Abstract
The lateral lemniscus encompasses processing stages for binaural hearing, suppressing spurious frequencies and frequency integration. Within the lemniscal fibres three nuclei can be identified, termed after their location as dorsal, intermediate and ventral nucleus of the lateral lemniscus (DNLL, INLL and VNLL). While the DNLL and VNLL have been functionally and anatomically characterized, less is known about INLL neurons. Here, we quantitatively describe the morphology, the cellular orientation and distribution of synaptic contact sites along dendrites in mature Mongolian gerbils. INLL neurons are largely non-inhibitory and morphologically heterogeneous with an overall perpendicular orientation regarding the lemniscal fibers. Dendritic ranges are heterogeneous and can extend beyond the nucleus border. INLL neurons receive VGluT1/2 containing glutamatergic and a mix of GABA- and glycinergic inputs distributed over the entire dendrite. Input counts suggest that numbers of excitatory exceed the inhibitory contact sites. Axonal projections indicate connectivity to ascending and descending auditory structures. Our data show that INLL neurons form a morphologically heterogeneous continuum and incoming auditory information is processed on thin dendrites of various length and biased to perpendicular orientation. Together with the different axonal projection patterns, this indicates that the INLL is a highly complex structure that might hold many unexplored auditory functions.
Similar content being viewed by others
Introduction
In Mammals, within the fiber bundle of the lateral lemniscus (LL), at least three major neuronal populations can anatomically be identified. These nuclei are termed according to their position dorsal (DNLL), intermediate (INLL) and ventral (VNLL) nucleus of the lateral lemniscus1,2. From these nuclei, the DNLL can be identified by its ventral location to the inferior colliculus (IC) and the fiber crossing of the LL with the commissure of Probst2,3. In gerbils, the DNLL appears roundish3,4. A thin layer of highly horizontally organized bipolar cells3,5 of unknown function defines the ventral boarder of the DNLL towards the INLL. Ventrally the INLL merges with the cell population of the VNLL. The VNLL stretches to the most ventral site of the LL and is composed of a globular and a heterogeneously shaped cell population2,3,6. While the anatomical border between the INLL and the VNLL appears less well defined, both populations differ in their transmitter content. Neurons in the VNLL are co-assigned to GABA and glycine transmitters and, in rodent and cat, the INLL to glutamate7,8, while in the bat INLL also glycinergic neurons were reported9,10,11. The transmitter content of INLL neurons in gerbils is so far undetermined.
The morphology of DNLL neurons was suggested to be multipolar and elongated2,3,12,13 with a large range of somatic sizes12. In the VNLL, multipolar or elongated neurons are distributed throughout the nucleus with dendrites ranging up to 400 µm14. In rodents, globular VNLL neurons are located at the ventro-lateral side3,6,14 and are presumably the targets of large endbulb synapses6,15,16,17,18. In the INLL, neurons were suggested to be multipolar or horizontally arranged3, but a detailed and quantitative description or single cell reconstructions have not been reported. Thus, it is unclear how they compare to the other neuronal populations in the lateral lemniscus.
In the VNLL, synaptic input structures are well-described for globular neurons receiving a large somatic, excitatory endbulb synapse6,15,16,17,18. Other synapses emerge from small and thick afferent fibers with different sized boutons18,19. For DNLL neurons, inputs arising from the contralateral side and lemniscal fibers form pericellular baskets20, suggesting that GABAergic synapses are localized over the entire cell surface, which is corroborated by identified GABAergic synapses at dendrites and soma21. At other locations in the LL the composition of synaptic structures and their location at distinct cellular compartments remains elusive. Again only in the DNLL and VNLL, the functional input patterns have been investigated. Globular VNLL neurons receive a large excitatory input6,16,22,23 while other VNLL neurons receive many small excitatory inputs6, and inhibitory currents are similar in size and kinetics throughout the VNLL6. In the DNLL, excitation is based on AMPA and NMDA receptor mediated currents4,24,25,26,27 and inhibition is largely GABAA receptor mediated28,29. In the INLL, in vivo pharmacology indicated that inhibition is conveyed by ionotropic GABA and glycine receptors30,31, while excitatory inputs are unexplored. However, one function that was assigned to a subset of INLL neurons is in connecting auditory information across different sound frequencies32.
Here we assay the transmitter content, the single cell morphology and the distribution of synaptic inputs for neurons in the INLL in mature Mongolian gerbils. As in other rodents, INLL neurons appear largely non-inhibitory. Cells exhibit different morphologies and a wide range of dendritic sizes and shapes. INLL dendrites are overall perpendicular organized to the LL and in some cases extend beyond the medial border of the INLL. Excitatory input appears driven by VGluT1 and VGluT2 positive terminals. Inhibition appears dominated by GlyT2 contact sites but GABAergic terminals are also present. Excitatory and inhibitory contact sites show little clustering.
Methods
Animals
Mongolian gerbils (Meriones unguiculatus) of both sexes, based on Charles River hereditary background, were kept in the institute’s animal facility under 12 h light/dark cycle, fed ad libitum and were used between postnatal day (P) 28 and 40. Experiments were approved by the University animal welfare committee and animal welfare officer under the license number TiHo-T-2019–4 and TiHo-T-2021–4 and were compliant with German local and federal laws in accord with ARRIVE guidelines. We have used gerbils in this study, as these animals are a well-documented animal model for hearing research. The data presented in this study was obtained from a total of 15 gerbils. For single cell electroporation eight (four transversal cuts and four sagittal cuts), for single cell electroporation with post-hoc immunofluorescence labelling four animals and for standard immunofluorescence three animals were used.
Single cell labelling
For single cell electroporation, acute slices containing the lateral lemniscus were prepared. Gerbils of P28-P45 were anesthetized with isoflurane and after decapitation, brains were rapidly removed in preparation solution. Preparation solution 1 for morphological reconstruction contained (in mM): 93 NMDG, 93 HCl, 30 NaHCO3, 1.2 NaH2PO4, 2.5 KCl, 25 glucose, 20 HEPES, 5 ascorbic acid, 3 myo-inositol, 3 sodium pyruvate, 10 MgCl2, 2 CaCl2 and was bubbled with O2/CO2 (95/5%) to achieve a pH of 7.4. Preparation solution 2 for electroporation with subsequent immunofluorescent labelling contained (in mM): 120 saccharose, 25 NaCl, 25 NaHCO3, 1.25 NaH2PO4, 2.5 KCl, 25 glucose, 0.4 ascorbic acid, 3 myo-inositol, 0.11 sodium pyruvate, 3 MgCl2, 0.1 CaCl2 and was bubbled with O2/CO2 (95/5%) to achieve a pH of 7.4. Brains were trimmed, fixed in the slice chamber and submerged with the respective preparation solution. Transversal or sagittal slices of 200 µm (for morphological reconstruction) or 400 µm (for contact site reconstruction) thickness that contained the lateral lemniscal nuclei were taken. Slices were incubated either for 30 min at 34°C in recording solution (after using preparation solution 1), or for 7 min at 36°C in recording solution (after using preparation solution 2). Extracellular recording solution contained (in mM): 125 NaCl, 25 NaHCO3, 1.25 NaH2PO4, 2.5 KCl, 25 glucose, 0.4 ascorbic acid, 3 myo-inositol, 0.11 sodium pyruvate, 1 MgCl2, 1.2 CaCl2 and was bubbled with O2/CO2 (95/5%) to achieve a pH of 7.4.
