recognition. For instance, the C-glycosyltransferase TcCGT1 from Trollius chinensis showed regiospecificity in the C-glycosylation of flavones and catalyzed the O-glycosylation of diverse phenolics with UDP-Glc. The mutation (E396A) in the PSPG motif of TcCGT1 abolished the catalytic activity21. Other cases also reported that mutations in this region lead to a complete loss of enzymatic activity17,50, but the single mutation of P361W in the PSPG motif of TcOGT4 switched the sugar donor preference from UDP-Gal to UDP-Glc48. Our studies revealed more key residues in this motif that formed hydrogen bonds with the specific hydroxyl groups of sugar moieties, i.e., M296 and Q318 interacted with O4 and O2 of the sugar, respectively. In addition to the PSPG-like motif, the “hot spots” in other motifs could also affect sugar donor selectivity. The triple mutant P67T/I112K/A242V of GT OleD51 and the quadruple mutations in S60/V100/T104/I152 of GT MiCGTb22 were capable of efficiently catalyzing a set of different sugar donors. Notably, this study also revealed a single “hot spot” (residue F231) in the C2 loop region of BsGT-1, and the untapped C2 loop region is jointly involved in the recognition of sugar donors with PSPG-like motifs. This knowledge will undoubtedly provide more insight into the engineering of GTs for the alteration of sugar donor specificity in the future.
In contrast to the C2 loop, which impacts sugar donor specificity, the N3 loop of BsGT-1 is involved in the recognition of acceptors. Structure-based systematic mutations in the N3 loop indicated that it contributed to the formation of a binding cavity for EpoB (Fig. 4d). This was well reflected by saturated mutagenesis at the I62 site, whereby replacement with the amino acid glycine or alanine with small sider chains was favorable for polysaccharification of EpoB (Supplementary Fig. 8). The polysaccharification capacity of the mutants (I62G and I62A) was very likely attributed to an increase in the volume of the binding pocket to accommodate a larger substrate and to release larger products. Polysaccharification could evidently improve water solubility and is sometimes crucial for the bioactivities of natural products52. As loops often undergo a disordered to ordered transition53, we further hypothesized that the N3 loop is related to the incredible “substrate promiscuity” of BsGT-1 that accepts various chemical scaffolds, such as macrolides epothilone A25 and rapamycin26, flavonoids, and coumarins24. Presumably, the flexible N3 loop adopts different conformations to adjust a spacious substrate-binding tunnel to accommodate different acceptors. This was reminiscent of loop dynamics of the glycosyltransferase GpgS that underwent multiple conformation changes along the catalytic cycle, wherein the open-to-closed motion of the L loop in GpgS was essential for assembling the reaction center53. The conformational plasticity of the N3 loop in BsGT-1 is probably commonplace in GT-B fold GTs. In addition to the evidence presented in Fig. 5c, residue P67 in the N3 loop of OleD has also been proven to be the most advantageous site for expanding its substrate spectrum54.
The severe side effects and poor bioavailability of EpoB8,9 necessitate chemical modifications, including glycosylation. Regretfully, the cytotoxicity and antitumor activity of the obtained glycosides in this study were dramatically decreased (Table 1). This is because the 7-OH of EpoB is a key functionality that forms hydrogen bonds with target microtubulin, and the bulky substituents at this position caused huge steric hindrances for ligand entry into the binding cavity of the protein (Supplementary Fig. 11). Hence, enzymatic glycosylation at other sites (such as 3-OH) instead of 7-OH should be tried in the future. As the O-glycosyltransferase BsGT-1 could recognize only the two innate hydroxyl groups at C-3 and C-7 of EpoB, characterization of potential C-glycosyltransferases21,47 that modify macrolide-class molecules could randomly glycosylate the backbone of EpoB. Alternatively, it is also tempting to develop cascade enzymatic reactions to regioglycodiversify EpoB: first, P450 monooxygenases55 are used to introduce extra hydroxyls onto EpoB, and then O-glycosyltransferases are used to install glycosyls. Meanwhile, we noticed that the activity of galactose-modified EpoB (1i) was almost 10-fold higher than that of glucose-modified EpoB (1a, Table 1), implying that further intensive enzymatic glycosylation studies with more variable sugar donors are still promising to deliver efficacious EpoB derivatives. This hypothesis seems reasonable because most sugar moieties in bioactive macrolides (i.e., erythromycin, avermectin) are uncommon deoxysugars (i.g. desosamine, oleandrose), and they are indeed crucial for interaction with target proteins56,57. The availability of unusual sugar donors could be problematic, which could be solved by a synthetic biology approach58. On the other hand, we revealed that the decline in the antitumor efficacy of current EpoB glycosides was also related to the lower cellular intake rate. Therefore, the combination of EpoB glycosides with other therapeutics to enhance the penetration of cell membranes will probably elevate the efficacy of EpoB glycosides.