For single cell electroporation, slices were transferred to a set up with an upright BX50WI Olympus microscope equipped with a TILL Photonics Imaging system composed of an Imago CCD camera, a Polychorme IV and a control unit managed via TILLvisION software. Neurons were visualized with a 60 × or 40 × water immersion objective. Slices were continuously perfused with recording solution and kept at ~ 34°C. Patch-pipettes with a tip-diameter of about 1 µm were filled with 10 mM Alexa568 hydrazide or Alexa594 hydrazide dye (ThermoFisher scientific) dissolved in water. The dye filled pipette was pressed onto a visually selected neuron and a single ~ 15 ms long voltage pulse of ~ 15 V was applied. The trigger for the voltage pulse was delivered from a HEKA EPC10/2 amplifier to a stimulator unit (Model 2100, A-M systems). For re-slicing, cells were filled under whole cell configuration for at least 10 min using an intracellular solution containing (in mM): 145 K-gluconate, 15 HEPES, 4.5 KCl, 7.5 Na2-phosphocreatine, 2 Mg-ATP, 2 K2-ATP, 0.3 Na2-GTP and 0.5 K-EGTA, 0.3 Alexa568 hydrazide. Cell loading was visually verified. Slices with dye loaded cells were fixed in 4% paraformaldehyde (PFA) (for morphological reconstruction) or 3.7% PFA with 0.3% glutaraldehyde (for contact site reconstruction) over night. After fixation, slices were washed three times for 5 min in PBS. 200 µm slices were mounted with Vectashield and prepared for microscopy. 400 µm thick slices were mounted and readily used for overview microscopy. Images with a 2.5 × 0.075NA and a 20 × 0.5NA objective were taken to identify the position of the cells within the INLL and the shape of their soma and primary dendrites. These images were required for final re-assembly, because these 400 µm thick sections were re-sliced in 50 µm thick sections and prepared for immunofluorescence labelling.
Immunofluorescence labelling
Gerbils were euthanized by inhalation of carbon dioxide and declared dead after 1–2 min of breathing arrest. Subsequently, they were transcardially perfused with cold Ringer solution containing heparin, followed by fixation with 3.7% PFA with 0.3% glutaraldehyde (for transmitter content labelling) or with 2% PFA containing 15% picric acid (for pre-synaptic labelling). Brains were removed, post-fixed overnight in PFA and washed with phosphate-buffered saline (PBS), adjusted to pH 7.4. Transversal slices of 50 μm were obtained with a Leica vibratome VT1200S. Sections from perfusions and from re-sliced electroporations were labelled in the same manner. Free-floating sections were washed in PBS, treated with sodium borohydride (1 mg/ml, for 10 min on ice) and subsequently with blocking solution (0.5% Triton, 1% bovine serum albumin and 0.1% saponin, diluted in PBS) for 30 min at room temperature. Sections were incubated for 2–3 days at 4°C in blocking solution containing primary antibodies (see Table 1). Sections were washed in 0.5% Triton and 0.1% saponin diluted in PBS and incubated in blocking solution containing secondary antibodies (see Table 2) for 4 h at room temperature. Sections were washed with PBS and mounted on microscope slides embedded in fluorescence mounting medium (VECTASHIELD©, Vector Laboratories).
Image acquisition, alignment and three-dimensional reconstruction
The appropriate localization of the soma of electroporated neurons within the INLL was verified by epifluorescence overview microscopy. The LL fiber bundle was apparent by an increased autofluorescence excited with 488 nm. Images for single cell reconstruction were acquired with a confocal laser microscope (Leica TCS SP5, Leica Microsystems GmBH, Wetzlar, Germany) using a 40 × 1.25 NA oil immersion objective with an optical zoom of 2.5-fold. The magnification led to a pixel size of 0.303 × 0.303 µm. Z-stacks were taken with 0.29 µm step size generating a nearly cubic voxel. AMCA-fluorescence was excited with a 405 nm laser, Alexa488-fluorescence with an Argon Laser with the excitation tuned to 488, Alexa568-fluorescence with a HeNe laser tuned to 594 nm. Emission filters were adjusted to 410 – 470 nm for AMCA, 495 – 540 nm for Alexa488 and 598 – 700 nm for Alexa568. Images were acquired sequentially with a line averaging of 6 or 7. Images taken from standard free floating immunofluorescence labelling were taken with the same confocal laser microscope with 20 × 0.75 NA, 40 × 1.25 NA objective or for higher magnifications 63 × 1.4 NA objective.
Care was taken that the gain during image acquisition was adjusted so that all small cell compartments could be detected within the image stack. In this approach, the image stack that contains the soma and proximal dendrites lead to a largely over-illuminated soma. Therefore a second soma stack was taken where the gain was adjusted to be non-over-illuminating the soma. From these image stacks, the soma volume was reconstructed and used for further analysis. All other z-stacks that contained parts of a given cell were stitched together with the Fiji plugin33 to form an image of the entire optical reconstruction of a cell. Stitched image stacks containing labelled neurons were imported to Neurolucida360 (MBFBioscience) with the voxel size defined from the confocal reconstruction. In this image volume, neurons were manually reconstructed in three dimensions. As a central reference point the soma was digitalized first. From this starting point, individual processes were traced manually towards their distal end and the diameter was continuously adjusted. Nissl stains were imaged with a bright field microscope (Axioskop) with a 2.5 × 0.075 NA objective. Images were acquired with a USB camera (DFK 23UX249, Imaging Source) controlled by IC capture software. Images were post-hoc edited in Fiji and pairwise stitched33.