Collectively, through structure-directed engineering (alanine scanning and/or saturated mutagenesis), we systematically explored the structural basis that determines the catalytic efficiency of BsGT-1 and identified the key “hot spots” that are critical for the selectivity of sugar donors and acceptors. This study provides much deeper insights into the catalytic mechanism of promiscuous GT-B fold glycosyltransferases, as represented by BsGT-1, and lays a foundation to make unnatural natural biocatalysts for the concise synthesis of various bioactive glycosides. Furthermore, in view of the obvious shortfalls in the clinical usage of EpoB, the EpoB glycosides obtained in this study differing in the categories and/or iteration pattern of glycosyl(s) have greatly enriched the glycochemistry of this druggable molecule. Although the antitumor activity of EpoB was generally attenuated, the fact that different types of sugar appendages exerted disparate influences on the activity underscored further intensive enzymatic glycosylation with more variable sugar donors to give glycosylated derivatives of EpoB with both appreciable potency and good water solubility.
Methods
Reagents, cells, and main media preparation
NDP-glucose (UDPG, ADPG, TDPG, and GDPG), UDP-N-acetylglucosamine (UDP-GlcNAc), UDP-galactose (UDP-Gal), and UDP-glucuronic acid (UDP-GlcA) were purchased from Sigma-Aldrich (St. Louis, MO, USA). UDP-GlcNAz, UDP-GalNAz, UDP-GlcNTFA, UDP-GalNTFA, UDP-GalNAc, UDP-GalA, CMP-Neu5Ac, GDP-Fuc and GDP-Man were kindly provided by Professor Junqiang Fang59,60,61,62,63. Epothilone B was purchased from Zhejiang Hisun Pharmaceutical Co., Ltd. The human liver carcinoma cell line HepG2 and human hepatocyte cell line HL-7702 were purchased from Gaining (Gaining Bio. Co., Ltd, Shanghai, China). The cells were cultivated in T-25 flasks in a Thermo Scientific CO2 incubator in a humidified environment at 37°C and 5% CO2. HepG2 cells were preserved in DMEM (Gibco, Thermo Fisher Scientific, Waltham, MA, USA), while HL-7702 cells were preserved in RPMI-1640 (Gibco, Thermo Fisher Scientific, Waltham, MA, USA). All cultures contained 10% fetal calf serum and 1% penicillin C/streptomycin. All chemicals and reagents are of analytical grade.
Protein expression, purification, and site-directed or saturated mutagenesis
The recombinant plasmids of BsGT-1, YjiC, BaGT, BgGT, and BpGT were transformed into E. coli BL21 (DE3) for protein expression and purification by using previously described protocols25. A Fast Mutagenesis Kit V2 (Vazyme Biotech Co., Ltd, Nanjing, China) was used for alanine scanning, deletion of H57–M71 and A156-F168, and saturated mutagenesis of BsGT-1. The primers were listed in Supplementary Tables 7–9. After confirmation by Sanger sequencing, the mutants were subjected to protein expression and purification, followed by glycosylation of epothilone B with the sugar donors displayed in Fig. 1.
In vitro glycosylation reactions and monitorization
The purified protein and mutants were subjected to glycosylation reactions in vitro. For the analytical scale reaction, a mixture of 50 μl containing 50 mM Tris–HCl buffer, 10 mM MgCl2, 2 mM sugar donors, 0.4 mM epothilone B (dissolved in DMSO), and 500 μg/mL of each protein was incubated at 37°C. To determine the conversion curves, an aliquot of 50 μl of the reaction mixture was sampled at different time points (0.2 h, 0.4 h, 0.6 h, 0.8 h, 1 h, 2 h, 4 h, 6 h, 8 h, and 10 h). The reaction was terminated with a 3-fold volume of methanol, and the suspensions were centrifuged at 14,000 rpm for 30 min. The samples were subjected to UPLC-PDA and HR-QTOF ESI-MS instruments equipped with a C18 column (Thermo Fisher Scientific, C18, 250 mm × 4.6 mm, 5 μm). Products were detected by UV absorbance at 249 nm. The mobile phase was isocratic 35% acetonitrile in water at a flow rate of 1 mL/min for 35 min.
For the preparative scale reaction, a 20 mL system containing BsGT-1 (500 μg/mL), 6 mM sugar donors, 1.2 mM EpoB (dissolved in DMSO), 50 mM Tris–HCl (pH 7.5) buffer, and 10 mM MgCl2 was used. The reaction was incubated for 12 h at 37°C and stopped by adding the same volume of chilled methanol. After centrifugation at 14,000 rpm and 4°C for 30 min, the supernatant was concentrated by evaporation and lyophilization. The dry powder was dissolved in methanol and then subsequently separated by reversed-phase semipreparative HPLC using a C18 column (YMC CO., LTD, Pack Pro C18, 250 × 10 mm, 5 μm) with isocratic 40% acetonitrile at a flow rate of 4 mL/min for 30 min. 1D and 2D NMR spectra were recorded on a Bruker Avance III-600 NMR spectrometer (Bruker, Billerica, MA, USA) equipped with CryoprobeTM.
Kinetic parameters determination
Kinetic parameters of BsGT-1 and its mutants for the glycosylation of epothilone B were measured in a reaction mixture (50 μl) containing 20 μg/mL protein, saturated sugar donors, different concentrations of epothilone B, and 50 mM Tris–HCl buffer (50 mM Tris–HCl, pH 7.5, 10 mM MgCl2) at 37°C for 10 min in triplicate. The kinetic values were determined by fitting the Michaelis–Menten curve to the data using the nonlinear regression method (Supplementary Figs. 13–14).
Modeling and docking analysis
The GT sequences were retrieved from the CAZy database and aligned using MAFFT (Version 7)64. The protein structure was remedied by the I-TASSER method with the high-quality crystal structure of BsGT-1 (PDB accession NO: 7BOV) as a reference, and fragment-guided molecular dynamics (FG-MD) simulations and TM-score assessment were used30,38,65. The I-TASSER server first identified structural templates from the PDB database by the multiple threading approach LOMETS, with full-length atomic models constructed by iterative template-based fragment assembly simulations66. The program AutoDock Vina was used to dock epothilone B and UDP-D-glucose to the 3D structure of BsGT-131. The optimal docking results with high absolute values of ΔG were selected based on program scoring (Supplementary Table 3).
The cocrystal structure of tubulin (β-tubulin subunit)-EpoB with 2.4 Å resolution (7DAE) was used as the model to predict the glycosylation impacts on the antitumor activities of EpoB. AutoDock Vina was also used to dock the representative epothilone B 7-O-β-D glucoside (1a) to the target cavity of the β-tubulin subunit of microtubules. The optimal docking results with high absolute values of ΔG are shown in Supplementary Table 10.
Molecular dynamics simulations
Molecular dynamics (MD) simulations of the BsGT-1-EpoB complex were performed using the GROMACS software package67. The protein was solvated using the SPC model68,69,70,71. A cubic box was constructed to perform MD calculations. Water molecules that overlapped with the protein-heavy atoms were removed. The total number of atoms in different solvent systems was ~74,000. To produce a neutral system with 0.1 mol/L ionic concentration, appropriate amounts of Na+ and Cl− were added by replacing water molecules with ions randomly. All MD simulations were performed under periodic boundary conditions. The CHARMM27 force field was used to describe the protein. To eliminate steric interference, the steepest energy minimization was performed for every system to give the maximum force below 1000 kJ mol−1 nm−2. System equilibration was performed at the NVT and NPT ensembles, during which position restraints were applied to protein-heavy atoms. Complete equilibration was assessed by the convergence of the potential energy and the temperatures of the systems. Finally, 100 ns production MD simulations with three replicas were performed in an isothermal−isobaric ensemble. The LINCS algorithm was used to constrain all bonds to hydrogen atoms in the protein, and the SETTLE algorithm was used for the water molecules72,73. The particle mesh Ewald (PME) method was used to evaluate the long-range electrostatic interactions74. The nonbonded pair list cut-off was 10.0 Å, and the pair list was updated every 10 fs.