Data analysis and statistics
Digitalized cells were transferred from NeuroLucida360 to Neurolucida Explorer for analysis. The neuron summary extracted the number of primary dendrites, number of nodes, total dendritic length, total dendritic volume and soma volume. A three-dimensional Sholl analysis with 10 µm concentric shell interval originating from soma center was carried out. We extracted the average diameter, cumulative length, cumulative area and number of intersections for each Sholl radii. For the wedge analysis in Neurolucida360, the reference angle was adjusted perpendicular to the lateral direction of the lateral lemniscus in image volumes and centered on the soma. The dendritic length within 30° wedges was extracted and summed in polar plots according to their orientation. Synaptic contact sites were visually detected by the overlay of dendritic fluorescence from cell labelling and VGluT1/2 or GlyT2 immunofluorescence. Here, we define a synapse as the sum of all contact sites between a single pre-synaptic axon with a post-synaptic partner. Thereby, a single contact site might be composed of multiple active zones. A contact site is therefore the close proximity, partial fluorescence overlap, of a single presynaptic marked junction with the post-synaptic element. For positive identification, at least two successive images of the z-stack were required to show this fluorescence overlay. Contact site location was determined in two ways. First, input location was determined as distance from soma center according to the Sholl distance of the concentric shells and the nearest neighbor tool of Neurolucida360 was used to extract the inter-contact sites distance. Second, the contact site location was mapped directly on the extent and shape of the dendrite. For this purpose, the number of pixels of the dendritic extent between soma and contact site location were measured and converted to µm. Each dendritic branch was mapped from the soma and contact sites on each branch were only counted once. The distance was expressed as relative distance of the length of the dendrite from soma to distal end.
Values obtained from Neurolucida360 and Neurolucida Explorer were exported to Excel or IgorPro and further processed. Statistical significance was assayed by t-tests. Data are shown as mean ± SEM and for inter-contact distance for the example neuron as medians. Average values for Sholl analysis was only derived from radii with a minimum of events from three cells. Correlations of morphological features were statistically analyzed in IgorPro with the linear correlation tool and their r and p values are given.
Results
To anatomically address the INLL in Mongolian gerbils, Nissl staining on serial sections were carried out. The three LL nuclei could be identified (Fig. 1a) with the following landmarks. The DNLL resides ventrally of the IC with roundish appearance. The INLL follows ventrally to the DNLL and reaches to the position where the brain tapers to the ventral edge. The VNLL is slightly thinner compared to the other LL nuclei and stretches far ventrally where dark Nissl staining is present. These landmarks correspond to differences in transmitter content (Fig. 1b). DNLL neurons showed positive immunoreactivity to GABA but not glycine. INLL neurons mostly lack somatic immunoreactivity for both inhibitory transmitters. VNLL neurons appeared positive for both GABA and glycine immunoreactivity in individual cells, indicating the described co-release34. These findings are illustrated by higher magnification images (Fig. 1c). To confirm the specificity of antibody labelling, we added a labelling of the medial nucleus of the trapezoid body (MNTB), where MNTB neurons show their well-known glycinergic, non-GABAergic transmitter content. Thus, the profile of specific transmitter types in the gerbil LL matches that in cat8 and other rodents7.
Position and transmitter content of nuclei in the lateral lemniscus. (a) Nissl staining of the lateral lemnisicus and inferior colliculus. Black circle indicates position of the DNLL, magenta circle the position of the INLL and pink square the position of the VNLL. Scale bar equals 1 mm. (b) GABA (green) and glycine (red) immunofluorescence in the DNLL, INLL and VNLL (from left to right). Scale bare equals 100 µm. (c) Co-labelling of MAP2 (blue), GABA (green) and glycine (red) immunofluorescence in the DNLL, INLL, VNLL and MNTB as indicated. Triple staining is given on the left, the single fluorescence of each channel is given in black in the indicated order: MAP-2, GABA and glycine. Scale bar equals 50 µm.
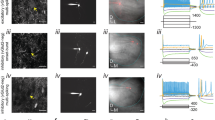
Neurons in the INLL were suggested to be horizontally oriented or multipolar2,3. To quantify the cellular extent, shape and orientation, we electroporated single neurons in acute brain slices and recovered their morphology (Fig. 2a). Images were acquired with the ventro-dorsal axis of the lateral lemniscus always being vertically oriented. The recovered neuron morphology was digitalized using Neurolucida 360 software (transversal: n = 25, sagittal: n = 17; Fig. 2ai, aii). The orientation of the reconstructed cells was analyzed to gain insight into the dendritic arrangement within the lemniscal fiber tract. Towards this aim, the total dendritic length in a 30° wedge (Fig. 2ai, aii) was cumulated. Some neurons were bipolar and horizontally orientated (Fig. 2aii), while others were multipolar with little specific orientation (Fig. 2ai). Since we took care that all images were aligned with the ventro-dorsal axis of the lateral lemniscus, we were able to average the orientation of our sampled neurons. The overall dendritic orientation appeared horizontally orientated perpendicular to the fiber bundle irrespectively of the section plane (Fig. 2b). In case cells were recovered from transversal cuts, the average polar graph indicated that the lateral processes of INLL neurons are more focused in the perpendicular orientation to the ascending fibers than the medial extent (Fig. 2b). Thus, despite the heterogeneity of dendritic orientation, the overall population of INLL neurons appeared to have their dendrites orthogonally spread out in respect to the passing lemniscal fibers. No specific pattern of cell orientation was observed for cell locations within the transversal INLL template (Fig. 2c). Moreover, placing all reconstructed cells into a transversal template of the INLL nucleus (Fig. 2d), indicated that some neurons spanned the entire horizontal width of the nucleus and beyond.
Neuronal orientation relative to the lemniscal fibers. (a) Examples of two electroporated INLL neurons. Scale bar equals 100 µm. (ai and aii) Left: Reconstructions from neurons shown in (a). Wedges used to extract dendritic length in a given orientation are indicated by gray lines. Right: Summed dendritic length of a given wedge presented as polar graph. Axis values are in µm. (b) Average dendritic length of all reconstructed INLL neurons (transversal: n = 25, black and sagittal: n = 17, gray) as a function of orientation. Axis values are in µm. (c) Polar plots of single cells at their position within a schematized INLL in the transversal plane. Sizes of polar plots are normalized to the maximal extension. (d) Overview of the position of reconstructed cells in the tranversal INLL. All labelled and reconstructed cells were added to the schematized INLL nucleus area (gray surrounding) and scaled to a similar proportion. Green compartments are axonal structures.
INLL neurons were further characterized by morphometric parameters extracted from the digitally reconstructed neurons (transversal: n = 25, sagittal: n = 17). INLL neurons had average somatic volumes of 766.3 ± 67.4 µm3 ranging from 396.7 to 1870.8 µm3 for transversal and 1232 ± 207 µm3 ranging from 424.0 to 3659.9 µm3 for sagittal sections. In average these neurons had 4.0 ± 0.3 and 3.8 ± 0.3 primary dendrites in transversal and sagittal slices respectively. The number of primary dendrites did not correlate with the soma volume (transversal: r = 0.094, p = 0.326; sagittal: r = 0.169, p = 0.259; Fig. 3a). The number of branch points, nodes, can approximate the elaboration of dendrites. INLL neurons showed a large heterogeneity in the node number (Fig. 3b). Some neurons had dendrites with little branching or side protrusions, while in some dendrites many small protrusions were observed (Fig. 3b). Since these protrusions were larger than 10 µm, they were not categorized as dendritic spines according to Hering and Sheng35. Neither the number of nodes (transversal: r = 0.201, p = 0.163, sagittal: r = 0.023, p = 0.465; Fig. 3b), nor the total dendritic length of INLL neurons correlated with soma volume (transversal: r = 0.174, p = 0.203, sagittal: r = 0.179, p = 0.245; Fig. 3c). However, the total dendritic volume correlated with the somatic volume (transversal: r = 0.674, p = 0.00011, sagittal: r = 0.646, p = 0.003; Fig. 3d), indicating that neurons with larger soma had overall thicker dendrites. These data of single cells did not indicate specific subpopulations, but rather appeared to form a continuum of morphological features. Therefore, no classification in specific neuronal morphologies was implemented.