Water solubility, antitumor, and cytotoxicity assays
The water solubility of EpoB and its glycosides was determined using the minimum shake-flask solubility method26,75.
By using the MTT colorimetric assay, the antitumor and cytotoxicity of EpoB and its glucosides were tested against the liver carcinoma cell line HepG2 and hepatocyte cell line HL-7702, respectively. The cells were seeded in a 96-well microtiter plate, and the wall-attached cells were treated with varying concentrations of drugs by dilution from 300 μM to 0.1 μM for EpoB glycosides (dissolved in DMSO). After 48 h of incubation with the test compounds, the cultures were treated with MTT (2 mg/mL) solution, and the cells were incubated for an additional 4 h at 37°C until a purple precipitate was visible. The purple formazan crystals were dissolved in DMSO, and the absorbance of each sample was measured at 492 nm using a microplate reader. All MTT assay experiments were performed in triplicate, and IC50 values were calculated using GraphPad Prism software (Version 6.01)26.
Immunofluorescence assays
Freshly trypsinized HepG2 cells (1 × 104−1 × 105) were seeded in a 12-well plate and cultivated for 24 h with DMEM (Gibco, Thermo Fisher Scientific, Waltham, MA, USA). Cells were treated with EpoB and 1a at different concentrations (50 nM, 500 nM, and 5 μM) according to the IC50 values for 24 h. The supernatants were removed, and the cells were washed with PBS buffer 2–3 times. The cells were further fixed with 4% paraformaldehyde for 1 h and then permeabilized with 0.2% Triton X-100 (diluted in PBS buffer) for 10 min. Next, the cells were blocked with bovine serum albumin (BSA) for 30 min and incubated with a primary antibody against β-tubulin (1:50, diluted in 2% BSA, β-tubulin Rabbit Monoclonal Antibody, Beyotime Biotechnology) for 1 h at room temperature. After that, the cells were treated with Alexa Fluor 488 goat anti-rabbit IgG (H + L) (secondary antibody, 1:500, diluted in 2% BSA, Beyotime Biotechnology) for another 1 h. The supernatants were removed, the cells were washed, and the nuclei were labeled with DAPI for 10 min. Images were collected using an Olympus BX81 fluorescence microscope system (Olympus, Tokyo, Japan) and a confocal microscope (UltraVIEW®VoX).
Cellular intake assays
HepG2 cells (1 × 105−1 × 106 cells/mL) were cultivated overnight at 37°C and then treated with 5 μM EpoB and glycosides for 1 h. After removing the supernatant, the cells were sequentially rinsed with PBS buffer, trypsinized, and then washed with DMEM. Next, the cells were suspended in acetone, disrupted by ultrasonication, and centrifuged at 12000 rpm for 10 min to remove the debris. The resulting supernatants were dried with nitrogen and then redissolved in 200 μL of methanol. Finally, the compounds were analyzed by HPLC-DAD at 249 nm. Each experiment was conducted in three replicates. The concentrations of EpoB and its glycosides (cellular intake) were calculated based on the linear fitting regression equations of the HPLC peak areas.
Statistics and reproducibility
Glycosylation, water solubility, MTT, and cellular intake assays were independently replicated three times and the individual data points are reported for each experiment in the main figures.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The protein sequences of glycosyltransferases are deposited in GenBank under the following accession numbers: YjiC (AAU40842), BsGT-1 (CUB50191), BaGT (ADP31706), BgGT (SCA85980), BpGT (ARA85718) Details are presented in the Supplementary Table 6. All source data underlying the graphs and charts presented in the main figures are available in the Supplementary Data 1. All other data that support the findings of this study are available from the corresponding authors upon reasonable request.