Basic morphological features of INLL neurons. (a) Number of primary dendrites emerging from a single soma as a function of the corresponding soma volume. Black symbols correspond to neurons recovered from transversal sections, gray symbols correspond to neurons recovered from sagittal sections. (b) Number of dendritic nodes as a function of soma volume. Insets show dendritic segments. Color code as in (a). One from a neuron without small dendritic protrusions and one from a neuron with such protrusions. Scale bars equals 10 µm. (c) Total dendritic length as a function of soma volume. Color code as in (a). (d) Total dendritic volume as a function of soma volume. Color code as in (a).
In 10 of our 42 single cell electroporated neurons (transversal: 7/25, sagittal: 3/17) an emerging axon was identified, by either an axonal bleb or its pathway to the DNLL or beyond. From these axons, 8 showed collaterals that not only indicated an ascending but also a descending information transfer, as exemplified in Fig. 4a and b. The reconstructed INLL neurons were further subjected to a Sholl analysis. Beside the electroporated neurons, where data obtained from transversal and sagittal sections were pooled, eight neurons were dye loaded under whole-cell conditions and their reconstructions were also used for Sholl analysis. Both approaches resulted in similar dendritic distributions along the Sholl radii. The average diameter of the dendrites dropped from the soma (electroporation: 1.68 ± 0.13 µm, n = 42; whole-cell filling: 1.17 ± 0.28 µm, n = 8; Fig. 4c) towards the distal dendritic end (electroporation 0.44 ± 0.05 µm, n = 33 and whole-cell filling: 0.58 ± 0.12 µm, n = 4 at 200 µm Sholl radius). Moreover, we found that dendrites in the INLL could extend up to 700 µm from the soma center. The dendritic diameter differed not from the extracted axonal diameter (electroporation dendrite 0.34 ± 0.04 µm, n = 11 and electroporation axon 0.45 ± 0.07µm, n = 8 at 350 µm Sholl radius; t-test, p = 0.36; Fig. 4c). Thus, the diameter of the different compartments did not allow their unambiguous categorization. The cumulated length and area of dendritic segments peaked (cumulated length electroporation: 109,4 ± 10.0 µm; whole-cell filling: 115.3 ± 27.8 µm; cumulated area: electroporation: 263.5 ± 30.9 µm2; whole-cell filling: 238.5 ± 54.3 µm2) at a Sholl radius of 60 – 70 µm (Fig. 4d and e). This peak indicated a local increase in arborisation and was similar to the stellate cells described in the VNLL14. The transient increase in arborisation was corroborated by the number of intersections within a Sholl radius (Fig. 4f). This value reached a maximal average of 7.9 ± 0.7 at 60 µm Sholl radius and 7.9 ± 1.6 at 50 µm for electroporated and whole-cell filled neurons respectively.
Sholl analysis of reconstructed INLL neurons. (a) Reconstructed example neuron. Dendrites colored in magenta, soma in cyan and axon in green. (b) Concentric shells of the Sholl analysis for the example neuron shown in (a). (c) Average diameter of neuronal processes within the Sholl radii. Dendritic data from whole-cell fillings colored in black (n = 8), dendritic data from single cell electroporation colored in magenta (n = 42), axonal data colored in green (n = 10). (d) Cumulative length of dendritic segments within the Sholl radii. Colors as in (c). (e) Cumulative area of the dendritic segment within the Sholl radii. Colors as in (c). (f) Number of intersections for each Sholl radius. Colors as in (c).
Our next aim was to quantify the distribution of synaptic contact sites along the dendrites of INLL neurons. From eight single neurons, we were able to determine the dendritic input pattern for VGluT1/2 and GlyT2 positive sites by immunofluorescence of resliced and realigned sections (Fig. 5a). Together with the Sholl analysis of these cells (Fig. 5b), the number of contact sites in each Sholl segment was counted. Thus, the number of inputs per dendritic surface area was obtained for VGluT1/2 and GlyT2 inputs for individual neurons (Fig. 5c). In the presented example (Fig. 5a to c), the number of glutamatergic inputs matched the distribution of the dendritic surface area closely. The glycinergic inputs were spread rather evenly over the dendritic extend. On this exemplary neuron, less GlyT2 than VGlut1/2-positive inputs were counted. In the next step, the distance between different VGluT1/2 and different GlyT2 contact siteswas extracted by the nearest neighbor tool of Neurlucida360 (Fig. 5d). For the example neuron, the excitatory distance was smaller (median 4.2 µm, 154 contact sites) compared to inhibition (median 10.2 µm, 37 contact sites, Fig. 5d). Similar to the exemplified cell, also the median inter-contact site distance for all analyzed cells was always smaller for VGluT1/2 (5.1 ± 0.4 µm) compared to GlyT2 (9.1 ± 1 µm) contact sites (Fig. 5e, t-test: p = 0.007). For all cells tested (n = 8), the number of VGluT1/2-positive sites (90.6 ± 23.1) was larger compared to the amount of GlyT2-positive (33.9 ± 5.9) sites (Fig. 5f, t-test: p = 0.038).
Distribution of synaptic input on dendrites of INLL neurons. (a) Single filled, reconstructed neuron (red) after re-slicing and labelling for VGlutT1/2 (green) and GlyT2 (blue). White squares indicate the position of the magnified insets. In the insets contact sites are marked with a white asterisk. Scale bars equal 50 µm and 5 µm for insets. (b) Digitalized neuron and the shells for the Sholl analysis for the neurons shown in (a). (c) Contact site number and dendritic surface within the Sholl radii. (d) Inter-contact distances for VGluT1/2 (VGluT) and GlyT2 labelled contact sites for the neuron shown in (a). Inter-contact distance is measured as the Euclidian distance between two contact sites on one dendrite. Inter-contact site distances across different dendrites were not taken into account. Box-plot shows median and quartiles. (e) Average inter-contact site distances. Each color corresponds to data from one neuron. (f) Number of VGluT1/2 and GlyT2 labelled contact sites along the sampled neurons (n = 8). Color code as in (e). (g) Top: Normalized contact site and dendritic surface distributions as a function of Sholl radius for the example neuron. Bottom: Difference between the normalized contact site (con.) and dendritic surface distributions as a function of Sholl radius. (h) Sum of the squared difference of contact site (con.) and dendritic surface distributions as shown in (g) bottom. Color code as in (e).
The difference in contact site numbers did not indicate whether the relative distribution profile of these inputs differ in respect to the dendritic surface distribution. Therefore, the normalized profiles along the radial distance from soma were calculated (Fig. 5g) and analyzed. To quantify possible differences of input distribution, the normalized VGluT1/2 and GlyT2 profiles were subtracted from the normalized dendritic surface distribution (Fig. 5g). Summing the squared values of these differences gave values for the deviance of input distribution from the dendritic surface distribution (Fig. 5h). There was no significant difference between the VGluT1/2 (1.64 ± 0.45) and GlyT2 (2.50 ± 0.54) distribution (p = 0.209, t-test), indicating that on average both input types follow the dendritic membrane distribution equally well.
So far, the position and distance between contact sites was determined based on distances of Sholl radii but not on actual dendritic distances. Therefore, we analyzed the input location based on the actual dendritic morphology. We determined the relative position of each contact site along the dendrite. The dendritic length was normalized between soma and distal end, where 0% equals the beginning of the dendrite at the soma and 100% its end. Each branch ends therefore with 100% distance and thus, the dendritic arborisation of a single cell can be averaged. In Fig. 6b the number of VGluT1/2 and GlyT2-positive contact sites is culminated over their position on the normalized dendritic extent. A steep increase indicates a higher local density of synaptic inputs. Mostly we observed a steady increase in the number of VGluT1/2 and GlyT2-positive contact sites suggesting a homogeneous distribution along the dendrite. To corroborate this notion, the normalized number of VGluT1/2 and GlyT2-positive contact sites was mapped to their distance along the normalized dendritic tree (Fig. 6b). Both, inhibitory and excitatory inputs were, apart from one case, present at the soma and ended within the last 15% of dendritic length. Thus, both input types are distributed over the entire dendrite. However, for most cells the steepness of the distribution is lower in about the first 25% of the dendrite compared to distances between 70 and 90% , indicating that the synaptic density was lower at proximal than on distal dendrites.
Synaptic location on dendrites of INLL neurons. (a) Total number of contact sites as a function of normalized dendritic location . Color code matches the data shown in Fig. 5. Solid lines correspond to VGluT1/2 and broken lines to GlyT2 positive contact sites. (b) Normalized number of contact sites as a function of normalized dendritic location. Color and line code as in (a). Black line indicates unity.
Finally, we seek to identify the molecular composition of the synaptic input types. Therefore, we immunofluorescently labelled sections for VGluT1 together with VGluT2 and VIAT together with GlyT2 to distinguish specific excitatory and inhibitory contact sites respectively. In sections containing INLL and DNLL the inhibitory inputs distributed differently (Fig. 7). As expected from the DNLL’s prominent reciprocal GABAergic connectivity1,36,37, VIAT was more present than GlyT2 in the DNLL. In the INLL, GlyT2 appeared the dominant marker for inhibitory contact sites (Fig. 7). This corroborated the different input nuclei of the INLL and DNLL1,36. The different input patterns in the DNLL and INLL were also present for glutamatergic inputs. VGluT1 prevails in the INLL and VGluT2 in the DNLL (Fig. 7). Thus, excitation appeared to arise from different sources in DNLL and INLL. Moreover, our estimated contact site distributions (Fig. 5 and 6) were based on the major synaptic input marker in the INLL.
Discussion
Here we characterize the anatomy of INLL neurons in Mongolian gerbils. We provide landmarks for the INLL in gerbils, confirming that these neurons were mainly non-inhibitory, and therefore classified as excitatory. Inhibitory inputs to INLL neurons were more glycinergic and less GABAergic compared to the DNLL. Both inhibitory and excitatory inputs to INLL neurons distribute along the dendrite with overall less inhibition compared to excitation. Neurons of the INLL come in manifold morphologies and the dendritic orientation of the overall population appears orthogonal to the lateral lemniscus.
In gerbils, the INLL can be addressed as the cell population ventral to the DNLL and H-cells3,5 stretching ventrally to the position where in transversal sections the edge of the brain begins to taper towards its ventral side. In this region, the neurons appear neither GABA- nor glycinergic in gerbils. This lack of inhibitory transmitter phenotype is consistent with the description in rodents7 and cat8, but contrasts bats, where at least a fraction of INLL neurons are marked for inhibitory transmitters9,10,11. Thus, we conclude that INLL neurons in gerbils are largely glutamatergic with possibly a fraction of dopaminergic neurons that recently have been described in this lemniscal area in rats38. INLL neurons receive glutamatergic inputs that exhibit predominantly VGluT1-positive contact sites. Thus, the origin of ascending excitation originates mainly from the cochlear nucleus and not the SOC, because medial and lateral superior olivary neurons are positive for VGluT2 expression7. This molecular pattern is consistent with the physiological data that present INLL neurons as largely monaural39,40. The presence of both GABA- and glycinergic contact sites in the INLL indicates that both transmitters play a role in sound processing. This interpretation agrees with in vivo pharmacology in bats that indicated both GABA- and glycinergic effects on sound evoked discharges in the INLL30,31. Moreover, the mixed presence of GABA- and glycinergic contact sites in the INLL contrasts the dominant GABAergic inputs in the DNLL.
INLL neurons were described as multipolar and horizontal3 or fusiform cells2. Our reconstructions does not indicate a clear cut off between bipolar or multipolar neurons or other morphological features. Rather it appears that there is a continuum of morphological variations. Since we relate the different morphologies to a continuum rather than to specific appearances, we do not introduce a categorization of different cell types, as has been presented for other LL nuclei13,14,20. Importantly, no clustering of specific neuronal morphologies occurs within the nucleus and cells of different shapes distributed throughout the INLL. In various cases, the dendrites of the reconstructed neurons progressed beyond the apparent medial or dorsal border of the INLL. This might imply that neurons in the INLL receive non-auditory and possibly auditory inputs not only within the lemnsical fiber bundle and therefore also from sources outside the auditory lemniscus pathways.
Somatic volumes of INLL neurons show a wide heterogeneity covering a whole magnitude, and therefore show a similar heterogeneity compared to DNLL neurons in cat12. The dendrites of rat VNLL neurons have been quantified by a Sholl analysis before14. A similar value of maximal Sholl radius between these VNLL neurons and our INLL neurons exists. Moreover, the number of intersections is similar between our INLL and these VNLL neurons, with the exception of more dendritic branches in bushy cells of the VNLL14. Thus, the neuronal size and morphology seems similar between the non-bushy cells of the VNLL, with neurons from the INLL. Whether these neurons have also biophysical or functional similarities remains to be elucidated. This similarity of cell morphology between VNLL and INLL neurons adds to the difficulty to delineate a border between both nuclei and requires the use of other landmarks.