References
Bollag, D. M. et al. Epothilones, a new class of microtubule-stabilizing agents with a taxol-like mechanism of action. Cancer Res. 55, 2325–2333 (1995).
Kowalski, R. J., Giannakakou, P. & Hamel, E. Activities of the microtubule-stabilizing agents epothilones A and B with purified tubulin and in cells resistant to paclitaxel (Taxol(R)).J. Biol. Chem. 272, 2534–2541 (1997).
Goodin, S., Kane, M. P. & Rubin, E. H. Epothilones: mechanism of action and biologic activity. J. Clin. Oncol. 22, 2015–2025 (2004).
Gerth, K., Bedorf, N., Hofle, G., Irschik, H. & Reichenbach, H. Epothilons A and B: antifungal and cytotoxic compounds from Sorangium cellulosum (Myxobacteria). Production, physico-chemical and biological properties. J. Antibiot. 49, 560–563 (1996).
Giannakakou, P. et al. A common pharmacophore for epothilone and taxanes: molecular basis for drug resistance conferred by tubulin mutations in human cancer cells. Proc. Natl. Acad. Sci. USA 97, 2904–2909 (2000).
Lee, F. Y. et al. BMS-247550: a novel epothilone analog with a mode of action similar to paclitaxel but possessing superior antitumor efficacy. Clin.Cancer Res. 7, 1429–1437 (2001).
Pronzato, P. New therapeutic options for chemotherapy-resistant metastatic breast cancer: the epothilones. Drugs 68, 139–146 (2008).
Cardoso, F., de Azambuja, E. & Lago, L. D. Current perspectives of epothilones in breast cancer. Eur. J. Cancer 44, 341–352 (2008).
Chiorazzi, A. et al. Experimental epothilone B neurotoxicity: results of in vitro and in vivo studies. Neurobiol. Dis. 35, 270–277 (2009).
Hancock, S. M., Vaughan, M. D. & Withers, S. G. Engineering of glycosidases and glycosyltransferases. Curr. Opin. Chem. Biol. 10, 509–519 (2006).
Chang, A., Singh, S., Phillips, G. N. Jr. & Thorson, J. S. Glycosyltransferase structural biology and its role in the design of catalysts for glycosylation. Curr. Opin. Biotechnol. 22, 800–808 (2011).
Liang, D. M. et al. Glycosyltransferases: mechanisms and applications in natural product development. Chem. Soc. Rev. 44, 8350–8374 (2015).
Gloster, T. M. Advances in understanding glycosyltransferases from a structural perspective. Curr. Opin. Struct. Biol. 28, 131–141 (2014).
Gupta, R. D. Recent advances in enzyme promiscuity. Sustain. Chem. Process. 4, 2 (2016).
Khersonsky, O. & Tawfik, D. S. in Annual Review of Biochemistry Vol. 79 (eds Kornberg,R. D. et al.) 471–505 (Annual Reviews, 2010).
Zhang, C. et al. Exploiting the reversibility of natural product glycosyltransferase-catalyzed reactions. Science 313, 1291–1294 (2006).
Bolam, D. N. et al. The crystal structure of two macrolide glycosyltransferases provides a blueprint for host cell antibiotic immunity. Proc. Natl. Acad. Sci. USA 104, 5336–5341 (2007).
Zhang, P., Zhang, Z., Zhang, L., Wang, J. & Wu, C. Glycosyltransferase GT1 family: Phylogenetic distribution, substrates coverage, and representative structural features. Comput. Struct. Biotechnol. J. 18, 1383–1390 (2020).
Masada, S., Terasaka, K. & Mizukami, H. A single amino acid in the PSPG-box plays an important role in the catalytic function of CaUGT2 (curcumin glucosyltransferase), a group D family 1 glucosyltransferase from Catharanthus roseus. FEBS Lett. 581, 2605–2610 (2007).
Shao, H. et al. Crystal structures of a multifunctional triterpene/flavonoid glycosyltransferase from Medicago truncatula. Plant Cell 17, 3141–3154 (2005).
He, J. B. et al. Molecular and structural characterization of a promiscuous C-Glycosyltransferase from Trollius chinensis. Angew. Chem. Int. Ed. Engl. 58, 11513–11520 (2019).