Our analysis shows that the sum of INLL dendrites orientats perpendicular to the fibers of the LL. Elongated neurons are mostly horizontally oriented, but also multipolar neurons emit their dendrites sometimes more in this orientation. The perpendicular orientation of dendrites to the lemniscal fiber bundle has been noted also in bats2 and is apparent in Golgi stains from gerbils3. In gerbils, the ascending lemniscal fiber bundle appears only partially tonotopically organized with lower frequencies located at the lateral and higher frequencies at more central positions41. Dendrites oriented perpendicularly across the entire INLL will be able to contact passing fibers carrier different frequency information. Therefore, it might be speculated that perpendicular oriented dendrites form synapses beyond the classical tonotopical arrangement and grab information across different frequency bands. Thus, the output of INLL neurons might not be narrowly tuned to a specific frequency. Indeed morphological evidence is provided from bats, where INLL neurons appeared less restricted to tonotopical projection patterns2. Moreover, the tontopical tuning of INLL neurons is less clearly conserved compared to nuclei of the superior olivary complex42, or the DNLL40,43,44,45,46. Again, in bats, the tonotopy of the INLL appears patchy17,39,47. In rodents, the tonotopy might be ventro-dorsally organized, however only a limited amount of in vivo data are present40 and do not fully agree with anatomical data41. In cat, again partially contradicting data about the tonotopy from in vivo recordings48 and anatomical studies are present49. The perpendicular orientation of INLL dendrites however might play a role in the functionality of INLL neurons and might underlie the weak tonotopical tuning. This speculation about a weak tonotopical arrangement of the INLL is consistent with reports of cross-frequency integration in INLL neurons32 and broad frequency tuning31. These functional features might be supported by information transfer across different frequencies from ascending frequency bands. Thus, a perpendicular orientation of dendrites to passing fibers might facilitate input detection across different ascending frequency areas and hence cross-frequency integration.
In INLL neurons, the synaptic density was larger for excitation compared to inhibition, judged by VGluT1/2 and GlyT2-positive co-localization with the dendrite or soma. However, it should be considered that each contact site might encompass more than a single active zone. Furthermore, GlyT2 will not cover all inhibitory inputs because of the presence of GABAergic synapses that are detected as VIAT/GlyT2-negative contact sites. Therefore, the absolute inhibitory density will be larger. Whether the total amount of inhibition is larger compared to the overall excitation remains unresolved. Because we detected more GlyT2 than VIAT/GlyT2-negative contact sites, we assume that GABAergic inputs are less pronounced.
Both, the density of excitation and GlyT2 mediated inhibition follow the size of the dendritic surface area. However, it appears that at the beginning of the dendrite GlyT2 positive contact sites are slightly more present compared to excitation, while at the far distal end of dendrites both excitation and inhibition are present in the same manner. A well-known example in the auditory pathways for somatic inhibition are the neurons of the medial superior olive (MSO). In MSO neurons, inhibition is more or less restricted to the soma50,51,52 and the restriction to the soma is species-dependent52, while excitation is distributed along the bipolar dendrites and soma50,51,53. This sub-cellular segregation is regarded to be functionally relevant for ultra-fast coincidence detection and binaural hearing54. Since in INLL neurons the somatic bias for inhibition is small, inhibition might serve other functions for postsynaptic integration compared to MSO neurons, irrespectively of neuronal morphology. In the auditory brainstem, synaptic distributions have been studied also in fusiform neurons of the dorsal cochlear nucleus55. In these cells the density of inputs increases with distance and larger inputs were observed at more basal dendrites and somata. The resolution of our images does not allow to estimate differences in synaptic size along the dendrite. Therefore, no direct comparison to fusiform cells can be drawn.
The distribution of contact sites along the dendritic extent highlights that sound processing in these neurons is based on dendritic integration. Since we do not observe a different distribution between distal or proximal dendritic branches, inhibitory and excitatory inputs are likely to be integrated in individual dendritic compartments. Therefore, dependent on the input pattern, dendritic integration might activate a full dendrite or only sub-regions that locally process input strength. The slight somatic bias of inhibition might serve as the standard shunt for suppressing dendritic evoked excitation. It remains to be seen, whether the large morphological heterogeneity of INLL neurons follows a specific structure–function relationship as proposed for VNLL neurons14 or similar to DNLL neurons where no structure–function relationship is indicated13.
In a small fraction of neurons, we could unambiguously identify axonal compartments. These axons projected dorsally and ventrally and in some cases appeared to have small local collaterals. Our experimental approach does not allow to trace projection axons to their final destination. Nevertheless, the initial projection patterns indicated that INLL neurons send their output to ascending structures, such as the DNLL and inferior colliculus. These ascending structures have been identified as targets of the INLL1,2,36. Our single cell labelling also identifies potential descending axonal compartments. Thus, INLL neurons are likely to be connected to descending structures of the superior olivary complex or cochlear nucleus. Since INLL neurons receive inputs from these structures1,36 and the inferior colliculus, it suggests that INLL neurons might form a central hub for looping sound information with other auditory nuclei. Together with the local axonal branching, this indicates that the INLL is a highly connected nucleus regarding its input and output targets. Such complex input–output structures suggest that INLL neurons are a center for computing complex monaural signals.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Glendenning, K. K., Brunso-Bechtold, J. K., Thompson, G. C. & Masterton, R. B. Ascending auditory afferents to the nuclei of the lateral lemniscus. J. Comp. Neurol. 197, 673–703. https://doi.org/10.1002/cne.901970409 (1981).
Schweizer, H. The connections of the inferior colliculus and the organization of the brainstem auditory system in the greater horseshoe bat (Rhinolophus ferrumequinum). J. Comp. Neurol. 201, 25–49. https://doi.org/10.1002/cne.902010104 (1981).
Mylius, J., Brosch, M., Scheich, H. & Budinger, E. Subcortical auditory structures in the Mongolian gerbil: I. Golgi architecture. J. Comp. Neurol. 521, 1289–1321. https://doi.org/10.1002/cne.23232 (2013).
Ammer, J. J., Grothe, B. & Felmy, F. Late postnatal development of intrinsic and synaptic properties promotes fast and precise signaling in the dorsal nucleus of the lateral lemniscus. J. Neurophysiol. 107, 1172–1185. https://doi.org/10.1152/jn.00585.2011 (2012).
Caicedo, A. & Herbert, H. Topography of descending projections from the inferior colliculus to auditory brainstem nuclei in the rat. J. Comp. Neurol. 328, 377–392. https://doi.org/10.1002/cne.903280305 (1993).
Caspari, F., Baumann, V. J., Garcia-Pino, E. & Koch, U. Heterogeneity of intrinsic and synaptic properties of neurons in the ventral and dorsal parts of the ventral nucleus of the lateral lemniscus. Front. Neural Circ. 9, 74. https://doi.org/10.3389/fncir.2015.00074 (2015).
Ito, T., Bishop, D. C. & Oliver, D. L. Expression of glutamate and inhibitory amino acid vesicular transporters in the rodent auditory brainstem. J. Comp. Neurol. 519, 316–340. https://doi.org/10.1002/cne.22521 (2011).
Saint Marie, R. L., Shneiderman, A. & Stanforth, D. A. Patterns of gamma-aminobutyric acid and glycine immunoreactivities reflect structural and functional differences of the cat lateral lemniscal nuclei. J. Comp. Neurol. 389, 264–276 (1997).
Ito, T. et al. Organization of subcortical auditory nuclei of Japanese house bat (Pipistrellus abramus) identified with cytoarchitecture and molecular expression. J. Comp. Neurol. 526, 2824–2844. https://doi.org/10.1002/cne.24529 (2018).
Winer, J. A., Larue, D. T. & Pollak, G. D. GABA and glycine in the central auditory system of the mustache bat: structural substrates for inhibitory neuronal organization. J. Comp. Neurol. 355, 317–353. https://doi.org/10.1002/cne.903550302 (1995).
Yavuzoglu, A., Schofield, B. R. & Wenstrup, J. J. Substrates of auditory frequency integration in a nucleus of the lateral lemniscus. Neuroscience 169, 906–919. https://doi.org/10.1016/j.neuroscience.2010.04.073 (2010).
Kane, E. S. & Barone, L. M. The dorsal nucleus of the lateral lemniscus in the cat: neuronal types and their distributions. J. Comp. Neurol. 192, 797–826. https://doi.org/10.1002/cne.901920412 (1980).
Wu, S. H. & Kelly, J. B. In vitro brain slice studies of the rat’s dorsal nucleus of the lateral lemniscus. II. Physiological properties of biocytin-labeled neurons. J. Neurophysiol. 73, 794–809. https://doi.org/10.1152/jn.1995.73.2.794 (1995).
Zhao, M. & Wu, S. H. Morphology and physiology of neurons in the ventral nucleus of the lateral lemniscus in rat brain slices. J. Comp. Neurol. 433, 255–271. https://doi.org/10.1002/cne.1139 (2001).
Adams, J. C. Projections from octopus cells of the posteroventral cochlear nucleus to the ventral nucleus of the lateral lemniscus in cat and human. Audit. Neurosci. 3, 335–350 (1997).
Berger, C., Meyer, E. M., Ammer, J. J. & Felmy, F. Large somatic synapses on neurons in the ventral lateral lemniscus work in pairs. J. Neurosci. 34, 3237–3246. https://doi.org/10.1523/JNEUROSCI.3664-13.2014 (2014).
Covey, E. & Casseday, J. H. Connectional basis for frequency representation in the nuclei of the lateral lemniscus of the bat Eptesicus fuscus. J. Neurosci. 6, 2926–2940. https://doi.org/10.1523/JNEUROSCI.06-10-02926.1986 (1986).
Stotler, W. A. An experimental study of the cells and connections of the superior olivary complex of the cat. J. Comp. Neurol. 98, 401–431 (1953).
Schofield, B. R. & Cant, N. B. Ventral nucleus of the lateral lemniscus in guinea pigs: cytoarchitecture and inputs from the cochlear nucleus. J. Comp. Neurol. 379, 363–385 (1997).
Iwahori, N. A Golgi study on the dorsal nucleus of the lateral lemniscus in the mouse. Neurosci. Res 3, 196–212 (1986).
Adams, J. C. & Mugnaini, E. Dorsal nucleus of the lateral lemniscus: a nucleus of GABAergic projection neurons. Brain Res Bull. 13, 585–590 (1984).
Kladisios, N., Fischer, L. & Felmy, F. Minimal number of required inputs for temporally precise action potential generation in auditory brainstem nuclei. Front. Cell Neurosci. 14, 592213. https://doi.org/10.3389/fncel.2020.592213 (2020).
Kladisios, N. et al. Synaptic mechanisms underlying temporally precise information processing in the VNLL, an auditory brainstem nucleus. J. Neurosci. 42, 6536–6550. https://doi.org/10.1523/JNEUROSCI.0948-22.2022 (2022).
Fu, X. W., Brezden, B. L., Kelly, J. B. & Wu, S. H. Synaptic excitation in the dorsal nucleus of the lateral lemniscus: whole-cell patch-clamp recordings from rat brain slice. Neuroscience 78, 815–827. https://doi.org/10.1016/s0306-4522(96)00580-5 (1997).
Kelly, J. B. & Kidd, S. A. NMDA and AMPA receptors in the dorsal nucleus of the lateral lemniscus shape binaural responses in rat inferior colliculus. J. Neurophysiol. 83, 1403–1414. https://doi.org/10.1152/jn.2000.83.3.1403 (2000).
Porres, C. P., Meyer, E. M., Grothe, B. & Felmy, F. NMDA currents modulate the synaptic input-output functions of neurons in the dorsal nucleus of the lateral lemniscus in Mongolian gerbils. J. Neurosci. 31, 4511–4523. https://doi.org/10.1523/JNEUROSCI.6054-10.2011 (2011).
Siveke, I. et al. Electrogenic N-methyl-D-aspartate receptor signaling enhances binaural responses in the adult brainstem. Eur. J. Neurosci. 47, 858–865. https://doi.org/10.1111/ejn.13859 (2018).
Pecka, M. et al. Inhibiting the inhibition: a neuronal network for sound localization in reverberant environments. J. Neurosci. 27, 1782–1790. https://doi.org/10.1523/JNEUROSCI.5335-06.2007 (2007).
Yang, L. & Pollak, G. D. GABA and glycine have different effects on monaural response properties in the dorsal nucleus of the lateral lemniscus of the mustache bat. J. Neurophysiol. 71, 2014–2024. https://doi.org/10.1152/jn.1994.71.6.2014 (1994).
Kutscher, A. & Covey, E. Functional role of GABAergic and glycinergic inhibition in the intermediate nucleus of the lateral lemniscus of the big brown bat. J. Neurophysiol. 101, 3135–3146. https://doi.org/10.1152/jn.00766.2007 (2009).
Xie, R., Meitzen, J. & Pollak, G. D. Differing roles of inhibition in hierarchical processing of species-specific calls in auditory brainstem nuclei. J. Neurophysiol. 94, 4019–4037. https://doi.org/10.1152/jn.00688.2005 (2005).
Portfors, C. V. & Wenstrup, J. J. Responses to combinations of tones in the nuclei of the lateral lemniscus. J. Assoc. Res. Otolaryngol. 2, 104–117. https://doi.org/10.1007/s101620010057 (2001).
Preibisch, S., Saalfeld, S. & Tomancak, P. Globally optimal stitching of tiled 3D microscopic image acquisitions. Bioinformatics 25, 1463–1465. https://doi.org/10.1093/bioinformatics/btp184 (2009).