Chen, D. W. et al. Probing and engineering key residues for bis-C-glycosylation and promiscuity of a C-glycosyltransferase. ACS Catal. 8, 4917–4927 (2018).
Akere, A. et al. Structure-based enzyme engineering improves donor-substrate recognition of Arabidopsis thaliana glycosyltransferases. Biochem. J. 477, 2791–2805 (2020).
Dai, L. et al. Exploiting the aglycon promiscuity of glycosyltransferase Bs-YjiC from Bacillus subtilis and its application in synthesis of glycosides. J. Biotechnol. 248, 69–76 (2017).
Zhang, P. et al. Phylogeny-guided characterization of glycosyltransferases for epothilone glycosylation. Microb. Biotechnol. 12, 763–774 (2019).
Zhang, P. et al. Effects of glycosylation on the bioactivity of rapamycin. Appl. Microbiol. Biotechnol. https://doi.org/10.1007/s00253-020-10895-3 (2020).
Gantt, R. W., Goff, R. D., Williams, G. J. & Thorson, J. S. Probing the aglycon promiscuity of an engineered glycosyltransferase. Angew. Chem. Int. Ed. Engl. 47, 8889–8892 (2008).
Zhang, C., Albermann, C., Fu, X. & Thorson, J. S. The in vitro characterization of the iterative avermectin glycosyltransferase AveBI reveals reaction reversibility and sugar nucleotide flexibility. J. Am. Chem. Soc. 128, 16420–16421 (2006).
Liu, B. et al. Structural and biochemical studies of the glycosyltransferase Bs-YjiC from Bacillus subtilis. Int. J. Biol. Macromol. https://doi.org/10.1016/j.ijbiomac.2020.10.238 (2020).
Zhang, Y. I-TASSER server for protein 3D structure prediction. BMC Bioinform. 9, 40 (2008).
Trott, O. & Olson, A. J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31, 455–461 (2010).
Abraham, M. J. et al. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1-2, 19–25 (2015).
Paquette, S., Møller, B. L. & Bak, S. On the origin of family 1 plant glycosyltransferases. Phytochemistry 62, 399–413 (2003).
Modolo, L. V., Escamilla-Trevino, L. L., Dixon, R. A. & Wang, X. Q. Single amino acid mutations of Medicago glycosyltransferase UGT85H2 enhance activity and impart reversibility. FEBS Lett. 583, 2131–2135 (2009).
Offen, W. et al. Structure of a flavonoid glucosyltransferase reveals the basis for plant natural product modification. EMBO J. 25, 1396–1405 (2006).
Osmani, S. A., Bak, S. & Moller, B. L. Substrate specificity of plant UDP-dependent glycosyltransferases predicted from crystal structures and homology modeling. Phytochemistry 70, 325–347 (2009).
Gantt, R. W., Peltier-Pain, P., Singh, S., Zhou, M. & Thorson, J. S. Broadening the scope of glycosyltransferase-catalyzed sugar nucleotide synthesis. Proc. Natl. Acad. Sci. USA 110, 7648–7653 (2013).
Zhang, Y. & Skolnick, J. TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 33, 2302–2309 (2005).
Altmann, K. H., Wartmann, M. & O’Reilly, T. Epothilones and related structures-a new class of microtubule inhibitors with potent in vivo antitumor activity. Biochim. Biophys. Acta 1470, M79–M91 (2000).
Senapati, S., Mahanta, A. K., Kumar, S. & Maiti, P. Controlled drug delivery vehicles for cancer treatment and their performance. Signal Transduct. Target. Ther. 3, 7 (2018).
Elshahawi, S. I., Shaaban, K. A., Kharel, M. K. & Thorson, J. S. A comprehensive review of glycosylated bacterial natural products. Chem. Soc. Rev. 44, 7591–7697 (2015).
Nidetzky, B., Gutmann, A. & Zhong, C. Leloir glycosyltransferases as biocatalysts for chemical production. ACS Catal. 8, 6283–6300 (2018).
Joshi, R. et al. Semirational design and engineering of grapevine glucosyltransferases for enhanced activity and modified product selectivity. Glycobiology 29, 765–775 (2019).