Moore, L. A. & Trussell, L. O. Corelease of Inhibitory Neurotransmitters in the Mouse Auditory Midbrain. J. Neurosci. 37, 9453–9464. https://doi.org/10.1523/JNEUROSCI.1125-17.2017 (2017).
Hering, H. & Sheng, M. Dendritic spines: structure, dynamics and regulation. Nat. Rev. Neurosci. 2, 880–888. https://doi.org/10.1038/35104061 (2001).
Kelly, J. B., van Adel, B. A. & Ito, M. Anatomical projections of the nuclei of the lateral lemniscus in the albino rat (Rattus norvegicus). J. Comp. Neurol. 512, 573–593. https://doi.org/10.1002/cne.21929 (2009).
Porres, C. P., Grothe, B. & Felmy, F. Breakdown of excitability by attenuated PRV-152 infection in auditory brainstem neurons of mongolian gerbils. Neuroscience 367, 1–9. https://doi.org/10.1016/j.neuroscience.2017.10.020 (2017).
Harris, S. et al. Dopamine in auditory nuclei and lemniscal projections is poised to influence acoustic integration in the inferior colliculus. Front. Neural Circ. 15, 624563. https://doi.org/10.3389/fncir.2021.624563 (2021).
Covey, E. & Casseday, J. H. The monaural nuclei of the lateral lemniscus in an echolocating bat: parallel pathways for analyzing temporal features of sound. J. Neurosci. 11, 3456–3470. https://doi.org/10.1523/JNEUROSCI.11-11-03456.1991 (1991).
Kelly, J. B., Buckthought, A. D. & Kidd, S. A. Monaural and binaural response properties of single neurons in the rat’s dorsal nucleus of the lateral lemniscus. Hear Res 122, 25–40. https://doi.org/10.1016/s0378-5955(98)00082-3 (1998).
Benson, C. G. & Cant, N. B. The ventral nucleus of the lateral lemniscus of the gerbil (Meriones unguiculatus): organization of connections with the cochlear nucleus and the inferior colliculus. J. Comp. Neurol. 510, 673–690. https://doi.org/10.1002/cne.21820 (2008).
Kandler, K., Clause, A. & Noh, J. Tonotopic reorganization of developing auditory brainstem circuits. Nat. Neurosci 12, 711–717. https://doi.org/10.1038/nn.2332 (2009).
Bajo, V. M., Merchan, M. A., Malmierca, M. S., Nodal, F. R. & Bjaalie, J. G. Topographic organization of the dorsal nucleus of the lateral lemniscus in the cat. J. Comp. Neurol. 407, 349–366. https://doi.org/10.1002/(sici)1096-9861(19990510)407:3%3c349::aid-cne4%3e3.0.co;2-5 (1999).
Friauf, E. Tonotopic order in the adult and developing auditory system of the rat as shown by c-fos immunocytochemistry. Eur. J. Neurosci. 4, 798–812. https://doi.org/10.1111/j.1460-9568.1992.tb00190.x (1992).
Merchan, M. A., Saldana, E. & Plaza, I. Dorsal nucleus of the lateral lemniscus in the rat: concentric organization and tonotopic projection to the inferior colliculus. J. Comp. Neurol. 342, 259–278. https://doi.org/10.1002/cne.903420209 (1994).
Ryan, A. F., Woolf, N. K. & Sharp, F. R. Tonotopic organization in the central auditory pathway of the Mongolian gerbil: a 2-deoxyglucose study. J. Comp. Neurol. 207, 369–380. https://doi.org/10.1002/cne.902070408 (1982).
Metzner, W. & Radtke-Schuller, S. The nuclei of the lateral lemniscus in the rufous horseshoe bat, Rhinolophus rouxi. A neurophysiological approach. J. Comp. Physiol. A 160, 395–411. https://doi.org/10.1007/BF00613029 (1987).
Aitkin, L. M., Anderson, D. J. & Brugge, J. F. Tonotopic organization and discharge characteristics of single neurons in nuclei of the lateral lemniscus of the cat. J. Neurophysiol. 33, 421–440. https://doi.org/10.1152/jn.1970.33.3.421 (1970).
Malmierca, M. S., Leergaard, T. B., Bajo, V. M., Bjaalie, J. G. & Merchan, M. A. Anatomic evidence of a three-dimensional mosaic pattern of tonotopic organization in the ventral complex of the lateral lemniscus in cat. J. Neurosci. 18, 10603–10618. https://doi.org/10.1523/JNEUROSCI.18-24-10603.1998 (1998).
Clark, G. M. The ultrastructure of nerve endings in the medial superior olive of the cat. Brain Res. 14, 293–305. https://doi.org/10.1016/0006-8993(69)90111-5 (1969).
Couchman, K., Grothe, B. & Felmy, F. Functional localization of neurotransmitter receptors and synaptic inputs to mature neurons of the medial superior olive. J. Neurophysiol. 107, 1186–1198. https://doi.org/10.1152/jn.00586.2011 (2012).
Kapfer, C., Seidl, A. H., Schweizer, H. & Grothe, B. Experience-dependent refinement of inhibitory inputs to auditory coincidence-detector neurons. Nat. Neurosci. 5, 247–253. https://doi.org/10.1038/nn810 (2002).
Callan, A. R., Hess, M., Felmy, F. & Leibold, C. Arrangement of excitatory synaptic inputs on dendrites of the medial superior olive. J. Neurosci. 41, 269–283. https://doi.org/10.1523/JNEUROSCI.1055-20.2020 (2021).
Grothe, B. New roles for synaptic inhibition in sound localization. Nat. Rev. Neurosci. 4, 540–550. https://doi.org/10.1038/nrn1136 (2003).
Salloum, R. H. et al. Mapping and morphometric analysis of synapses and spines on fusiform cells in the dorsal cochlear nucleus. Front. Syst. Neurosci. 8, 167. https://doi.org/10.3389/fnsys.2014.00167 (2014).
Acknowledgements
We like to thank Alexandra Benn, Dr. Christina Pätz-Warncke for technical support with the immunofluorescence labelling and confocal microscopy.
Funding
Open Access funding enabled and organized by Projekt DEAL. DFG789/10–1.
Author information
Authors and Affiliations
Contributions
K.D.W: data acquisition and analysis, writing; L.O.: data acquisition and analysis; C.G.: data acquisition and analysis; A.K.S: data acquisition and analysis; F.F.: study design and concept, data acquisition and analysis, first draft, writing and editing.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wicke, K.D., Oppe, L., Geese, C. et al. Neuronal morphology and synaptic input patterns of neurons in the intermediate nucleus of the lateral lemniscus of gerbils. Sci Rep 13, 14182 (2023). https://doi.org/10.1038/s41598-023-41180-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-41180-8
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.