Li, J. et al. Efficient O-glycosylation of triterpenes enabled by protein engineering of plant glycosyltransferase UGT74AC1. ACS Catal. 10, 3629–3639 (2020).
Li, X. H. et al. Site-saturation mutagenesis of proline 176 in cyclodextrin glucosyltransferase from Bacillus sp. Y112 effects product specificity and enzymatic properties. Process Biochem. 94, 180–189 (2020).
Wang, L., Xia, Y., Su, L. & Wu, J. Modification of bacillus clarkii γ-cyclodextrin glycosyltransferase and addition of complexing agents to increase γ-cyclodextrin production. J. Agric. Food Chem. 68, 12079–12085 (2020).
Zhang, M. et al. Functional characterization and structural basis of an efficient di-C-glycosyltransferase from Glycyrrhiza glabra. J. Am. Chem. Soc. 142, 3506–3512 (2020).
Liu, S. et al. Characterization of a Highly Selective 2″-O-Galactosyltransferase from Trollius chinensis and structure-guided engineering for improving UDP-glucose selectivity. Org. Lett. https://doi.org/10.1021/acs.orglett.1c02581 (2021).
Wang, Z. et al. Highly promiscuous flavonoid 3- O-glycosyltransferase from Scutellaria baicalensis. Org. Lett. 21, 2241–2245 (2019).
Liu, M. et al. Crystal structures of the C-glycosyltransferase UGT708C1 from buckwheat provide insights into the mechanism of C-glycosylation. Plant Cell 32, 2917–2931 (2020).
Williams, G. J., Goff, R. D., Zhang, C. & Thorson, J. S. Optimizing glycosyltransferase specificity via “hot spot” saturation mutagenesis presents a catalyst for novobiocin glycorandomization. Chem. Biol. 15, 393–401 (2008).
Zhang, C. et al. Biochemical and structural insights of the early glycosylation steps in calicheamicin biosynthesis. Chem. Biol. 15, 842–853 (2008).
Albesa-Jové, D. et al. Structural snapshots and loop dynamics along the catalytic cycle of glycosyltransferase GpgS. Structure. 25, 1034–1044.e1033 (2017).
Williams, G. J., Zhang, C. & Thorson, J. S. Expanding the promiscuity of a natural-product glycosyltransferase by directed evolution. Nat. Chem. Biol. 3, 657–662 (2007).
Zhang, X. & Li, S. Expansion of chemical space for natural products by uncommon P450 reactions. Nat. Prod. Rep. 34, 1061–1089 (2017).
Kuo, M. S. et al. Microbial glycosylation of erythromycin-A. Antimicrob. Agents Chemother. 33, 2089–2091 (1989).
Burg, R. W. et al. Avermectins, new family of potent anthelmintic agents: producing organism and fermentation. Antimicrob. Agents Chemother. 15, 361–367 (1979).
Zhang, G., Li, Y., Fang, L. & Pfeifer, B. A. Tailoring pathway modularity in the biosynthesis of erythromycin analogs heterologously engineered in E. coli. Sci. Adv. 1, e1500077 (2015).
Zhai, Y. et al. Enhancing GDP-fucose production in recombinant Escherichia coli by metabolic pathway engineering. Enzyme Microb. Technol. 69, 38–45 (2015).
Guan, W., Cai, L., Fang, J., Wu, B. & George Wang, P. Enzymatic synthesis of UDP-GlcNAc/UDP-GalNAc analogs using N-acetylglucosamine 1-phosphate uridyltransferase (GlmU). Chem. Commun. (Camb.) https://doi.org/10.1039/b917573c (2009).
Fang, J. et al. Systematic study on the broad nucleotide triphosphate specificity of the pyrophosphorylase domain of the N-acetylglucosamine-1-phosphate uridyltransferase from Escherichia coli K12. Bioorg. Med. Chem. Lett. 19, 6429–6432 (2009).
Guo, Y. et al. Comparing substrate specificity of two UDP-sugar pyrophosphorylases and efficient one-pot enzymatic synthesis of UDP-GlcA and UDP-GalA. Carbohydr. Res. 411, 1–5 (2015).
Martin, T. J., Braun, H. & Schmidt, R. R. A convenient synthesis of nucleoside monophosphate-N-acetylneuraminic acids (NMP-Neu5Ac). Bioorg. Med. Chem. 2, 1203–1208 (1994).
Katoh, K., Rozewicki, J. & Yamada, K. D. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Briefings in bioinformatics https://doi.org/10.1093/bib/bbx108 (2017).
Zhang, J., Liang, Y. & Zhang, Y. Atomic-level protein structure refinement using fragment-guided molecular dynamics conformation sampling. Structure. 19, 1784–1795 (2011).
Yang, J. & Zhang, Y. I-TASSER server: new development for protein structure and function predictions. Nucleic Acids Res. 43, W174–W181 (2015).
Pronk, S. et al. GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics 29, 845–854 (2013).
Zielkiewicz, J. Structural properties of water: comparison of the SPC, SPCE, TIP4P, and TIP5P models of water. J. Chem. Phys. 123, 104501 (2005).
Chen, L. et al. Thermal stability and unfolding pathways of hyperthermophilic and mesophilic periplasmic binding proteins studied by molecular dynamics simulation. J. Biomol. Struct. Dyn. 34, 1576–1589 (2016).
Paul, M., Hazra, M., Barman, A. & Hazra, S. Comparative molecular dynamics simulation studies for determining factors contributing to the thermostability of chemotaxis protein “CheY”. J. Biomol. Struct. Dyn. 32, 928–949 (2014).
Kundu, S. & Roy, D. Temperature-induced unfolding pathway of a type III antifreeze protein: insight from molecular dynamics simulation. J. Mol. Graph. Model 27, 88–94 (2008).
Miyamoto, S. & Kollman, P. A. Settle: An analytical version of the SHAKE and RATTLE algorithm for rigid water models. J. Comput. Chem. 13, 952–962 (1992).
Hess, B., Bekker, H., Berendsen, H. J. & Fraaije, J. G. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 18, 1463–1472 (1997).
Abraham, M. J. & Gready, J. E. Optimization of parameters for molecular dynamics simulation using smooth particle‐mesh Ewald in GROMACS 4.5. J. Comput. Chem. 32, 2031–2040 (2011).
Veseli, A., Žakelj, S. & Kristl, A. A review of methods for solubility determination in biopharmaceutical drug characterization. Drug Dev. Ind. Pharm. 45, 1717–1724 (2019).
Acknowledgements
This work was financially supported by the National Key Research and Development Programs of China (nos. 2018YFA0901704, 2021YFC2101000, 2018YFA0900400, and 2019YFA0905700), the National Natural Science Foundation of China (NSFC) (nos. 31670076, 31471183, 31900042, and 81973215), and the Excellent Youth Program of Shandong Natural Science Foundation (no. ZR2020YQ62). The scientific calculations in this paper were performed on the HPC Cloud Platform of Shandong University. We would like to thank Haiyan Sui, Jing Zhu, and Zhifeng Li from State Key Laboratory of Microbial Technology of Shandong University for NMR and LC-MS measurements.
Author information
Authors and Affiliations
Contributions
P.Z., C.W., and Y.L. designed studies; P.Z., L.Z., X.D., S.L., and D.L. performed studies; P.Z., L.Z., X.J., Z.Z., D.W., C.W., and Y.L. analyzed data; X.J., J.F., and Y.T. contributed analytical tools; P.Z., C.W., and Y.L. wrote the paper; Y.L. and C.W. provided funds for the project. All authors read and approved the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editor: Anam Akhtar.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhang, P., Zhang, L., Jiang, X. et al. Docking-guided rational engineering of a macrolide glycosyltransferase glycodiversifies epothilone B. Commun Biol 5, 100 (2022). https://doi.org/10.1038/s42003-022-03047-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-022-03047-y
This article is cited by
-
Improving the Catalytic Properties of Xylanase from Alteromones Macleadii H35 Through Sequence Analysis
Applied Biochemistry and Biotechnology (2024)
-
Unnatural activities and mechanistic insights of cytochrome P450 PikC gained from site-specific mutagenesis by non-canonical amino acids
Nature Communications (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.